Abstract
The development of a new strategy for antibody humanization is described. This strategy incorporates key recognition sequences from the parental rodent antibody into a phage display-based selection strategy. The original sequences of the third complementarity-determining regions (CDRs) of heavy and light chains, HCDR3 and LCDR3, were maintained and all other sequences were replaced by human sequences selected from phage-displayed antibody libraries. This approach was applied to the humanization of mouse mAb LM609 that is directed to human integrin αvβ3 and has potential applicability in cancer therapy as an antiangiogenic agent. We demonstrate this approach (i) provides a rapid route for antibody humanization constraining the content of original mouse sequences in the final antibodies to the most hypervariable of the CDRs; (ii) generates several humanized versions with different sequences at the same time; (iii) results in affinities as high as or higher than the affinity of the original antibody; and (iv) retains the antigen and epitope specificity of the original antibody. The production of multiple humanized variants may present advantages in the selection of antibodies that are more readily expressed on a large scale and could be important in therapeutic regimens that call for long-term treatment with antibodies in which antiidiotypic responses might be avoided by administration of alternative antibodies.
Since the development of the hybridoma approach (1), a large number of rodent mAbs with specificity for antigens of therapeutic interest have been generated and characterized. The fact that rodent antibodies are highly immunogenic in humans, however, severely limits their clinical applications, especially when repeated administration is required for therapy. As a means of circumventing this limitation, several strategies have been developed to convert rodent antibody sequences into human antibody sequences, a process termed antibody humanization. Ideally, antibody humanization must not diminish specificity and affinity toward the antigen whereas immunogenicity must be completely eliminated. It has become apparent that the accomplishment of both aims is usually a time-consuming and costly undertaking with even the most current humanization strategies. Here, we report the development of a new humanization strategy that combines rational design with combinatorial selections using phage display. We demonstrate that this approach provides a rapid route to antibody humanization and demonstrate its application to the humanization of mouse mAb LM609 which is directed against the human integrin αVβ3. We chose LM609 as a model antibody for our humanization strategy because of its clinical potential. Recent findings by Brooks et al. (2–4) in a chorioallantoic membrane model and a severe combined immunodeficient mouse/human skin chimeric model have shown that LM609, when administered i.v., is able to reduce growth and metastasis of human tumors due to the inhibition of angiogenesis induced by the tumors. These findings suggest that integrin αVβ3 may be a target and LM609 a tool for cancer therapy.
MATERIALS AND METHODS
Proteins and Cell Lines.
Human integrin αvβ3 was purified from human placenta as described (5). Human integrin αIIbβ3 was purchased from Enzyme Research Laboratories (South Bend, IN). mAb LM609 was described previously (6) and mAb AP3 was kindly provided by P. Newman (Milwaukee Blood Center, Milwaukee, WI). LM609 Fab was generated from IgG by digestion with immobilized papain using the ImmunoPure Fab Preparation kit from Pierce and separated from Fc and undigested IgG by three consecutive runs on a protein A column. CS-1 hamster cells were transfected with either human β3 or β5 cDNA as described (7) and maintained in RPMI 1640 supplemented with 10% fetal calf serum and 500 μg/ml G-418 (Life Technologies, Gaithersburg, MD) at 37°C and in 7% CO2.
cDNA Cloning of LM609.
Total RNA was prepared from 108 LM609 hybridoma cells (6) using the RNA Isolation kit from Stratagene. Reverse transcription and PCR amplification of the Fd fragment- and light chain-coding sequences were performed essentially as described (8). Fd fragment- and light chain-coding PCR products were cut with XhoI/SpeI and SacI/XbaI, respectively, and ligated sequentially into the appropriately digested phagemid vector pComb3H (9). The ligation products were introduced into Escherichia coli strain XL1-Blue by electrotransformation and subsequent steps were as described (10) to produce phage displaying Fab on their surface. Phage were selected by panning (10) against immobilized human integrin αVβ3. After two panning rounds, single clones were analyzed for LM609 Fab expression. Supernatants from cultures that had been induced by the addition of isopropyl β-d-thiogalactopyransoside (10) were tested for binding to αVβ3 by ELISA using goat anti-mouse F(ab′)2 conjugated to alkaline phosphatase (Pierce) as secondary antibody. The sequence of Fd fragment- and light chain-coding sequences of positive clones was determined by DNA sequencing.
Amplification of Human Light Chain and Fd Fragment Sequences.
