Abstract
Th17 cells are a distinct lineage of effector CD4+ T cells characterized by their production of interleukin (IL)-17. We demonstrate that Th17 cells also expressed IL-22, an IL-10 family member, at substantially higher amounts than T helper (Th)1 or Th2 cells. Similar to IL-17A, IL-22 expression was initiated by transforming growth factor β signaling in the context of IL-6 and other proinflammatory cytokines. The subsequent expansion of IL-22–producing cells was dependent on IL-23. We further demonstrate that IL-22 was coexpressed in vitro and in vivo with both IL-17A and IL-17F. To study a functional relationship among these cytokines, we examined the expression of antimicrobial peptides by primary keratinocytes treated with combinations of IL-22, IL-17A, and IL-17F. IL-22 in conjunction with IL-17A or IL-17F synergistically induced the expression of β-defensin 2 and S100A9 and additively enhanced the expression of S100A7 and S100A8. Collectively, we have identified IL-22 as a new cytokine expressed by Th17 cells that synergizes with IL-17A or IL-17F to regulate genes associated with skin innate immunity.
IL-17A and IL-17F are cytokines that induce multiple proinflammatory mediators, including chemokines, cytokines, and metalloproteinases, from epithelial and fibroblast cells (1). Both IL-17A and IL-17F are produced by Th17 cells, a distinct lineage of CD4+ effector cells (2–8). Th17 cell differentiation is initiated by TGF-β signaling in the context of proinflammatory cytokines, particularly IL-6 as well as IL-1β and TNF-α. Maintenance and survival of Th17 cells, in contrast, are dependent on IL-23. In vivo studies in murine disease and infection models indicate that the Th17 lineage plays a pathogenic role in autoimmune disease and a protective role in immunosurveillance (3, 5, 7–11).
We sought to identify other ILs that are produced by Th17 cells. We demonstrate that IL-22, an IL-10 family member, was preferentially expressed by this T cell lineage. IL-22 acts directly on epithelial and some fibroblast cells in peripheral tissues and induces an acute phase response in vivo and chemokines and metalloproteinases in vitro (12–16). We show that IL-22 acted cooperatively with IL-17A or IL-17F to enhance expression of antimicrobial peptides associated with host defense. Our data reveal IL-22 as an effector cytokine of the Th17 lineage.
RESULTS AND DISCUSSION
IL-22 transcript is more highly expressed in Th17 cells than Th1 or Th2
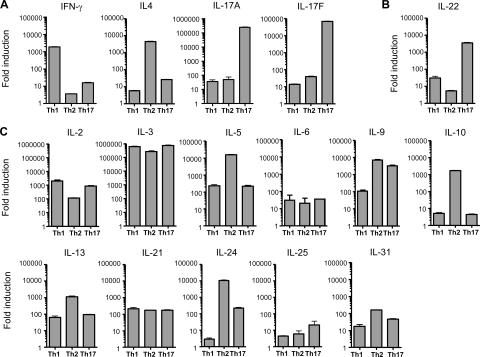
To identify potential Th17 cytokines, we differentiated naive (CD62LhiCD4+) T cells purified from DO11.10 (DO11) mice to the Th1 (IL-12 and anti–IL-4), Th2 (IL-4 and anti–IFN-γ), and Th17 (TGF-β, IL-6, IL-1β, TNF-α, IL-23, anti–IFN-γ, and anti–IL-4) lineages for 7 d. We then examined the expression of cytokine transcripts by quantitative PCR after restimulation of T cells with PMA and ionomycin. Th1 cells expressed the highest amounts of IFN-γ transcript, Th2 cells had the highest abundance of IL-4, and Th17 cells produced the greatest abundance of IL-17A and IL-17F, demonstrating that these cells were successfully differentiated (Fig. 1 A). Of 22 additional ILs examined, IL-22 transcript expression was higher in Th17 cells relative to Th1 by 120-fold or Th2 by 700-fold (Fig. 1 B). In contrast, expression of IL-2, IL-3, IL-5, IL-6, IL-9, IL-10, IL-13, IL- 21, IL-24, IL-25, and IL-31 transcript was equivalent or more abundant in Th1 or Th2 cells relative to Th17 (Fig. 1 C). Non–T cell–derived cytokines, including IL-1, IL-7, IL-11, IL-15, IL-16, IL-18, IL-19, IL-20, IL-27, and IL-28, were not expressed highly in any of the T cell lineages (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20061308/DC1). In summary, IL-22 was expressed at higher amounts by Th17 cells than by Th1 or Th2.
Figure 1.
