Abstract
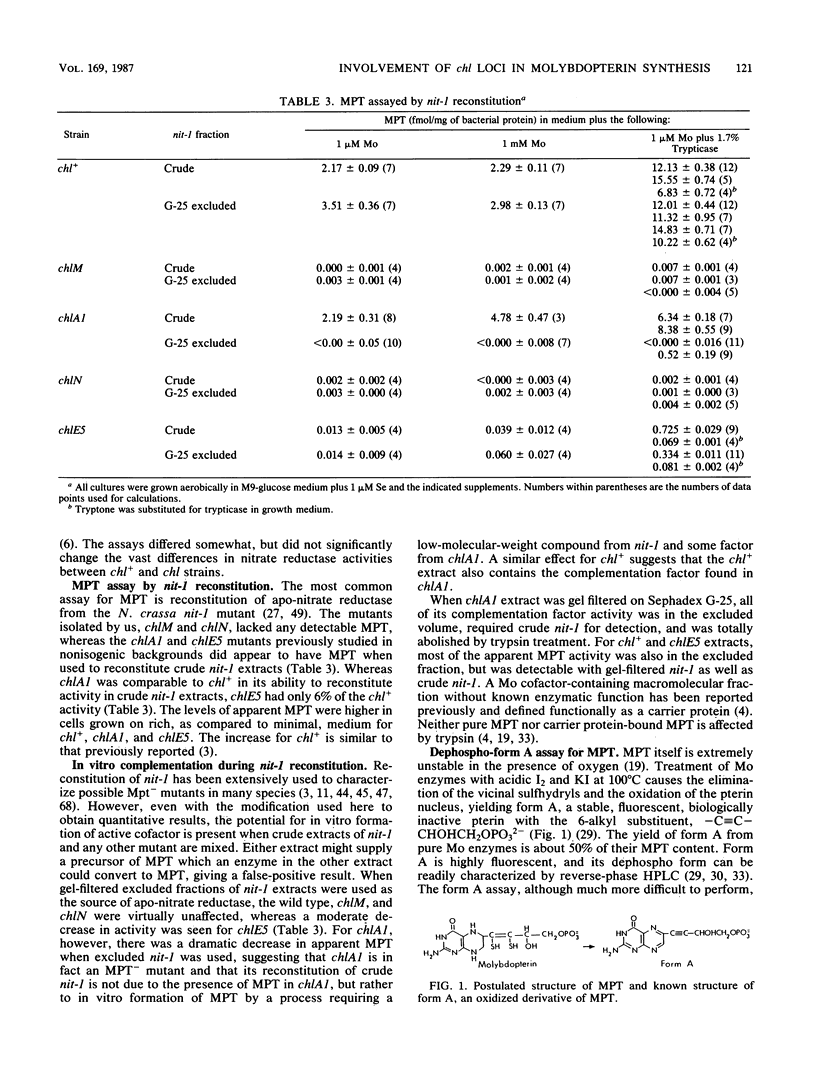
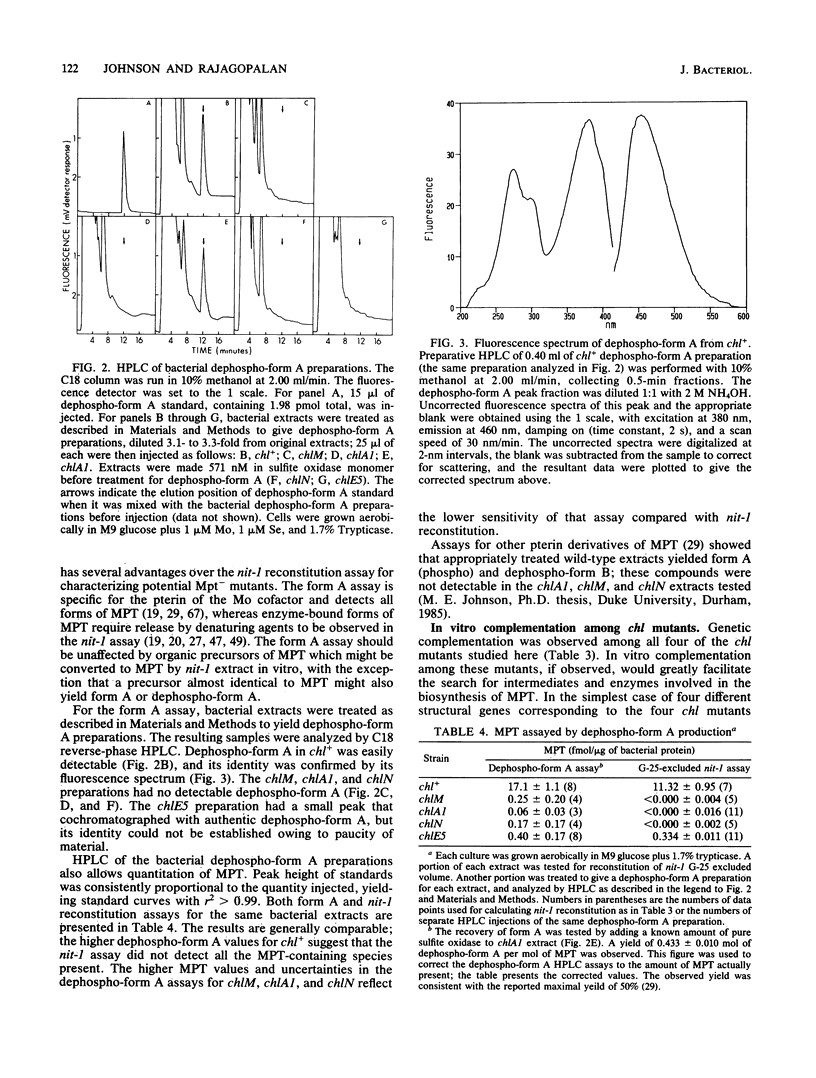
All molybdenum enzymes except nitrogenase contain a common molybdenum cofactor, whose organic moiety is a novel pterin called molybdopterin (MPT). To assist in elucidating the biosynthetic pathway of MPT, two MPT-deficient mutants of Escherichia coli K-12 were isolated. They lacked activities of the molybdenum enzymes nitrate reductase and formate dehydrogenase, did not reconstitute apo nitrate reductase from a Neurospora crassa nit-1 strain, and did not yield form A, a derivative of MPT. By P1 mapping, these two mutations mapped to chlA and chlE, loci previously postulated but never definitely shown to be involved in MPT biosynthesis. The two new mutations are in different genetic complementation groups from previously isolated chlA and chlE mutations and have been designated as chlM and chlN (closely linked to chlA and chlE, respectively). The reported presence of Mo cofactor activity in the chlA1 strain is shown to be due to in vitro synthesis of MPT through complementation between a trypsin-sensitive macromolecule from the chlA1 strain and a low-molecular-weight compound from the nit-l strain.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Cleary P., Campbell A. A deletion analysis of prophage lambda and adjacent genetic regions. Proc Natl Acad Sci U S A. 1968 Nov;61(3):956–962. doi: 10.1073/pnas.61.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper M. D., Ames B. N. Positive selection of mutants with deletions of the gal-chl region of the Salmonella chromosome as a screening procedure for mutagens that cause deletions. J Bacteriol. 1975 Jan;121(1):259–266. doi: 10.1128/jb.121.1.259-266.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amy N. K. Identification of the molybdenum cofactor in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1981 Oct;148(1):274–282. doi: 10.1128/jb.148.1.274-282.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amy N. K., Rajagopalan K. V. Characterization of molybdenum cofactor from Escherichia coli. J Bacteriol. 1979 Oct;140(1):114–124. doi: 10.1128/jb.140.1.114-124.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E. L., Riggs D. L. Evidence of a second nitrate reductase activity that is distinct from the respiratory enzyme in Salmonella typhimurium. J Bacteriol. 1982 May;150(2):563–571. doi: 10.1128/jb.150.2.563-571.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassen V. P., Oltmann L. F., Bus S., v 't Riet J., Stouthamer A. H. The influence of growth conditions on the synthesis of molybdenum cofactor in Proteins mirabilis. Arch Microbiol. 1981 Sep;130(1):44–49. doi: 10.1007/BF00527070. [DOI] [PubMed] [Google Scholar]
- Cronan J. E., Jr Mini-Mu-duction as a test for genetic complementation in Escherichia coli. J Bacteriol. 1984 Apr;158(1):357–361. doi: 10.1128/jb.158.1.357-361.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett R. H., Cove D. J. Formation of NADPH-nitrate reductase activity in vitro from Aspergillus nidulans niaD and cnx mutants. Mol Gen Genet. 1976 Dec 8;149(2):179–186. doi: 10.1007/BF00332887. [DOI] [PubMed] [Google Scholar]
