Abstract
The T cell receptor for antigen (TCR) complex is organized into two functional domains: the antigen-binding clonotypic heterodimer and the signal-transducing invariant CD3 and TCRζ chains. In most vertebrates, there are two different clonotypic heterodimers (TCRαβ and TCRγδ) that define the αβ and γδ T cell lineages, respectively. αβ- and γδTCRs also differ in their invariant chain subunit composition, in that αβTCRs contain CD3γɛ and CD3δɛ dimers, whereas γδTCRs contain only CD3γɛ dimers. This difference in subunit composition of the αβ- and γδTCRs raises the question of whether the stoichiometries of these receptor complexes are different. As the stoichiometry of the murine γδTCR has not been previously investigated, we used two quantitative immunofluorescent approaches to determine the valency of TCRγδ heterodimers and CD3γɛ dimers in surface murine γδTCR complexes. Our results support a model of murine γδTCR stoichiometry in which there are two CD3γɛ dimers for every TCRγδ heterodimer.
The multimeric TCR is composed of an antigen-binding clonotypic heterodimer (TCRαβ or TCRγδ) and a signal-transducing complex, consisting of the CD3 dimers (CD3γɛ and/or CD3δɛ) and a TCRζ homodimer. TCR signaling is required for lineage commitment and repertoire selection during development, for maintenance of the peripheral T cell pool, and for differentiation of naive T cells into effector and memory cell populations during an immune response. Despite the fact that many of the components of the TCR-coupled signaling pathways have been elucidated, it is not precisely known how these signaling events are initially triggered. Two models have been proposed to provide a mechanism for the initiation of TCR signal transduction, with each implicating a distinct stoichiometry for the TCR (for reviews see references 1 and 2). In the first model, the surface TCR complex contains one TCR heterodimer, two CD3 dimers, and one TCRζ homodimer (3–6). According to this monovalent TCR model, ligand engagement would initiate signaling by conformational changes in the subunits and/or by oligomerization of individual TCR complexes. The second model, known as the bivalent TCR model, proposes that the surface TCR complex contains two clonotypic heterodimers, two CD3 dimers, and one TCRζ homodimer (7–10). Triggering of the bivalent TCR complex may still require a conformational change in the subunits, but the presence of two TCR heterodimers within a single TCR complex would preclude the need for receptor oligomerization.
Most studies of TCR stoichiometry have been performed on the αβTCR and, consequently, little is known about the stoichiometry of γδTCR. We recently provided evidence to suggest that the stoichiometry of the γδTCR differs from that of the αβTCR. Specifically, whereas αβTCRs contain both CD3δɛ and CD3γɛ dimers, most γδTCRs were found to contain only CD3γɛ dimers (11). However, these experiments did not resolve whether one or two CD3γɛ dimers are incorporated into the fully assembled γδTCR complex. Interestingly, signal transduction by the γδTCR was shown to be superior to that of the αβTCR after cross-linking of CD3 alone (11). A multivalent γδTCR complex could explain this enhanced signaling capacity of the γδTCR. To determine the stoichiometry of the γδTCR, we developed two quantitative immunofluorescence techniques to measure (a) the ratio of CD3 dimers to TCRγδ heterodimers and (b) the relative percentage of CD3γɛ dimers on the surface of polyclonal γδ T cells. In this paper, we report findings that favor a monovalent model for γδTCR stoichiometry.