Total RNA was prepared from bone marrow of five healthy donors supplied by Poietic Technologies (Germantown, MD) shortly after aspiration using TRI REAGENT (Molecular Research Center, Cincinnati, OH) and was further purified by lithium chloride precipitation (11). First-strand cDNA was synthesized using the SUPERSCRIPT Preamplification System for First Strand cDNA Synthesis kit with oligo(dT) priming (Life Technologies). The generated five first-strand cDNAs were subjected to separate PCR amplifications. Vκ, Vλ, and VH sequences of each of the first-strand cDNAs were amplified using the primers listed below. All amplifications were performed under standard PCR conditions using Taq polymerase (Pharmacia). While the sense primers hybridize to sequences that encode the N-terminal amino acids of the various Vκ, Vλ, and VH families, the antisense primers hybridize to sequences that encode the C-terminal amino acids of framework region 3 (FR3) of Vκ, Vλ, or VH, respectively, which are highly conserved (12). The primers used for the amplification of human antibody sequences are Vκ sense primers: HSCK1-F, 5′-GGGCCCAGGCGGCCGAGCTCCAGATGACCCAGTCTCC-3′; HSCK24-F, 5′-GGGCCCAGGCGGCCGAGCTCGTGATGACYCAGTCTCC-3′; HSCK3-F, 5′-GGGCCCAGGCGGCCGAGCTCGTGWTGACRCAGTCTCC-3′; and HSCK5-F, 5′-GGGCCCAGGCGGCCGAGCTCACACTCACGCAGTCTCC-3′; Vκ antisense primers: BKFR3UN, 5′-CAGTAATACACTGCAAAATCTTC-3′; BK2FR3UN and 5′-CAGTAATAAACCCCAACATCCTC-3′; Vλ sense primers: HSCLam1a, 5′-GGGCCCAGGCGGCCGAGCTCGTGBTGACGCAGCCGCCCTC-3′; HSCLam1b, 5′-GGGCCCAGGCGGCCGAGCTCGTGCTGACTCAGCCACCCTC-3′; HSCLam2, 5′-GGGCCCAGGCGGCCGAGCTCGCCCTGACTCAGCCTCCCTCCGT-3′; HSCLam3, 5′-GGGCCCAGGCGGCCGAGCTCGAGCTGACTCAGCCACCCTCAGTGTC-3′; HSCLam4, 5′-GGGCCCAGGCGGCCGAGCTCGTGCTGACTCAATCGCCCTC-3′; HSCLam6, 5′-GGGCCCAGGCGGCCGAGCTCATGCTGACTCAGCCCCACTC-3′; HSCLam70, 5′-GGGCCCAGGCGGCCGAGCTCGGGCAGACTCAGCAGCTCTC-3′; HSCLam78, 5′-GGGCCCAGGCGGCCGAGCTCGTGGTGACYCAGGAGCCMTC-3′; and HSCLam9, 5′-GGGCCCAGGCGGCCGAGCTCGTGCTGACTCAGCCACCTTC-3′; Vλ antisense primer: BLFR3UN, 5′-GCAGTAATAATCAGCCTCRTC-3′; VH sense primers: HFVH1-F, 5′-GCTGCCCAACCAGCCATGGCCCAGGTGCAGCTGGTGCAGTCTGG-3′; HFVH2-F, 5′-GCTGCCCAACCAGCCATGGCCCAGATCACCTTGAAGGAGTCTGG-3′; HFVH35-F, 5′-GCTGCCCAACCAGCCATGGCCGAGGTGCAGCTGGTGSAGTCTGG-3′; and HFVH4-F, 5′-GCTGCCCAACCAGCCATGGCCCAGGTGCAGCTGCAGGAGTCGGG-3′; VH antisense primer: BFR3UN, 5′-CGCACAGTAATACACGGCCGTGTC-3′.
Construction of a Chimeric Mouse/Human Fd Fragment by Fusing VH of LM609 to Human CH1.
The phagemid vector pComb3H containing the LM609 Fab sequence was used as a template for amplification of the sequence encoding the N-terminal FR1 through FR3 fragment of the LM609 VH by the PCR primer pair PELSEQ (5′-ACCTATTGCCTACGGCAGCCG-3′)/BFR3UN (5′-CGCACAGTAATACACGGCCGTGTC-3′). By overlap-extension PCR (13), the PELSEQ/BFR3UN product was fused to a PCR fragment encoding the HCDR3 of LM609, FR4 of VH, and the entire CH1 domain of the human anti-gp120 antibody b8 (14). This fragment was generated from the PCR primer pair CR501 (5′-GACACGGCCGTGTATTACTGTGCGCGTCATAACTACGGCAGTTTTGCTTACTGGGGCCAGGGAACCCTG-3′)/CR301 (5′-GAGGAGGAGGAGGAGACTAGTTTTGTCACAAGATTTGGGCTC-3′). FR4 of b8 was chosen because it is identical to FR4 of the LM609 VH, with the exception of the C-terminal amino acid, which is A for LM609 and S for b8. The product of the overlap-extension PCR was cut with XhoI/SpeI, ligated into the appropriately digested phagemid vector pComb3H, cloned, and the correct sequence was confirmed by DNA sequencing.
Substitution of the LM609 Light Chain by a Human Light Chain That Contains the LCDR3 of LM609.
Using overlap-extension PCR, the amplified human sequences encoding the N-terminal FR1 through FR3 fragment of Vκ and Vλ were fused to PCR fragments encoding the LCDR3 of LM609 coupled to FR4 of human Vκ or Vλ and the human Cκ or Cλ domain. Two κ fragments were generated by the PCR primer pairs CR503 (5′-GAAGATTTTGCAGTGTATTACTGCCAACAGAGTAACAGCTGGCCTCACACGTTTGGCCAGGGGACCAAGCTG-3′)/T7B (5′-AATACGACTCACTATAGGGCG-3′) and CR508 (5′-GAGGATGTTGGGGTTTATTACTGCCAACAGAGTAACAGCTGGCCTCACACGTTTGGCCAGGGGACCAAGCTG-3′)/T7B using the sequence of the anti-gp120 antibody b11 in pComb3 as a template (14). FR4 of b11 was chosen because it is identical to FR4 of the LM609 Vκ, with the exception of the third amino acid, which is G in LM609 and Q in b11. The 23-bp overlap of CR503 with BKFR3UN and CR508 with BK2FR3UN allowed the fusion of the corresponding PCR products by overlap-extension PCR. A λ fragment was generated by the PCR primer pair CR510 (5′-GAYGAGGCTGATTATTACTGCCAACAGAGTAACAGCTGGCCTCACACGTTCGGCGGAGGGACCAAGCTG-3′)/CLext (5′-AGAGAGAGAGAGAGAGAGAGCGCCGTCTAGAATTATGAACATTCTGTAGG-3′) using CLext-primed, first-strand cDNA from human bone marrow as a template. The 21-bp overlap of CR510 with BLFR3UN allowed the fusion of the corresponding PCR products by overlap-extension PCR. The generated light chain-coding sequences were cut with SacI/XbaI and ligated into the appropriately digested phagemid vector pComb3H that contained the chimeric mouse/human Fd fragment. Electrotransformation of the ligation products into E. coli strain ER 2537 (New England Biolabs) resulted in a light chain library consisting of 1.5 × 108 independent transformants. DNA sequencing revealed the correct assembly of the fused fragments. Four rounds of panning against immobilized human integrin αVβ3 were carried out using 200 ng of protein in 25 μl of metal buffer [25 mM Tris⋅HCl (pH 7.5), 137 mM NaCl, 1 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 1 mM MnCl2] for coating, 0.05% Tween 20 in Tris-buffered saline for washing, and 10 mg/ml trypsin (Difco) in Tris-buffered saline for elution. Trypsinization was for 30 min at 37°C. The washing steps were increased from 5 in the first round to 10 in the second round and 15 in the third and fourth rounds. The output phage pool of each round was monitored by phage ELISA using sheep anti-M13 conjugated to horseradish peroxidase (Pharmacia) as secondary antibody. After the fourth round, phage were produced from single clones and tested for binding to αVβ3 by phage ELISA. Light chain-coding sequences of positive clones were analyzed by DNA sequencing using the primer OMPSEQ (5′-AAGACAGCTATCGCGATTGCAG-3′).