- Giordano G., Grillet L., Pommier J., Terriere C., Haddock B. A., Azoulay E. Precursor forms of the subunits of nitrate reductase in chlA and chlB mutants of Escherichia coli K12. Eur J Biochem. 1980 Apr;105(2):297–306. doi: 10.1111/j.1432-1033.1980.tb04501.x. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Phenotypic restoration by molybdate of nitrate reductase activity in chlD mutants of Escherichia coli. J Bacteriol. 1971 Nov;108(2):854–860. doi: 10.1128/jb.108.2.854-860.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Hawkes T. R., Bray R. C. Quantitative transfer of the molybdenum cofactor from xanthine oxidase and from sulphite oxidase to the deficient enzyme of the nit-1 mutant of Neurospora crassa to yield active nitrate reductase. Biochem J. 1984 Apr 15;219(2):481–493. doi: 10.1042/bj2190481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Flanders P., Boyce R. P., Theriot L. Three loci in Escherichia coli K-12 that control the excision of pyrimidine dimers and certain other mutagen products from DNA. Genetics. 1966 Jun;53(6):1119–1136. doi: 10.1093/genetics/53.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingledew W. J., Poole R. K. The respiratory chains of Escherichia coli. Microbiol Rev. 1984 Sep;48(3):222–271. doi: 10.1128/mr.48.3.222-271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V., Arison B. H. The pterin component of the molybdenum cofactor. Structural characterization of two fluorescent derivatives. J Biol Chem. 1984 May 10;259(9):5414–5422. [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V. Characterization of the molybdenum cofactor of sulfite oxidase, xanthine, oxidase, and nitrate reductase. Identification of a pteridine as a structural component. J Biol Chem. 1980 Mar 10;255(5):1783–1786. [PubMed] [Google Scholar]
- Johnson J. L., Rajagopalan K. V. Structural and metabolic relationship between the molybdenum cofactor and urothione. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6856–6860. doi: 10.1073/pnas.79.22.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. E., Rajagopalan K. V. In vitro system for molybdopterin biosynthesis. J Bacteriol. 1987 Jan;169(1):110–116. doi: 10.1128/jb.169.1.110-116.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. W., Garland P. B. Sites and specificity of the reaction of bipyridylium compounds with anaerobic respiratory enzymes of Escherichia coli. Effects of permeability barriers imposed by the cytoplasmic membrane. Biochem J. 1977 Apr 15;164(1):199–211. doi: 10.1042/bj1640199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S., Hageman R. V., Rajagopalan K. V. In vitro reconstitution of nitrate reductase activity of the Neurospora crassa mutant nit-1: specific incorporation of molybdopterin. Arch Biochem Biophys. 