Abstract
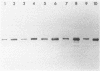
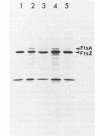
The ftsZ (sulB) gene of Escherichia coli codes for a 40,000-dalton protein that carries out a key step in the cell division pathway. The presence of an ftsZ gene protein in other bacterial species was examined by a combination of Southern blot and Western blot analyses. Southern blot analysis of genomic restriction digests revealed that many bacteria, including species from six members of the family Enterobacteriaceae and from Pseudomonas aeruginosa and Agrobacterium tumefaciens, contained sequences which hybridized with an E. coli ftsZ probe. Genomic DNA from more distantly related bacteria, including Bacillus subtilis, Branhamella catarrhalis, Micrococcus luteus, and Staphylococcus aureus, did not hybridize under minimally stringent conditions. Western blot analysis, with anti-E. coli FtsZ antiserum, revealed that all bacterial species examined contained a major immunoreactive band. Several of the Enterobacteriaceae were transformed with a multicopy plasmid encoding the E. coli ftsZ gene. These transformed strains, Shigella sonnei, Salmonella typhimurium, Klebsiella pneumoniae, and Enterobacter aerogenes, were shown to overproduce the FtsZ protein and to produce minicells. Analysis of [35S]methionine-labeled minicells revealed that the plasmid-encoded gene products were the major labeled species. This demonstrated that the E. coli ftsZ gene could function in other bacterial species to induce minicells and that these minicells could be used to analyze plasmid-endoced gene products.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler H. I., Fisher W. D., Cohen A., Hardigree A. A. MINIATURE escherichia coli CELLS DEFICIENT IN DNA. Proc Natl Acad Sci U S A. 1967 Feb;57(2):321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K. J., Donachie W. D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985 Aug;163(2):615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Better M., Helinski D. R. Isolation and characterization of the recA gene of Rhizobium meliloti. J Bacteriol. 1983 Jul;155(1):311–316. doi: 10.1128/jb.155.1.311-316.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Davie E., Sydnor K., Rothfield L. I. Genetic basis of minicell formation in Escherichia coli K-12. J Bacteriol. 1984 Jun;158(3):1202–1203. doi: 10.1128/jb.158.3.1202-1203.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Warrick R. M., Elkana Y., Ehrlich S. D., Lederberg J. Electrophoretic separation of Bacillus subtilis genes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2207–2211. doi: 10.1073/pnas.72.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. A., Holland I. B. Inactivation of essential division genes, ftsA, ftsZ, suppresses mutations at sfiB, a locus mediating division inhibition during the SOS response in E. coli. EMBO J. 1984 May;3(5):1181–1186. doi: 10.1002/j.1460-2075.1984.tb01948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C., Holland I. B. Role of the SulB (FtsZ) protein in division inhibition during the SOS response in Escherichia coli: FtsZ stabilizes the inhibitor SulA in maxicells. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6045–6049. doi: 10.1073/pnas.82.18.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener S. L., McNamee K. P., McEntee K. Cloning and characterization of recA genes froM Proteus vulgaris, Erwinia carotovora, Shigella flexneri, and Escherichia coli B/r. J Bacteriol. 1984 Oct;160(1):153–160. doi: 10.1128/jb.160.1.153-160.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Love P. E., Yasbin R. E. Genetic characterization of the inducible SOS-like system of Bacillus subtilis. J Bacteriol. 1984 Dec;160(3):910–920. doi: 10.1128/jb.160.3.910-920.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett C. M., Jr, Roberts J. W. Purification of a RecA protein analogue from Bacillus subtilis. J Biol Chem. 1985 Mar 25;260(6):3305–3313. [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wolf-Watz H., Donachie W. D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol. 1980 May;142(2):615–620. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J., Sanjanwala B., Lowe M. Overproduction of FtsZ suppresses sensitivity of lon mutants to division inhibition. J Bacteriol. 1986 Jun;166(3):756–762. doi: 10.1128/jb.166.3.756-762.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin E., Lutkenhaus J., D'Ari R. Reversibility of SOS-associated division inhibition in Escherichia coli. J Bacteriol. 1986 Jun;166(3):733–738. doi: 10.1128/jb.166.3.733-738.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. Use of minicells for bacteriophage-directed polypeptide synthesis. Methods Enzymol. 1979;68:493–503. doi: 10.1016/0076-6879(79)68038-2. [DOI] [PubMed] [Google Scholar]
- Robinson A. C., Kenan D. J., Hatfull G. F., Sullivan N. F., Spiegelberg R., Donachie W. D. DNA sequence and transcriptional organization of essential cell division genes ftsQ and ftsA of Escherichia coli: evidence for overlapping transcriptional units. J Bacteriol. 1984 Nov;160(2):546–555. doi: 10.1128/jb.160.2.546-555.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupprecht K. R., Markovitz A. Conservation of capR (lon) DNA of Escherichia coli K-12 between distantly related species. J Bacteriol. 1983 Aug;155(2):910–914. doi: 10.1128/jb.155.2.910-914.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnier J., Faist G. Comparative studies on the structural gene for the ribosomal protein S1 in ten bacterial species. Mol Gen Genet. 1985;200(3):476–481. doi: 10.1007/BF00425734. [DOI] [PubMed] [Google Scholar]
- Siede W., Eckardt F. Inducibility of error-prone DNA repair in yeast? Mutat Res. 1984 Oct;129(1):3–11. doi: 10.1016/0027-5107(84)90116-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sullivan N. F., Donachie W. D. Overlapping functional units in a cell division gene cluster in Escherichia coli. J Bacteriol. 1984 Jun;158(3):1198–1201. doi: 10.1128/jb.158.3.1198-1201.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. E., Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985 Oct;42(3):941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- West S. C., Little J. W. P. mirabilis RecA protein catalyses cleavage of E. coli LexA protein and the lambda repressor in vitro. Mol Gen Genet. 1984;194(1-2):111–113. doi: 10.1007/BF00383505. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Lutkenhaus J. The nucleotide sequence of the essential cell-division gene ftsZ of Escherichia coli. Gene. 1985;36(3):241–247. doi: 10.1016/0378-1119(85)90179-9. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Rockenbach S., Ward J. E., Jr, Lutkenhaus J. Structure and expression of the cell division genes ftsQ, ftsA and ftsZ. J Mol Biol. 1985 Aug 5;184(3):399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]