Substitution of the LM609 Fd Fragment by a Human Fd Fragment That Contains the HCDR3 of LM609.
Three PCR fragments were fused in one step by overlap-extension PCR. Using the selected phagemids from the light chain panning as a template, fragment 1 was amplified with the PCR primer pair RSC-F (5′-GAGGAGGAGGAGGAGGAGGCGGGGCCCAGGCGGCCGAGCTC-3′)/lead-B (5′-GGCCATGGCTGGTTGGGCAGC-3′). While the sense primer RSC-F hybridizes to a sequence upstream of the light chain-coding sequence, the antisense primer lead-B hybridizes to a sequence upstream of the Fd fragment-coding sequence. The amplified human sequences encoding FR1 through FR3 of the VH fragment (see above) were used as fragment 2. Fragment 3 was amplified with the PCR primer pair CR501/HIgG1-B (5′-GCAGAGCCCAAATCTTGTGACACTAGTGGCCAGGCCGGCCAG-3′) using the hybrid mouse/human Fd fragment (see above) as a template. The antisense primer HIgG1-B hybridizes to the 3′ end of the CH1-coding sequence. Using the 21-bp overlap of lead-B with the HFVH-F primers and the 24-bp overlap of BFR3UN with CR501, the three fragments were fused and amplified with the PCR primer pair RSC-F/RSC-B (5′-GAGGAGGAGGAGGAGGAGCCTGGCCGGCCTGGCCACTAGTG-3′). The antisense primer RSC-B overlaps with HIgG1-B. RSC-F and RSC-B introduce two asymmetric SfiI sites. To maintain high complexity, separate PCRs were performed for each selected phagemid from the light chain panning (fragment 1) and for each of the five VH fragment pools derived from the five first-strand cDNA sources (fragment 2). The generated fragments encoding the selected human light chains linked to human Fd fragments were cut with SfiI and ligated into the appropriately digested phagemid vector pComb3H, generating a library of 3 × 107 independent transformants. DNA sequencing revealed the correct assembly of the fused DNA fragments. Four rounds of panning against immobilized human integrin αVβ3 were carried out as described for the light chain panning. The output phage pool of each round was monitored by phage ELISA. After the fourth round, soluble Fab was produced from single clones as described (10) and tested for binding to immobilized αVβ3 by ELISA using goat anti-human F(ab′)2 conjugated to alkaline phosphatase (Pierce) as secondary antibody. Light chain- and Fd fragment-coding sequences of positive clones were analyzed by DNA sequencing using the primers OMPSEQ and PELSEQ, respectively.
Flow Cytometry.
Flow cytometry was performed using a FACSscan instrument from Becton Dickinson. For each determination, 5 × 103 untransfected hamster CS-1 cells or hamster CS-1 cells transfected with either human β3 or β5 cDNA were analyzed. Indirect immunofluorescence staining was performed with 2 μg/ml Fab in 1% BSA, 25 mM Hepes, and 0.05% sodium azide in PBS supplemented with 1% nonimmune goat serum. A 1:100 dilution of fluorescein isothiocyanate-conjugated goat anti-human F(ab′)2 (Jackson ImmunoResearch Laboratories) was used for detection. Incubation with primary antibodies was for 1 h, with secondary antibodies for 30 min, at room temperature. Competition experiments were performed by adding a fourfold molar excess of LM609 or AP3 IgG to the incubation mixture with the primary antibodies.
Surface Plasmon Resonance.
Association (kon) and dissociation (koff) rate constants for binding of mouse and humanized LM609 Fab to human integrin αvβ3 were determined by surface plasmon resonance on a Biacore instrument (Pharmacia). The sensor chip was activated for immobilization with N-hydroxysuccinimide and N-ethyl-N′-(3-diethyl aminopropyl)carbodiimide according to the methods outlined by Pharmacia. Human integrin αVβ3 was coupled to the surface by injection of 6–9 μl of a 50 ng/μl sample in 10 mM sodium acetate (pH 3.5). Between 5,000 and 10,000 resonance units were immobilized. Subsequently, the sensor chip was deactivated with 1 M ethanolamine (pH 8.5). Binding of Fab to αVβ3 was studied by injection of Fab in a range of concentrations (10–600 nM), using PBS as a running buffer. The sensor chip was regenerated with 10 mM HCl and remained active for at least 50 measurements. Based on five measurements at different Fab concentrations, the kon and koff values were calculated using Biacore kinetics evaluation software (Pharmacia) and the equilibrium dissociation constant, Kd, was calculated from koff/kon. The reliability of the data was validated by analyzing the binding of each Fab on at least two different sensor chips and by applying the internal consistency tests suggested by Schuck and Minton (15). In addition, the rough range of the Kd values was independently confirmed by competition ELISA using a procedure described by Friguet et al. (16).