IL-22 transcript expression is high in Th17 cells. Purified CD62LhiCD4+ (naive) DO11 T cells were activated with irradiated splenocytes, OVA323–339 (OVAp), and various cytokines and antibodies to differentiate cells to Th1 (IL-12, anti–IL-4), Th2 (IL-4, anti–IFN-γ), and Th17 (TGF-β, IL-6, IL-1β, TNF-α, IL-23, anti–IFN-γ, anti–IL-4) lineages. On day 7 of culture, CD4+ T cells were repurified and rested overnight. Cells were then restimulated with PMA and ionomycin under differentiation conditions for 6 h. RNA was prepared and quantitative PCR was performed in A to ensure that cells were properly differentiated. (B and C) ILs known to be expressed in T cells were evaluated by quantitative PCR. All transcript amounts were normalized to HPRT, and fold induction was calculated relative to unactivated, naive DO11 T cells. Data shown are representative of two independent experiments. Error bars are SD.
Th17 cells are the main producers of IL-22
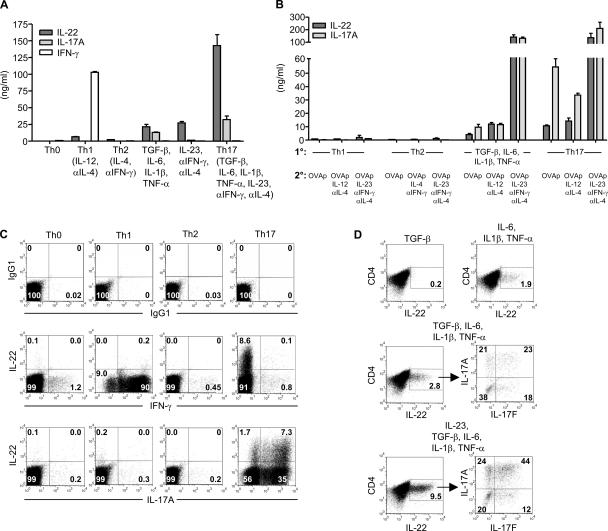
IL-22 is a member of the IL-10 family, along with IL-10, IL-19, IL-20, IL-24, and IL-26 (16). Activation of human CD4+ T cells with IL-12 and anti–IL-4 enhances IL-22 transcript expression, suggesting that IL-22 is a Th1 cytokine (17). However, the expression of IL-22 protein from T cells has not been reported. To study this, we generated monoclonal antibodies to murine IL-22. Naive DO11 T cells activated with irradiated splenocytes and OVA323–339 (OVAp) (Th0) produced minimal amounts of IL-22 (<100 pg/ml) as determined by ELISA (Fig. 2 A). Although IL-22 expression was enhanced during Th1 (110-fold) and Th2 (40-fold) differentiation as compared with Th0, activation with IL-17–inducing conditions resulted in an even greater increase in IL-22 production. TGF-β, IL-6, IL-1β, and TNF-α enhanced IL-22 expression by 360-fold, whereas activation with IL-23, anti–IFN-γ, and anti–IL-4 increased IL-22 production by 460-fold. A combination of these conditions (Th17) yielded the greatest expression of IL-22, 2,400-fold over Th0 and 22-fold higher than Th1. These data demonstrate that IL-22 protein is expressed most abundantly during Th17 differentiation.
Figure 2.
Th17 cells express IL-22 protein. (A) Naive DO11 T cells were activated with irradiated splenocytes, OVAp, and various cytokines and antibodies. IL-22, IL-17A, and IFN-γ concentrations were measured on day 5. (B) Naive DO11 cells were differentiated under Th1, Th2, or Th17 conditions, or with TGF-β, IL-6, IL-1β, and TNF-α for 7 d. After resting overnight, cells were then restimulated with OVAp, IL-2, irradiated splenocytes, and combinations of cytokines and antibodies. IL-22 and IL-17A concentrations were measured on day 5. (C) Intracellular cytokine staining was performed on cells from A on day 5. Plots are gated on KJ126+CD4+ cells. (D) Naive DO11 T cells were activated as in A with the indicated cytokines. Intracellular cytokine staining was performed on day 5. Data shown are representative of three independent experiments. Error bars are SD.
Because some IL-22 was induced under Th1 and Th2 conditions during primary T cell activation, we examined whether a secondary stimulation of these cells could enhance IL-22 production. Naive DO11 T cells were activated for 7 d with Th1, Th2, and Th17 conditions, or with TGF-β, IL-6, IL-1β, and TNF-α. Upon restimulation with just OVAp, IL-2, and irradiated splenocytes, cells originally differentiated with TGF-β, IL-6, IL-1β, and TNF-α or with Th17 conditions produced at least fivefold more IL-22 than Th1 or Th2 cells (Fig. 2 B). The continued differentiation of Th17 cells along the Th17 lineage by restimulating in the presence of IL-23, anti–IFN-γ, and anti–IL-4 enhanced IL-22 production by at least 12-fold over restimulation of cells with OVAp alone or with IL-12, anti–IL-4. In contrast, IL-22 production was not enhanced by restimulation of Th1 cells with IL-12, anti–IL-4 or of Th2 cells with IL-4, anti–IFN-γ. These results show that further differentiation toward Th1 or Th2 does not enhance IL-22 production. In addition, restimulation of Th1 and Th2 cells with IL-23, anti–IFN-γ, and anti–IL-4 did not enhance IL-22 production to that observed with Th17 cells activated under the same condition. These data demonstrate that IL-23 is more potent than IL-12 in stimulating IL-22 expression and that Th17 cells are the major producers of IL-22.