RESULTS AND DISCUSSION
Quantifying the ratio of CD3ɛ dimers to TCRγδ heterodimers on the surface of murine γδ T cells
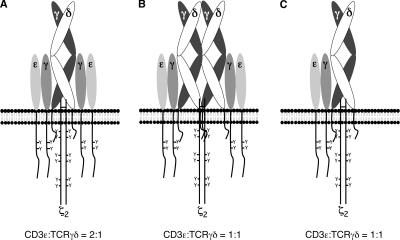
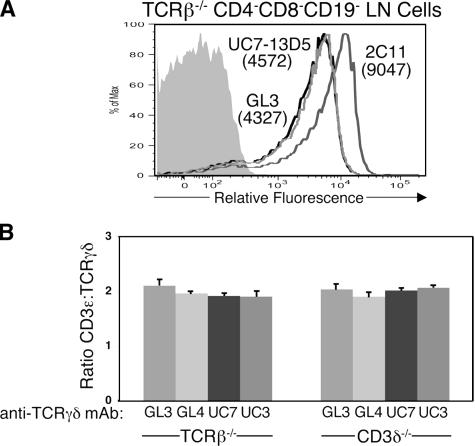
There are three possible models for the stoichiometry of the murine γδTCR. Two of these (Fig. 1, A and B) are based on the present models proposed for the stoichiometry of the αβTCR. The configuration in Fig. 1 A is based on the monovalent αβTCR model (3–6) and depicts the surface γδTCR complex with one TCRγδ heterodimer, two CD3γɛ dimers, and one TCRζ homodimer, for a total of eight subunits. The configuration in Fig. 1 B is based on the alternative bivalent αβTCR model (7–10) and depicts the surface γδTCR complex with 2 TCRγδ heterodimers, 2 CD3γɛ dimers, and 1 TCRζ homodimer, for a total of 10 subunits. It is also conceivable that the rules of γδTCR assembly and surface expression differ from those of the αβTCR, such that a γδTCR complex containing only one CD3γɛ dimer is transported to and stably expressed on the cell surface. This surface complex would contain one TCRγδ heterodimer, one CD3γɛ dimer, and one TCRζ homodimer, for a total of six subunits (Fig. 1 C). As the CD3ɛ/TCRγδ ratio varies in the configurations shown in Fig. 1, quantifying this ratio is the first step in solving the stoichiometry of the murine γδTCR. To this end, we developed a flow cytometric approach similar to those used by others to quantify the CD3ɛ/TCR ratio on primary mouse and human T cells (4, 12). This method takes advantage of the fact that the mAbs against CD3γɛ/δɛ dimers (2C11) and TCRγδ heterodimers (GL3, GL4, UC7-13D5, and UC3-10A6) are all hamster IgG antibodies containing κ light chains. As each primary antibody can be detected with the same anti–hamster Igκ secondary antibody, the relative expression levels of CD3 dimers and TCRγδ heterodimers on the surface of γδ T cells can be measured if saturating amounts of mAb are used. Our approach differs from those of previous studies in that we used a monoclonal anti–hamster antibody instead of polyclonal anti–hamster IgG antibodies, thereby restricting recognition to a single epitope on each primary antibody. A representative staining profile for anti-CD3γɛ/δɛ (2C11) and two anti-TCRγδ (GL3 and UC7-13D5) mAbs on gated CD4−CD8−CD19− LN cells from TCRβ−/− mice is shown in Fig. 2 A. Note that the relative fluorescence of 2C11 mAb staining is approximately twice that of anti-TCRγδ mAb staining, regardless of which anti-TCRγδ mAb was used (GL3, GL4, UC7-13D5, or UC3-10A6; Fig. 2, A and B). These results indicate that there are two CD3 dimers for every TCRγδ heterodimer on the surface of γδ T cells. Importantly, loss of CD3δ expression does not affect this ratio, because we also observed two CD3 dimers for every TCRγδ heterodimer on the surface of CD4−CD8− TCRβ−CD19− LN cells from CD3δ−/− mice (Fig. 2 B). This finding is consistent with previous results demonstrating that neither TCRδ nor TCRγ pairs efficiently to a CD3δɛ dimer (11). The observed 2:1 ratio of CD3 dimers to TCRγδ heterodimers favors the monovalent TCR model shown in Fig. 1 A, in which there is one TCRγδ heterodimer and two CD3 dimers in each γδTCR complex. Thus, our findings indicate that the γδTCR has a signal transducing complex that is similar to that of the αβTCR, in that it contains two CD3 dimers.
Figure 1.
Possible configurations for the murine γδTCR complex. (A) Monovalent γδTCR model. (B) Bivalent γδTCR model. (C) One CD3γɛdimer model.
Figure 2.