RESULTS
cDNA Cloning of LM609.
cDNAs encoding the Fd fragment and entire light chain were cloned by PCR from LM609-expressing hybridoma cells (6). The PCR products were cloned into the phage display vector pComb3H (9), which is derived from pComb3 (10), and engineered to facilitate the expression of Fab on the surface of M13 filamentous phage. Phage displaying LM609 Fab were selected by panning against immobilized human integrin αVβ3 and the corresponding cDNA sequences were determined. The cloned LM609 Fab purified from E. coli demonstrated specific binding to αVβ3 using the ELISA.
Humanization of the Light Chain of LM609.
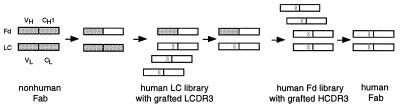
Our humanization strategy is outlined in Fig. 1. It involves two selection steps for the sequential humanization of the light chain and the Fd fragment of the heavy chain. Throughout these selections the only preserved sequences in the variable domains of light chain (VL) and heavy chain (VH) are two of six CDRs, LCDR3 and HCDR3.
Figure 1.
Humanization of nonhuman monoclonal antibodies by a combination of CDR grafting and V gene shuffling. Nonhuman sequences are shown in gray, human sequences in white. In the first step, a chimeric nonhuman/human Fd fragment is used as a template for the selection of a human light chain that contains the grafted LCDR3 loop of the nonhuman light chain. In the second step, a human Fd fragment that contains the grafted HCDR3 loop of the nonhuman Fd fragment is selected. The sequential V gene shuffling procedure is based on phage display. LCDR3, complementarity-determining region 3 of light chain; HCDR3, complementarity-determining region 3 of heavy chain.
For the humanization of the light chain, the mouse Fd fragment was substituted by a chimeric Fd fragment composed of mouse VH linked to the human constant domain 1 of the heavy chain, CH1. A single residue in the mouse FR4 was converted to the corresponding human residue, resulting in complete humanization of the FR4 region. Humanization of the light chain began by substituting the Vκ gene segment of LM609 by a human Vκ and Vλ gene library joined at the LCDR3 junction. As described for the Fd fragment, the FR4 region was humanized by one amino acid change and appended to a human Cκ region. To ensure a highly diverse V gene library, the human antibody sequences were amplified from cDNA prepared from the bone marrow of five healthy individuals using a variety of oligonucleotides that were designed to amplify most of the known human antibody sequences (see Materials and Methods). The corresponding phage libraries displaying hybrid Fab were combined and selected by four rounds of panning against immobilized human integrin αVβ3. Analysis of the output phage pool from each round for binding to αVβ3 by phage ELISA revealed an increasing signal. After the fourth round of selection, six clones that demonstrated strong reactivity to the antigen were studied. DNA sequence analysis of these clones revealed three different light chain sequences. Two light chains (Fig. 2A), found in five of six positive clones, differed in only four amino acids (i.e., they were 96% identical), whereas a third light chain sequence shared about 80% identity with the other two (data not shown). This latter sequence consisted of two parts, each of which could be aligned to germ-line genes from different Vκ families; thus, this light chain sequence probably arose from PCR crossover, which has been reported to occur frequently in the amplification of antibody sequences (17).
Figure 2.
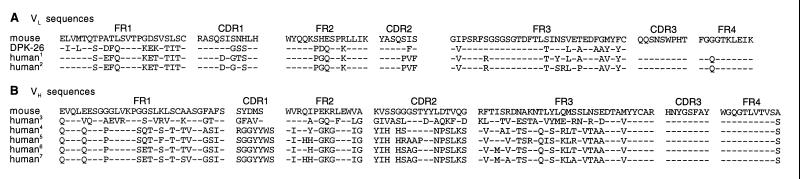
Amino acid sequence alignment of mouse and humanized LM609. Shown are FRs and CDRs. Dashes indicate identical amino acids. Note that due to our grafting procedure CDR3 is identical in the original mouse and the selected human sequences. (A) Alignment of the selected human VL sequences. Databank screening revealed that the two selected human VL sequences are derived from germ-line DPK-26 of the Vκ3 family. (B) Alignment of the selected human VH sequences. Five different human VH sequences were selected. Four of them, represented by clones 7, 4, 24, and 2, are highly related in amino acid sequence; they share an identical VL domain and an amino acid sequence identity of at least 85% in their VH domains. The VH sequences are all derived from germ-line DP-65 or the highly related DP-78. In contrast, clone 11 represents a humanized version with a VH domain that is derived from a different germ-line family. This humanized version also contains a different VL domain which is 96% identical and derived from the same germ-line. Germ-lines were determined by nucleic acid sequence alignment using DNAPLOT software provided by the VBASE Directory of Human V Gene Sequences (http://www.mrc-cpe.cam.ac.uk/imt-doc/). 1clone 11, germ-line DPK-26 (Vκ6 family); 2clones 2, 4, 7, 24, germ-line DPK-26 (Vκ6 family); 3clone 11, germ-line DP-10 (VH1 family); 4clone 7, germ-line DP-78 (VH4 family); 5clone 4, germ-line DP-65 (VH4 family); 6clone 24, germ-line DP-65 (VH4 family); 7clone 2, germ-line DP-65 (VH4 family).