1984 Sep;233(2):821–829. doi: 10.1016/0003-9861(84)90511-3. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lambden P. R., Guest J. R. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol. 1976 Dec;97(2):145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- Lambdren P. R., Guest J. R. A novel method for isolating chlorate-resistant mutants of Escherichia coli K12 by anaerobic selection on a lactate plus fumarate medium. J Gen Microbiol. 1976 Mar;93(1):173–176. doi: 10.1099/00221287-93-1-173. [DOI] [PubMed] [Google Scholar]
- Lester R. L., DeMoss J. A. Effects of molybdate and selenite on formate and nitrate metabolism in Escherichia coli. J Bacteriol. 1971 Mar;105(3):1006–1014. doi: 10.1128/jb.105.3.1006-1014.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A. Reconstitution of nitrate reductase activity and formation of membrane particles from cytoplasmic extracts of chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1973 Jun;114(3):1164–1176. doi: 10.1128/jb.114.3.1164-1176.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil T., MacNeil D., Roberts G. P., Supiano M. A., Brill W. J. Fine-structure mapping and complementation analysis of nif (nitrogen fixation) genes in Klebsiella pneumoniae. J Bacteriol. 1978 Oct;136(1):253–266. doi: 10.1128/jb.136.1.253-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer O., Rajagopalan K. V. Molybdopterin in carbon monoxide oxidase from carboxydotrophic bacteria. J Bacteriol. 1984 Feb;157(2):643–648. doi: 10.1128/jb.157.2.643-648.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. B., Amy N. K. Molybdenum cofactor in chlorate-resistant and nitrate reductase-deficient insertion mutants of Escherichia coli. J Bacteriol. 1983 Aug;155(2):793–801. doi: 10.1128/jb.155.2.793-801.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nason A., Lee K. Y., Pan S. S., Ketchum P. A., Lamberti A., DeVries J. Invitro formation of assimilatory reduced nicotinamide adenine dinucleotide phosphate: nitrate reductase from a Neurospora mutant and a component of molybdenum-enzymes. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3242–3246. doi: 10.1073/pnas.68.12.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijkamp H. J., De Haan P. G. Genetic and biochemical studies of the guanosine 5'-monophosphate pathway in Escherichia coli. Biochim Biophys Acta. 1967 Aug 22;145(1):31–40. doi: 10.1016/0005-2787(67)90651-x. [DOI] [PubMed] [Google Scholar]
- Ordal G. W., Adler J. Isolation and complementation of mutants in galactose taxis and transport. J Bacteriol. 1974 Feb;117(2):509–516. doi: 10.1128/jb.117.2.509-516.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PATEMAN J. A., COVE D. J., REVER B. M., ROBERTS D. B. A COMMON CO-FACTOR FOR NITRATE REDUCTASE AND XANTHINE DEHYDROGENASE WHICH ALSO REGULATES THE SYNTHESIS OF NITRATE REDUCTASE. Nature. 1964 Jan 4;201:58–60. doi: 10.1038/201058a0. [DOI] [PubMed] [Google Scholar]