Comparison of CD3 and TCRγδ surface levels on murine γδ T cells. (A) Histogram showing the relative fluorescence of 2C11, GL3, and UC7-13D5 (UC7) mAb staining on gated CD4−CD8−CD19− LN cells from TCRβ−/− mice. The 2C11 mAb (dark gray line) recognizes both CD3γɛ and CD3δɛ dimers, and the GL3 (bold line) and UC7 (light gray line) mAbs recognize TCRγδ heterodimers. Staining with a hamster isotype control is also shown (shaded histogram). The number in parentheses represents the mean fluorescence intensity for each mAb minus that of the hamster isotype control. (B) Ratio of CD3ɛ/TCRγδ (for calculation information see Materials and methods) for each anti-TCRγδ mAb on the surface of CD4−CD8−CD19− LN cells from TCRβ−/− mice (GL3, n = 10; GL4, n = 3; UC7, n = 10; UC3, n = 3) and CD4−CD8−TCRβ−CD19− LN cells from CD3δ−/− mice (GL3, n = 8; GL4, n = 4; UC7, n = 8; UC3, n = 3). Bars represent the means ± standard deviation.
Quantifying the relative percentage of CD3γɛ dimers on the surface of murine γδ T cells
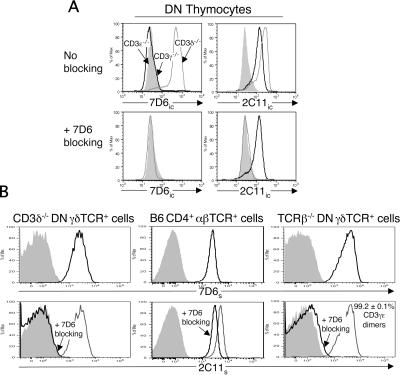
Biochemical analysis suggests that a small percentage of surface γδTCR complexes contain CD3δɛ dimers (11, 13). The γδTCRs that contain CD3δɛ dimers could be restricted to a distinct subpopulation of γδ T cells or may represent a minor subset of TCRs on each γδ T cell. To discern between these two possibilities, we developed a second flow cytometric assay that uses an anti-CD3γɛ mAb (7D6), which has been reported to block the binding of the 2C11 mAb to CD3γɛ dimers but not to CD3δɛ dimers (14). To confirm the specificity of the 7D6 mAb and its ability to block 2C11 mAb staining of CD3γɛ dimers, we assayed double-negative (DN) thymocytes from CD3δ−/−, CD3γ−/−, and CD3ɛ −/− mice (Fig. 3 A). The 7D6 mAb detected the intracellular CD3γɛ dimers present in CD3δ−/− DN thymocytes but not the intracellular CD3δɛ dimers present in CD3γ−/− DN thymocytes. In addition, pretreatment with the 7D6 mAb completely blocked 2C11 mAb staining of intracellular CD3γɛ dimers in CD3δ−/− DN thymocytes but had no effect on 2C11 mAb staining of intracellular CD3δɛ dimers in CD3γ−/− DN thymocytes. Next, we tested the efficacy of this flow cytometric approach. We first assayed γδ T cells from CD3δ−/− mice, which express only CD3γɛ dimers on their cell surface (13), and found that the 7D6 mAb was indeed able to completely block 2C11 mAb surface staining (Fig. 3 B). We then assayed CD4+ αβ T cells from B6 mice, which express both CD3γɛ and CD3δɛ dimers on their cell surface (for review see reference 15) and found, as expected, that the 7D6 mAb only partially blocked 2C11 mAb surface staining (Fig. 3 B). If expression of γδTCRs containing CD3δɛ dimers were limited to a subpopulation of γδ T cells, then pretreatment with purified 7D6 mAb should partially block 2C11 staining on this CD3δ+ subset and completely block 2C11 staining on the CD3δ− subset. However, if CD3δɛ containing γδTCRs were a minor subset of TCRs expressed on each γδ T cell, then pretreatment with purified 7D6 mAb should almost completely block 2C11 staining on all γδ T cells. We found that when γδ T cells from TCRβ−/− mice were assayed, the 7D6 mAb almost completely blocked the staining of the 2C11 mAb, indicating that the TCRs containing CD3δɛ dimers represent a minor subset of TCRs expressed on each γδ T cell (Fig. 3 B). In fact, the relative percentage of CD3γɛ dimers was calculated to be 99.2 ± 0.1% of all CD3 dimers.