The selected human light chains are κ light chains as was the original mouse light chain. Databank screening revealed that the selected human light chains are derived from the same germ-line gene, namely, DPK-26, which belongs to the Vκ6 family (Fig. 2A). This result supports a strong selection for the light chains since the Vκ6 family represents only a small fraction of the expressed human Vκ repertoire (18, 19). An obvious reason for this strong selection is the strong sequence similarity between the selected human light chains and the original mouse light chain. Limited sequencing of the unselected library confirmed its diversity and did not reveal any Vκ6-containing clones. Moreover, in contrast to the unselected sequences, both LCDR1 and LCDR2 of the selected human light chains are highly similar to the corresponding mouse sequence (Fig. 2A). The C-terminal amino acid of FR2 (Kabat position 49; ref. 12) of the original mouse light chain sequence is a lysine, which is an unusual amino acid at this position and, thus, may be involved in the formation of the antigen-binding site. Interestingly, this lysine is conserved in our selected sequences (Fig. 2A) as well as in the two known human germ-line Vκ6 sequences, whereas all of the unselected sequences that were analyzed contained a tyrosine instead. The Vκ6 family is the only human Vκ family that contains a lysine at this position. To study whether the selected human light chains are derived from germ-line V genes that are most similar to the original mouse light chain, the VBASE directory of human V gene sequences (maintained by I. M. Tomlinson, @ http://www.mrc-cpe.cam.ac.uk/imt-doc/) was searched for the highest sequence similarity with the original VL. Indeed, germ-lines DPK-26 and DPK-25, the only two members of the Vκ6 family, were determined to be most similar to the original mouse light chain. Thus, library and databank screening yielded the same result.
Three clones from the light chain selection demonstrated weaker binding to αVβ3 than the six clones discussed above, but still gave significant binding above background. DNA sequencing revealed three unrelated Vλ sequences that had no apparent similarity to the original mouse Vκ sequence. The Vλ sequences, along with the selected Vκ sequences (and with the exception of the clone containing the PCR crossover), were used as templates in the humanization of the heavy chain of LM609.
Humanization of the Heavy Chain of LM609.
A library of Fd fragments was prepared by stitching human VH gene libraries onto the chimeric Fd fragment described above. These libraries were paired with the four selected human light chains and were selected by four rounds of panning against immobilized human integrin αVβ3. As seen for the human light chain selection, analysis of the output phage pool from each round for binding to αVβ3 by phage ELISA revealed an increasing signal. After the fourth round, Fab was produced from single clones and tested for binding to αVβ3 by ELISA. Light chain- and Fd fragment-coding sequences from 14 binding clones were determined by DNA sequencing and revealed five different sequences (Fig. 2). The two human κ light chains with the highest sequence homology to the LM609 light chain were reselected. One of them was found to pair with four different Fd fragments that were closely related to each other (85–96% sequence identity) and derived from germ-line DP-65 or DP-78 of the VH4 family. The other human κ light chain was found to pair with a Fd fragment that was derived from germ-line DP-10 of the VH1 family. Neither germ- lines from the VH4 nor the VH1 family show high sequence homology with the VH of LM609. Indeed, databank screening yielded a germ-line from the VH3 family as the best human match for the VH of LM609. Phylogenetic analysis has shown that VH1, VH3, and VH4 not only form separate families but belong to different clans of the human VH germ-lines (20). In retrospect, Vλ selections should have been performed independent of Vκ selections to ensure the retention of highly diverse light chains in the humanized antibodies. The Vλ antibodies may have been lost in the selections for reasons other than antibody affinity, such as their relative toxicity to E. coli.
Binding Specificity and Affinity of Humanized LM609.
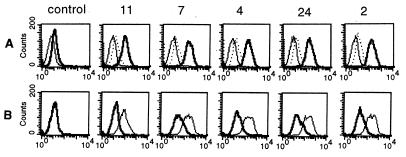
Five humanized LM609 versions, represented by clones 11, 7, 4, 24, and 2 were produced as soluble Fab by E. coli, purified by affinity chromatography, and their binding specificity and affinity was analyzed. Humanized LM609, which had been selected by binding to immobilized and thus potentially denatured human integrin αvβ3, was tested for binding to native human integrin αvβ3 expressed on the cell surface. For this, binding of humanized LM609 to untransfected CS-1 hamster cells and CS-1 hamster cells transfected with either human β3 or β5 cDNA (7) was analyzed by flow cytometry. By recruiting the endogenous hamster αv polypeptide, the human β3 and β5 polypeptides form functional integrins on the cell surface (7). Like mouse LM609, and in contrast to unrelated human Fab fragments that were used as controls (data not shown), all five humanized versions of LM609 revealed specific binding to CS-1 hamster cells transfected with human β3 (Fig. 3A). Binding of all five humanized LM609 versions to CS-1 cells transfected with human β3 cDNA could be blocked by an excess of LM609. As shown in Fig. 3B, a fourfold molar excess of mouse LM609 IgG blocked binding of humanized LM609 almost completely, whereas the same concentration of mouse AP3 IgG directed to a different epitope on human integrin αvβ3 had no effect. A 20-fold molar excess of LM609 Fab derived from IgG by papain digestion also blocked the binding of humanized LM609 (data not shown). Control experiments revealed that both LM609 and AP3 bound to αvβ3 expressed on the cell surface. Potential cross-reactivity of humanized LM609 with human integrin αIIbβ3 was analyzed by ELISA. Whereas an engineered RGD peptide mimetic antibody Fab-9 with known cross-reactivity (21) bound to both immobilized human integrin αvβ3 and αIIbβ3, cross-reactivity with αIIbβ3 was not detected for mouse LM609 nor its five humanized versions.
Figure 3.
Flow cytometry histograms demonstrating that humanized LM609 binds specifically to human integrin αvβ3 and can be blocked by mouse LM609. (A) Binding of humanized LM609 Fab to untransfected CS-1 hamster cells (fine line) and CS-1 hamster cells transfected with either human β3 (bold line) or β5 cDNA (dotted line). (B) Binding of humanized LM609 Fab to CS-1 hamster cells transfected with human β3 cDNA in the presence of a fourfold molar excess of mouse AP3 IgG (fine line) or mouse LM609 IgG (bold line). Clones 11, 7, 4, 24, and 2 represent the five humanized LM609 versions (cf. Fig. 2). Controls in A and B are based on identical experiments using buffer instead of humanized LM609 Fab. The y axis gives the number of events in linear scale, the x axis the fluorescence intensity in logarithmic scale.