- Pascal M. C., Burini J. F., Ratouchniak J., Chippaux M. Regulation of the nitrate reductase operon: effect of mutations in chlA, B, D and E genes. Mol Gen Genet. 1982;188(1):103–106. doi: 10.1007/BF00333001. [DOI] [PubMed] [Google Scholar]
- Puig J., Azoulay E. Etude génétique et biochimique des mutants résistant au Clo minus 3 (gènes chl A, chl B, chl C) C R Acad Sci Hebd Seances Acad Sci D. 1967 Apr 10;264(15):1916–1918. [PubMed] [Google Scholar]
- Reeve E. C., Doherty P. Linkage relationships of two genes causing partial resistance to chloramphenicol in Escherichia coli. J Bacteriol. 1968 Oct;96(4):1450–1451. doi: 10.1128/jb.96.4.1450-1451.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. L., Bedbrook J. R., Grant F. J., Kleinhofs A. Reconstitution of plant nitrate reductase by Escherichia coli extracts and the molecular cloning of the chlA gene of Escherichia coli K12. J Mol Appl Genet. 1983;2(3):261–271. [PubMed] [Google Scholar]
- Tomsett A. B., Garrett R. H. Biochemical analysis of mutants defective in nitrate assimilation in Neurospora crassa: evidence for autogenous control by nitrate reductase. Mol Gen Genet. 1981;184(2):183–190. doi: 10.1007/BF00272903. [DOI] [PubMed] [Google Scholar]
- Tomsett A. B., Garrett R. H. The isolation and characterization of mutants defective in nitrate assimilation in Neurospora crassa. Genetics. 1980 Jul;95(3):649–660. doi: 10.1093/genetics/95.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Venables W. A. Genetic studies with nitrate reductase-less mutants of Escherichia coli. I. Fine structure analysis of the narA, narB and narE loci. Mol Gen Genet. 1972;114(3):223–231. doi: 10.1007/BF01788891. [DOI] [PubMed] [Google Scholar]
- Venables W. A., Guest J. R. Transduction of nitrate reductase loci of Escherichia coli by phages P-1 and lambda. Mol Gen Genet. 1968;103(2):127–140. doi: 10.1007/BF00427140. [DOI] [PubMed] [Google Scholar]
- Wadman S. K., Duran M., Beemer F. A., Cats B. P., Johnson J. L., Rajagopalan K. V., Saudubray J. M., Ogier H., Charpentier C., Berger R. Absence of hepatic molybdenum cofactor: an inborn error of metabolism leading to a combined deficiency of sulphite oxidase and xanthine dehydrogenase. J Inherit Metab Dis. 1983;6 (Suppl 1):78–83. doi: 10.1007/BF01811328. [DOI] [PubMed] [Google Scholar]
- Wahl R. C., Hageman R. V., Rajagopalan K. V. The relationship of Mo, molybdopterin, and the cyanolyzable sulfur in the Mo cofactor. Arch Biochem Biophys. 1984 Apr;230(1):264–273. doi: 10.1016/0003-9861(84)90107-3. [DOI] [PubMed] [Google Scholar]
- Warner C. K., Finnerty V. Molybdenum hydroxylases in Drosophila. II. Molybdenum cofactor in xanthine dehydrogenase, aldehyde oxidase and pyridoxal oxidase. Mol Gen Genet. 1981;184(1):92–96. doi: 10.1007/BF00271201. [DOI] [PubMed] [Google Scholar]
- del Campillo-Campbell A., Campbell A. Molybdenum cofactor requirement for biotin sulfoxide reduction in Escherichia coli. J Bacteriol. 1982 Feb;149(2):469–478. doi: 10.1128/jb.149.2.469-478.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]