Figure 3.
Quantifying the relative percentage of CD3γɛ dimers on the surface of murine γδ T cells. (A) Intracellular staining (ic) of CD3 dimers in gated DN thymocytes from CD3γ−/− (bold line) and CD3δ−/− mice (gray line) using FITC-conjugated 2C11 (anti-CD3γɛ/δɛ) and Alexa 488–conjugated 7D6 (anti-CD3γɛ) mAbs. DN thymocytes are defined as CD4−CD8−CD19−TCRγδ−TCRβ−NK1.1−DX5−. Gated DN thymocytes from CD3ɛ −/− mice were used as a negative staining control (shaded histogram). Staining without (top) and with (bottom) 7D6 mAb blocking is shown. (B) Surface (s) staining of CD3 dimers on gated γδTCR+ CD4−CD8− LN cells from CD3δ−/− and TCRβ−/− mice and on gated αβTCR+ CD4+ LN cells from B6 mice using 2C11–FITC and 7D6–Alexa 488 mAbs. (top) Surface staining of 7D6–Alexa 488 mAb alone (bold line) and after blocking with purified 7D6 mAb (shaded histogram). (bottom) Surface staining of 2C11–FITC mAb alone (gray line), after blocking with purified 7D6 mAb (bold line), and after blocking with purified 2C11 mAb (shaded histogram). The mean relative percentage of CD3γɛ dimers on γδTCR+ CD4−CD8− LN cells from TCRβ−/− mice (n = 4; for calculation see Materials and methods) ± standard deviation is shown.
CD3γ is absolutely required for the assembly and expression of the γδTCR
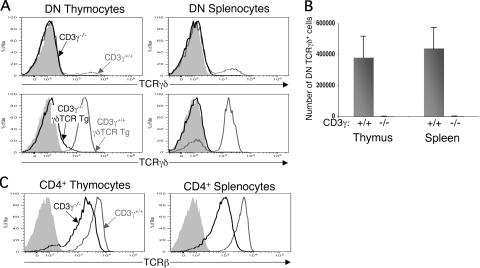
If two CD3γɛ dimers are found in each surface γδTCR complex, then the loss of CD3γ should have profound effects on γδTCR assembly and surface expression. Indeed, Haks et al. have reported that γδ T cell development is severely affected in CD3γ−/− mice (16). We sought to expand these earlier experiments by performing a more detailed analysis of γδTCR surface expression on thymocytes and splenocytes from CD3γ−/− mice and from CD3γ−/− mice carrying a γδTCR transgene. Virtually no γδTCR+ cells were detected in the thymus and spleen of CD3γ−/− mice (Fig. 4, A and B). Moreover, introduction of a γδTCR transgene into CD3γ−/− mice did not increase the number of γδTCR+ thymocytes and splenocytes as it does when introduced into CD3γ+/+ mice, indicating that the absence of γδTCR+ cells in CD3γ−/− mice cannot solely be caused by a failure to express productively rearranged TCRγ and -δ genes (Fig. 4 A). These findings demonstrate that γδTCR assembly and surface expression are absolutely dependent on the presence of CD3γ. Remarkably, unlike the γδTCR, the αβTCR can still be expressed on the surface of CD3γ−/− thymocytes and splenocytes, albeit at reduced levels compared with CD3γ+/+ cells (Fig. 4 C) (16). Therefore, CD3γ−/− mice reveal a difference in the requirement for CD3γ in αβ- and γδTCR assembly and surface expression. Importantly, this difference is consistent with the supposition that TCRγ and -δ chains each pair with a CD3γɛ dimer but not a CD3δɛ dimer.
Figure 4.