The kinetic parameters of Fab binding to human integrin αvβ3 binding were determined using surface plasmon resonance and the affinities were calculated from these kinetic parameters (Table 1). Analysis of these binding data revealed that the four humanized LM609 versions with the high sequence similarity, represented by clones 7, 4, 24, and 2, also show similar affinities for αvβ3. Clones 4, 24, and 2, which are derived from the same germ-lines, have almost identical Kd values in the range of 7–9 nM. Clone 7, which shares the same light chain but contains a Fd fragment that is derived from a different, though highly related VH germ-line, has a moderately higher affinity with a Kd value of 3 nM. In contrast, clone 11 with the unrelated Fd fragment has a much weaker affinity, which is mainly caused by a lower association rate constant (Table 1). LM609 Fab was analyzed for comparison. As Table 1 shows, Fab that was generated by papain digestion from LM609 IgG revealed an affinity with a Kd value of about 3 nM, whereas the affinity of Fab produced by E. coli was weaker by a factor of three. It is likely that this discrepancy is partly due to a lower concentration of functional Fab in preparations from E. coli. However, the higher dissociation rate constant of E. coli-derived LM609 Fab, which is independent from the concentration, indicates that the quality of the antigen-binding site might be affected as well. In any case, the humanized LM609 versions were derived from E. coli as well and, thus, should be compared with the corresponding LM609 preparation. Such a comparison (Table 1) shows that four of five humanized antibodies have an affinity that is as good as or better than the original mouse antibody.
Table 1.
Binding kinetics of mouse LM609 and humanized LM609 Fab
| Clone | kon/104, M−1 s−1 | koff/10−4, s−1 | Kd, nM |
|---|---|---|---|
| LM609* | 14 | 4.6 | 3.3 |
| LM609 | 8.6 | 8.6 | 10 |
| 11 | 1.0 | 16 | 160 |
| 7 | 18 | 5.4 | 3.0 |
| 4 | 6.8 | 5.8 | 8.5 |
| 24 | 13 | 9.9 | 7.6 |
| 2 | 11 | 7.5 | 6.8 |
Binding kinetics were determined using surface plasmon resonance. Human integrin αvβ3 was immobilized on the sensor chip. The Kd value was calculated from koff/kon. Clones 11, 7, 4, 24, and 2 represent the five humanized LM609 versions (cf. Fig. 2). Fab was produced by E. coli except LM609* which was prepared from IgG by papain digestion.
DISCUSSION
Though rodent mAbs have long been regarded as powerful therapeutic agents, a major obstacle for clinical applications has been their immunogenicity in humans. Two routes in antibody engineering have been taken to overcome the immunogenicity of mAbs, either the humanization of rodent mAbs or the direct generation of human mAbs. The latter route has recently gained importance with the development of new methodologies that allow the selection of human mAbs from immune, naive, and synthetic human antibody libraries displayed on phage (22, 23) as well as from transgenic mice (24). More than 20 yr of mAb generation by the classical hybridoma technology, however, has yielded a number of promising pharmaceutical candidates and their humanization compares well to the de novo generation and characterization of human mAb for accessing clinical applications in the coming years.
Currently, CDR grafting is the most frequently used strategy for the humanization of rodent mAbs (25). In this approach the six CDR loops comprising the antigen-binding site of the rodent mAb are grafted into corresponding human framework regions. CDR grafting takes advantage of the conserved structure of the variable Ig domains, with the four framework regions serving as a scaffold that supports the CDR loops. CDR grafting often yields humanized antibodies with much lower affinity because framework residues are involved in antigen binding, either indirectly, by supporting the conformation of the CDR loops, or directly, by contacting the antigen (26). Therefore, it is usually necessary to replace certain framework residues in addition to CDR grafting. The fact that about 30 framework residues potentially contribute to antigen binding (26) makes this fine-tuning step very laborious. Another humanization strategy is the method of resurfacing (27). In this approach only the surface residues of a rodent antibody are humanized.
Though both CDR grafting and resurfacing are based on rational design strategies and iterative optimization (i.e., site-directed mutagenesis of framework residues aided by computer modeling), selective approaches (i.e., randomization of a small set of framework residues and subsequent selection from phage display libraries) have been reported recently in humanization strategies (28, 29).
In vitro selection and evolution of antibodies derived from phage display libraries have become a powerful tool in antibody engineering (for recent reviews cf. refs. 9 and 30). An entirely selective humanization strategy based on phage display libraries has been reported by Jespers et al. (31). In two steps, each polypeptide of the rodent antibody, either light chain or heavy chain, is replaced by a corresponding human polypeptide library and the resulting hybrid antibody library is selected by panning against the particular antigen. Though this strategy may compete with CDR grafting because the arduous fine-tuning steps are unnecessary, the lack of other successful applications of this approach—and our failed attempts to humanize LM609 by this approach for comparative studies—suggest that in contrast to CDR grafting, the general applicability of this approach for antibody humanization is uncertain. Also, since this approach is a sequential chain shuffling procedure, it may lead to the production of a humanized antibody that recognizes a slightly different epitope (32–34). It has been observed that antibodies consisting of the same heavy chain paired with light chains that differ in LCDR3 and elsewhere in VL may bind different epitopes on the same antigen. It is conceivable that this is an interesting feature in particular cases; however, alteration of antigen specificity following antibody humanization is not desired in general.
Based on these general considerations, we designed a strategy that recognizes the key roles of HCDR3 and LCDR3 in antigen recognition and combined this with a selective approach that eliminates the arduous fine-tuning steps associated with CDR grafting as well as all mouse sequence. This strategy for humanizing antibodies is presented in Fig. 1. In the first step a human light chain library with the grafted original LCDR3 replaces the original light chain and a chimeric Fd fragment consisting of the original VH Ig domain fused to a human CH1 Ig domain replaces the original Fd fragment. FR4 of both chains is directly humanized by simple point mutations prior to the first selective step. Sequence changes required in this region should in general be minimal given the substantial homology between mouse and human J genes and should have little effect on the affinity. Human constant regions are preferred to stabilize the hybrid Fab of the first selection step by the interaction of two matching human constant domains Cκ and CH1. In addition, Fab carrying human constant regions are often better expressed in E. coli (35, 36; C.R. and C.F.B. III, unpublished observations), a prerequisite for phage display. In the second step, the selected human light chains from the first step are paired with a human Fd fragment library containing the original HCDR3. Since the heavy chain typically plays the most dominant role in antigen recognition, it is conserved in the first selection step.
By preserving the original LCDR3 and HCDR3 sequences of LM609 while subjecting the remaining sequence to selection, our humanization strategy was designed to ensure antigen specificity and epitope conservation. LCDR3 and HCDR3 contain the hypervariable joints of the V/J and V/D/J gene rearrangements that participate in direct antigen contact in all studied antigen/antibody complexes (37). Unlike the other CDR regions, both LCDR3 and HCDR3 interact with all three CDRs of the other variable domain (38). Thus, although generalizations might be misleading (37), LCDR3 and HCDR3 can be considered to make the most significant contributions to affinity and specificity. Given the tremendous sequence diversity displayed by human antibodies in these regions and the mechanism of its generation, it is difficult, if not impossible in most cases, to classify sequences of these regions as either mouse or human. Thus, from the perspective of sequence these antibodies may be considered completely human. Human HCDR3s are, however, on average longer than mouse HCDR3s and encompass the full range of lengths utilized by mice (39). The HCDR3 length of eight amino acids found in LM609 is well represented in both mouse and human antibodies (39). HCDR3 length will not likely be a significant issue in mouse to human conversions.
Antigen specificity and epitope conservation are critical demands in the humanization of LM609. LM609 binds to a conformational epitope on human integrin αvβ3. Importantly, by binding to this epitope LM609 induces apoptosis in vascular cells expressing αvβ3 (3). In contrast to antibodies that are engineered RGD peptide mimics (21), LM609 does not recognize the related human integrin αIIbβ3. A cross-reactivity with human integrin αIIbβ3, which is expressed on platelets, would preclude the use of LM609 as a tool in cancer therapy. The five humanized LM609 versions were analyzed for antigen specificity in terms of cross-reactivity with the human integrins αIIbβ3 and αVβ5, which are closely related to αVβ3 in sequence and function (40). Neither αIIbβ3 nor αVβ5 was recognized by the five humanized versions of LM609. In addition, all appeared to bind to the same epitope on αVβ3 as LM609, as their binding was specifically blocked in the presence of a molar excess of LM609 but not of AP3, a mAb that binds a different epitope on human integrin αvβ3. In addition to the binding specificity, the binding affinity provides evidence that epitope conservation was obtained; three of five humanized LM609 versions bound αvβ3 with an affinity very similar to that of LM609. Yet, the contribution of the selected CDRs to the antigen-binding site is obvious from the fact that one humanized LM609 version binds αvβ3 with higher, and another with slightly lower, affinity.
The generation of different humanized versions of the parental antibody is an attractive result of this methodology. CDR grafting, in contrast, generates a single humanized version. It is anticipated that an antiangiogenic strategy for cancer therapy would require long-term administration of antibody. It is conceivable that repeated therapeutic application may produce an antiidiotypic response that ablates the efficacy of the antibody. Administration of an equally potent antibody that is unreactive to the antiidiotypic response generated by the first would allow therapy to continue. Indeed, introduction of modest changes within the variable domain of an antibody can dramatically alter its reactivity to an antiidiotypic response (41). Finally, a practical advantage of producing multiple humanized antibodies is that the expression level of antibodies is antibody dependent. Thus, one or more of the humanized antibodies may be more suitable for large-scale production.
Acknowledgments
We thank Terri Jones, Kris Bower, and Jori Sutton for technical assistance and Dr. Peter Steinberger for assistance with flow cytometry. This study was supported by National Institutes of Health Grants AI 37470 and AI 41944 (to C.F.B.). C.R. was supported by postdoctoral fellowships from the Swiss National Science Foundation and the Krebsliga des Kantons Zürich.