Phenotypic analysis of CD3γ−/− and γδTCR Tg CD3γ−/− mice. (A) Comparison of γδTCR surface levels on DN thymocytes and splenocytes from CD3γ−/− (bold line) and CD3γ+/+ (gray line) mice (top) and from γδTCR Tg CD3γ−/− (bold line) and γδTCR Tg CD3γ+/+ (gray line) mice (bottom). DN thymocytes and splenocytes are defined as CD4−CD8−CD19−. Staining with a hamster isotype control is also shown (shaded histogram). (B) Number of γδTCR+ CD4−CD8−CD19− cells in the thymus and spleen of CD3γ+/+ (n = 4) and CD3γ−/− (n = 6) mice. Bars represent means ± standard deviation. (C) Comparison of αβTCR surface levels on CD4+ thymocytes and splenocytes from CD3γ+/+ (gray line) and CD3γ−/− (bold line) mice. Staining with a hamster isotype control is also shown (shaded histogram).
Concluding remarks
Using quantitative immunofluorescence techniques, we have addressed the issue of murine γδTCR stoichiometry. We observed a 2:1 ratio of CD3 dimers to TCRγδ heterodimers on the surface of peripheral γδ T cells, a ratio that supports the monovalent TCR model (Fig. 1 A). We also present new evidence, in accordance with previously reported biochemical data (11, 13), demonstrating that the two CD3 dimers contained within the γδTCR are almost exclusively CD3γɛ dimers. Lastly, an analysis of γδTCR surface expression on CD3γ−/− thymocytes and splenocytes revealed an absolute requirement for CD3γɛ dimers in γδTCR assembly. Together, these data strongly support the idea that, during γδTCR assembly, both TCRγ and TCRδ pair with a CD3γɛ dimer. In this study, the ratio of TCRζ homodimers to CD3 dimers or TCRγδ heterodimers was not measured and, therefore, the number of TCRζ homodimers contained within a surface γδTCR complex cannot be determined. However, based on the conservation of positively charged residues in the transmembrane regions of all four TCR chains that are required for association with the invariant TCR chains (for review see reference 15), we propose that, like the αβTCR, the γδTCR contains one TCRζ homodimer.
The vast majority of murine γδTCRs, whether expressed on naive or activated γδ T cells, contain only CD3γɛ dimers (Fig. 3 and not depicted) (11, 13). It is not clear, however, whether human γδTCRs share the same bias for CD3γɛ dimers. Biochemical analysis of surface γδTCR complexes on primary human γδ T cells detected some CD3δɛ dimers but considerably less than the amount observed in surface αβTCR complexes on primary human αβ T cells (11). Interestingly, biochemical analysis of surface γδTCR complexes on activated and expanded human γδ T cell clones detected CD3δɛ dimers in amounts comparable to those seen in surface αβTCRs (unpublished data) (17). Unfortunately, CD3δ deficiency in humans does not resolve the issue of whether CD3δ is required for human γδTCR surface expression, because it is not known whether the absence of peripheral γδ T cells (18, 19) is caused by the loss of CD3δ or by the markedly reduced levels of the other invariant subunits that accompany CD3δ deficiency in the patients analyzed (18). Nevertheless, these findings suggest that there may be important differences in the subunit requirements for murine and human γδTCR assembly. It is believed, based on sequence homology, that TCRδ is the counterpart to TCRα and, consequently, that TCRδ should pair preferentially with CD3δɛ dimers (for review see reference 15). Accordingly, the inconsistency in murine and human γδTCR assembly can be explained by a difference in either the binding affinities of the respective TCRδ chains for CD3δɛ dimers or the binding affinities of the respective CD3δɛ dimers for TCRδ chains. Murine TCRδ pairs to a CD3γɛ dimer but not to a CD3δɛ dimer (Figs. 2 and 3) (11, 13). However, this is not the case for human TCRδ, as a metabolic labeling study using TCRαβ− Jurkat cells transfected with a human TCRδ gene shows that human TCRδ can associate with either human CD3γɛ or CD3δɛ (20). Remarkably, in the same study, when a murine TCRδ gene was transfected into the TCRαβ− Jurkat variant, the murine TCRδ was also shown to pair with either human CD3γɛ or CD3δɛ. Collectively, these data indicate that murine and human CD3δɛ dimers differ in their ability to bind to TCRδ chains.
Of the current models of TCR stoichiometry, the observed 2:1 ratio of CD3 dimers to TCRγδ heterodimers favors the monovalent TCR model for γδTCR stoichiometry (Fig. 1 A). However, we cannot rule out the possibility that monovalent γδTCR complexes cluster or aggregate on the cell surface to form higher order complexes. If these higher order complexes exist, they may provide an explanation for how signal transduction by the γδTCR is superior to that of the αβTCR in the absence of coreceptor involvement. The difference in the subunit composition of the αβ- and γδTCR signal transducing complexes may also explain the increased signaling proficiency of the γδTCR. As the amino acid sequence of the immunoreceptor tyrosine-based activation motif in each CD3 chain is unique (for review see reference 21), it is conceivable that αβ- and γδTCR complexes recruit distinct signaling molecules. In addition, or alternatively, intrinsic differences in the signaling pathways coupled to αβ- and γδTCRs may provide a mechanism by which the γδTCR is capable of signaling better than the αβTCR.
MATERIALS AND METHODS
Mice.
B6.129P2-TCRβ−/− (TCRβ−/−) mice (22) were purchased from the Jackson Laboratory. C57BL/6-CD3δ−/− (CD3δ−/−) mice (23) were provided by D. Kappes (Fox Chase Cancer Center, Philadelphia, PA), and 129-CD3γ−/− (CD3γ−/−) mice (16) were provided by D. Wiest (Fox Chase Cancer Center). C57BL/6-Vγ6/Vδ1 γδTCR transgenic (Tg) mice (line 134) (24) were provided by B.J. Fowlkes (National Institutes of Health [NIH], Bethesda, MD). B6.129-CD3ɛ −/− (CD3ɛ −/−) (25) and C57BL/6 (B6) mice were generated in our colony. Mice were bred and maintained in an NIH Research Animal Facility in accordance with the specifications of the Association for Assessment and Accreditation of Laboratory Animal Care. Mouse protocols were approved by the NIH Animal Care and Use Committee. All mice were killed at 8–12 wk of age.
Antibodies and reagents.
mAbs used for flow cytometric analysis included anti-CD4 (RM4-5), anti-CD8α (53-6.7), anti-TCRγδ (GL3, GL4, and UC7-13D5), anti-Vγ4 (UC3-10A6), anti-TCRβ (H57-597), anti-CD3ɛ (145-2C11), anti-CD19 (1D3), and a hamster IgG isotype control, all of which were purchased from BD Biosciences. The secondary reagent, biotin-conjugated anti–hamster Igκ (HIG-29), was also purchased from BD Biosciences. The anti-CD3γɛ (7D6) hybridoma (14) was obtained from A. Singer (NIH, Bethesda, MD) and D. Wiest and was used to produce ascites. Protein A/G–purified 7D6 mAb was conjugated to AlexaFluor 488 according to the manufacturer's instructions (Invitrogen). AlexaFluor 488 conjugated to streptavidin was also purchased from Invitrogen.
Flow cytometry.
Flow cytometric analysis for surface antigens was performed as previously described (26). Intracellular staining for CD3 dimers was performed (Cytofix/Cytoperm kit; BD Biosciences) according to the manufacturer's instructions. The ratio of CD3 dimers to TCRγδ heterodimers was determined using an assay previously described for determining the ratio of CD3 dimers to TCRαβ heterodimers (4, 12). In brief, 1.5 × 106 lymph node cells were incubated with saturating amounts of purified anti-TCRγδ (GL3, GL4, UC7-13D5, or UC3-10A6) and anti-CD3ɛ (145-2C11) mAbs for 30 min on ice. Saturating amounts of antibody are defined as the concentration of purified antibody required to completely block the binding of the same antibody conjugated to a fluorochrome. All five mAbs are hamster IgG that use the Igκ light chain. Accordingly, their relative binding intensities can be assayed using saturating amounts of the same anti–hamster secondary mAb, biotin-conjugated anti–hamster Igκ (HIG-29). The CD3ɛ/TCRγδ ratio was calculated using the following equation, where MFI stands for mean fluorescence intensity:
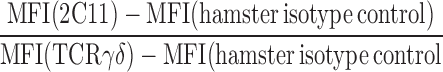 |
The relative percentage of CD3γɛ dimers in CD3 dimers on the surface of γδ T cells was determined using saturating amounts of anti-CD3γɛ (7D6; 300 μg/ml for intracellular staining and 200 μg/ml for surface staining) mAb to block the binding of CD3γɛ dimers by anti-CD3ɛ (145-2C11) mAb (14). The percentage of CD3γɛ dimers was calculated using the following equation:
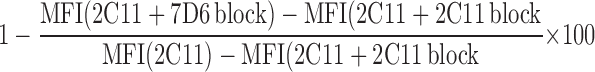 |
For all experiments, 2–4 × 105 cells were collected (FACSCalibur; Becton Dickinson) using CellQuest software or LSR II using FACSDiva software (BD Immunocytometry Systems) and analyzed using FlowJo software (Tree Star, Inc.). Dead cells were excluded from analysis based on forward and side scatter profiles.