ABBREVIATIONS
- CDR
complementarity-determining region
- FR
framework region
References
- 1.Köhler G, Milstein C. Nature (London) 1975;256:503–519. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 2.Brooks P C, Clark R A F, Cheresh D A. Science. 1994;264:569–571. doi: 10.1126/science.7512751. [DOI] [PubMed] [Google Scholar]
- 3.Brooks P C, Montgomery A M P, Rosenfeld M, Reisfeld R A, Hu T, Klier G, Cheresh D A. Cell. 1994;79:1157–1164. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 4.Brooks P C, Strömblad S, Klemke R, Visscher D, Sarkar F H, Cheresh D A. J Clin Invest. 1995;96:1815–1822. doi: 10.1172/JCI118227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith J W, Cheresh D A. J Biol Chem. 1988;263:18726–18731. [PubMed] [Google Scholar]
- 6.Cheresh D A. Proc Natl Acad Sci USA. 1987;84:6471–6475. doi: 10.1073/pnas.84.18.6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filardo E J, Cheresh D A. J Biol Chem. 1994;269:4641–4647. [PubMed] [Google Scholar]
- 8.Huse W D, Sastry L, Iverson S A, Kang A S, Alting-Mees M, Burton D R, Benkovic S J, Lerner R A. Science. 1989;246:1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- 9.Rader C, Barbas C F., III Curr Opin Biotechnol. 1997;8:503–508. doi: 10.1016/s0958-1669(97)80075-4. [DOI] [PubMed] [Google Scholar]
- 10.Barbas C F, III, Kang A S, Lerner R A, Benkovic S J. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. E.15. [Google Scholar]
- 12.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of Proteins of Immunological Interest. 5th Ed. Public Health Service, Natl. Inst. Health, Bethesda: U.S. Department of Health and Human Services; 1991. [Google Scholar]
- 13.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease R M. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 14.Barbas C F, III, Collet T A, Amberg W, Roben P, Binley J M, Hoekstra D, Cababa D, Jones T M, Williamson R A, Pilkington G R, et al. J Mol Biol. 1993;230:812–823. doi: 10.1006/jmbi.1993.1203. [DOI] [PubMed] [Google Scholar]
- 15.Schuck P, Minton A P. Trends Biochem Sci. 1996;21:458–460. doi: 10.1016/s0968-0004(96)20025-8. [DOI] [PubMed] [Google Scholar]
- 16.Friguet B, Chaffotte A F, Djavadi-Ohaniance L, Goldberg M E. J Immunol Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 17.Tomlinson I M, Walter G, Marks J D, Llewelyn M B, Winter G. J Mol Biol. 1992;227:776–798. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- 18.Straubinger B, Thiebe R, Huber C, Osterholzer E, Zachau H G. Biol Chem Hoppe-Seyler. 1988;369:601–607. doi: 10.1515/bchm3.1988.369.2.601. [DOI] [PubMed] [Google Scholar]
- 19.Cox J P L, Tomlinson I M, Winter G. Eur J Immunol. 1994;24:827–836. doi: 10.1002/eji.1830240409. [DOI] [PubMed] [Google Scholar]
- 20.Walter G, Tomlinson I M. In: Antibody Engineering: A Practical Approach. McCafferty J, Hoogenboom H R, Chiswell D J, editors. Oxford: Oxford Univ. Press; 1996. pp. 119–145. [Google Scholar]
- 21.Barbas C F, III, Languino L R, Smith J W. Proc Natl Acad Sci USA. 1993;90:10003–10007. doi: 10.1073/pnas.90.21.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaughan T J, Williams A J, Pritchard K, Osbourn J K, Pope A R, Earnshaw J C, McCafferty J, Hodits R A, Wilton J, Johnson K S. Nat Biotechnol. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 23.Barbas C F., III Nat Med. 1995;1:837–839. doi: 10.1038/nm0895-837. [DOI] [PubMed] [Google Scholar]
- 24.Mendez M J, Green L L, Corvalan J R F, Jia X-C, Maynard-Currie C E, Yang X-D, Gallo M L, Louie D M, Lee D V, Erickson K L, et al. Nat Genet. 1997;15:146–156. doi: 10.1038/ng0297-146. [DOI] [PubMed] [Google Scholar]
- 25.Riechmann L, Clark M, Waldmann H, Winter G. Nature (London) 1988;332:323–327. doi: 10.1038/332323a0. [DOI] [PubMed] [Google Scholar]
- 26.Foote J, Winter G. J Mol Biol. 1992;224:487–499. doi: 10.1016/0022-2836(92)91010-m. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen J T, Henry A H, Searle S J, Guild B C, Roguska M, Rees A R. J Mol Biol. 1994;235:959–973. doi: 10.1006/jmbi.1994.1050. [DOI] [PubMed] [Google Scholar]
- 28.Rosok M J, Yelton D E, Harris L J, Bajorath J, Hellström K-E, Hellström I, Cruz G A, Kristensson K, Lin H, Huse W D, Glaser S M. J Biol Chem. 1996;271:22611–22618. doi: 10.1074/jbc.271.37.22611. [DOI] [PubMed] [Google Scholar]
- 29.Baca M, Presta L G, O’Connor S J, Wells J A. J Biol Chem. 1997;272:10678–10684. doi: 10.1074/jbc.272.16.10678. [DOI] [PubMed] [Google Scholar]
- 30.Hoogenboom H R. Trends Biotechnol. 1997;15:62–70. doi: 10.1016/S0167-7799(97)84205-9. [DOI] [PubMed] [Google Scholar]
- 31.Jespers L S, Roberts A, Mahler S M, Winter G, Hoogenboom H R. Bio/Technology. 1994;12:899–903. doi: 10.1038/nbt0994-899. [DOI] [PubMed] [Google Scholar]
- 32.Kang A S, Jones T M, Burton D M. Proc Natl Acad Sci USA. 1991;88:11120–11123. doi: 10.1073/pnas.88.24.11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zebedee S L, Barbas C F, III, Hom Y-L, Caothien R H, Graff R, DeGraw J, Pyati J, LaPolla R, Burton D R, Lerner R A, et al. Proc Natl Acad Sci USA. 1992;89:3175–3179. doi: 10.1073/pnas.89.8.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohlin M, Owman H, Mach M, Borrebaeck C A K. Mol Immunol. 1996;33:47–56. doi: 10.1016/0161-5890(95)00123-9. [DOI] [PubMed] [Google Scholar]
- 35.Carter P, Kelley R F, Rodrigues M L, Snedecor B, Covarrubias M, Velligan M D, Wong W L T, Rowland A M, Kotts C E, Carver M E, et al. Bio/Technology. 1992;10:163–167. doi: 10.1038/nbt0292-163. [DOI] [PubMed] [Google Scholar]
- 36.Ulrich H D, Patten P A, Yang P L, Romesberg F E, Schultz P G. Proc Natl Acad Sci USA. 1995;92:11907–11911. doi: 10.1073/pnas.92.25.11907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson I A, Stanfield R L. Curr Opin Struct Biol. 1993;3:113–118. [Google Scholar]
- 38.Padlan E A. Mol Immunol. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 39.Wu T T, Johnson G, Kabat E A. Proteins. 1993;16:1–7. doi: 10.1002/prot.340160102. [DOI] [PubMed] [Google Scholar]
- 40.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 41.Glaser S M, Yelton D E, Huse W D. J Immunol. 1992;149:3903–3913. [PubMed] [Google Scholar]