Acknowledgments
We thank Dalal El-Khoury for excellent technical assistance. We also thank Drs. Dietmar Kappes, B.J. Fowlkes, and David Wiest for mice, and Drs. Alfred Singer and David Wiest for the 7D6 hybridoma.
The authors have no conflicting financial interests.
References
- 1.Alarcón, B., D. Gil, P. Delgado, and W.W. Schamel. 2003. Initiation of TCR signaling: regulation with CD3 dimers. Immunol. Rev. 191:38–46. [DOI] [PubMed] [Google Scholar]
- 2.Call, M.E., and K.W. Wucherpfennig. 2005. The T cell receptor: critical role of the membrane environment in receptor assembly and function. Annu. Rev. Immunol. 23:101–126. [DOI] [PubMed] [Google Scholar]
- 3.Manolios, N., F. Letourneur, J.S. Bonifacino, and R.D. Klausner. 1991. Pairwise, cooperative and inhibitory interactions describe the assembly and probable structure of the T-cell antigen receptor. EMBO J. 10:1643–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Punt, J.A., J.L. Roberts, K.P. Kearse, and A. Singer. 1994. Stoichiometry of the T cell antigen receptor (TCR) complex: each TCR/CD3 complex contains one TCRα, one TCRβ, and two CD3ɛ chains. J. Exp. Med. 180:587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearse, K.P., J.L. Roberts, and A. Singer. 1995. TCRα-CD3δɛ association is the initial step in αβ dimer formation in murine T cells and is limiting in immature CD4+CD8+ thymocytes. Immunity. 2:391–399. [DOI] [PubMed] [Google Scholar]
- 6.Call, M.E., J. Pyrdol, M. Wiedmann, and K.W. Wucherpfennig. 2002. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 111:967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Exley, M., T. Wileman, B. Mueller, and C. Terhorst. 1995. Evidence for multivalent structure of T-cell antigen receptor complex. Mol. Immunol. 32:829–839. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs, H. 1997. Pre-TCR/CD3 and TCR/CD3 complexes: decamers with differential signaling properties? Immunol. Today. 18:565–569. [PubMed] [Google Scholar]
- 9.San Jose, E., A.G. Sahuquillo, R. Bragado, and B. Alarcón. 1998. Assembly of the TCR/CD3 complex: CD3ɛ/δ and CD3ɛ/γ dimers associate indistinctly with both TCRα and β chains. Evidence for a double TCR heterodimer model. Eur. J. Immunol. 28:12–21. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Miguel, G., B. Alarcón, A. Iglesias, H. Bluethmann, M. Alvarez-Mon, E. Sanz, and A. de la Hera. 1999. Multivalent structure of an αβ T cell receptor. Proc. Natl. Acad. Sci. USA. 96:1547–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes, S.M., and P.E. Love. 2002. Distinct structure and signaling potential of the γδTCR complex. Immunity. 16:827–838. [DOI] [PubMed] [Google Scholar]
- 12.Thibault, G., and P. Bardos. 1995. Compared TCR and CD3ɛ expression on αβ and γδ T cells. Evidence for the association of two TCR heterodimers with three CD3ɛ chains in the TCR/CD3 complex. J. Immunol. 154:3814–3820. [PubMed] [Google Scholar]
- 13.Hayes, S.M., K. Laky, D. El-Khoury, D.J. Kappes, B.J. Fowlkes, and P.E. Love. 2002. Activation-induced modification in the CD3 complex of the γδ T cell receptor. J. Exp. Med. 196:1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulie, P.G., C. Uyttenhove, P. Wauters, N. Manolios, R.D. Klausner, L.E. Samelson, and J. van Snick. 1991. Identification of a murine monoclonal antibody specific for an allotypic determinant on mouse CD 3. Eur. J. Immunol. 21:1703–1709. [DOI] [PubMed] [Google Scholar]
- 15.Klausner, R.D., J. Lippincott-Schwartz, and J.S. Bonifacino. 1990. The T cell antigen receptor: insights into organelle biology. Annu. Rev. Cell Biol. 6:403–431. [DOI] [PubMed] [Google Scholar]
- 16.Haks, M.C., P. Krimpenfort, J. Borst, and A.M. Kruisbeek. 1998. The CD3γ chain is essential for development of both the TCRαβ and TCRγδ lineages. EMBO J. 17:1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Neerven, J., J.E. Coligan, and F. Koning. 1990. Structural comparison of αβ and γδ T cell receptor-CD3 complexes reveals identical subunit interactions but distinct cross-linking patterns of T cell receptor chains. Eur. J. Immunol. 20:2105–2111. [DOI] [PubMed] [Google Scholar]
- 18.Dadi, H.K., A.J. Simon, and C.M. Roifman. 2003. Effect of CD3δ deficiency on maturation of α/β and γ/δ T-cell lineages in severe combined immunodeficiency. N. Engl. J. Med. 349:1821–1828. [DOI] [PubMed] [Google Scholar]
- 19.de Saint Basile, G., F. Geissmann, E. Flori, B. Uring-Lambert, C. Soudais, M. Cavazzana-Calvo, A. Durandy, N. Jabado, A. Fischer, and F. Le Deist. 2004. Severe combined immunodeficiency caused by deficiency in either the δ or ɛ subunit of CD 3. J. Clin. Invest. 114:1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alibaud, L., J. Arnaud, R. Llobera, and B. Rubin. 2001. On the role of CD3δ chains in TCRγδ/CD3 complexes during assembly and membrane expression. Scand. J. Immunol. 54:155–162. [DOI] [PubMed] [Google Scholar]
- 21.Love, P.E., and E.W. Shores. 2000. ITAM multiplicity and thymocyte selection: how low can you go? Immunity. 12:591–597. [DOI] [PubMed] [Google Scholar]
- 22.Mombaerts, P., E. Mizoguchi, M.J. Grusby, L.H. Glimcher, A.K. Bhan, and S. Tonegawa. 1993. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 75:274–282. [DOI] [PubMed] [Google Scholar]
- 23.Dave, V.P., Z. Cao, C. Browne, B. Alarcon, G. Fernandez-Miguel, J. Lafaille, A. de la Hera, S. Tonegawa, and D.J. Kappes. 1997. CD3δ deficiency arrests development of the αβ but not γδ T cell lineage. EMBO J. 16:1360–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sim, G.-K., C. Olsson, and A. Augustin. 1995. Commitment and maintenance of the αβ and γδ T cell lineages. J. Immunol. 154:5821–5831. [PubMed] [Google Scholar]
- 25.DeJarnette, J.B., C.L. Sommers, K. Huang, K.J. Woodside, R. Emmons, K. Katz, E.W. Shores, and P.E. Love. 1998. Specific requirement for CD3ɛ in T cell development. Proc. Natl. Acad. Sci. USA. 95:14909–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shores, E.W., M. Ono, T. Kawabe, C.L. Sommers, T. Tran, K. Lui, M.C. Udey, J. Ravetch, and P.E. Love. 1998. T cell development in mice lacking all T cell receptor ζ family members (ζ, η, and FcɛRIγ). J. Exp. Med. 187:1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]