We also examined whether IL-22 could modulate proliferation or IFN-γ, IL-4, and IL-17A production from naive, Th1, Th2, and Th17 cells. We did not observe any changes when T cells were treated with exogenous IL-22 (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20061308/DC1). The transcript for the high affinity receptor subunit, IL-22R1, was not detected in any T cell population. IL-17A or IL-17F also did not induce IL-22 expression from naive, Th1, Th2, or Th17 cells (not depicted). These results demonstrate that IL-22 and IL-17A/IL-17F do not modulate each other's expression by CD4+ T cells.
We next performed intracellular cytokine staining to determine if IL-22 can be coexpressed with IL-17A. Cells activated under Th0, Th1, or Th2 conditions had minimal expansion of IL-22+ cells (≤0.2%; Fig. 2 C). Activation under Th17 conditions generated a substantial population of IL-22–expressing cells (8.7%), with 81% of IL-22+ cells expressing IL-17A and only 1% expressing IFN-γ. We further examined the roles of individual cytokines under Th17 differentiation conditions. Only 0.2% of cells activated with exogenous TGF-β expressed IL-22 (Fig. 2 D). Activation with IL-6, IL-1β, and TNF-α enhanced IL-22+ cells (1.9%). The use of a neutralizing antibody to TGF-β indicated that endogenous TGF-β is important for optimal expression of IL-22 induced by IL-6, IL-1β, and TNF-α (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20061308/DC1). The addition of exogenous TGF-β to IL-6, IL-1β, and TNF-α further increased IL-22+ cells (2.8%), with 62% of IL-22+ cells expressing IL-17A or IL-17F (Fig. 2 D). Activation with IL-23, along with TGF-β, IL-6, IL-1β, and TNF-α, led to a threefold increase in IL-22+ cells (9.5%). 80% of IL-22+ cells produced either IL-17A or IL-17F, with 44% expressing both IL-17A and IL-17F. In summary, these data demonstrate that IL-22 protein is produced at greater amounts by Th17 cells and that IL-22 is coexpressed with both IL-17A and IL-17F during Th17 differentiation.
IL-23 enhances the expansion of IL-22–producing cells during Th17 differentiation
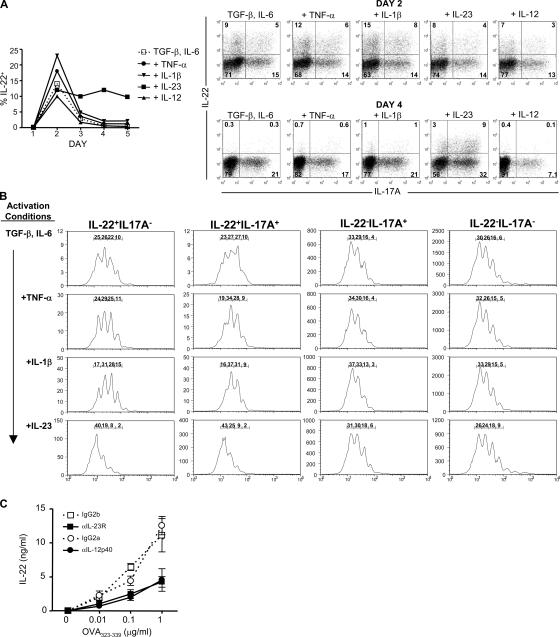
To further examine how IL-23 enhances IL-22 expression, we differentiated CFSE-labeled naive DO11 T cells to Th17 with various cytokines and analyzed the expression of IL-22 from days 1 to 5 of culture. Cells activated with only TGF-β and IL-6 peaked in IL-22 (14%) expression on day 2 and decreased substantially by day 3 (Fig. 3 A). Neither TNF-α, IL-1β, nor IL-12 addition prevented the decrease in expression of IL-22 observed after day 2. In contrast, cells activated with IL-23 as well as TGF-β and IL-6 expressed at least fivefold more IL-22 on day 4. To examine if IL-23 was inducing the expansion of IL-22–producing cells, we analyzed the CFSE dilution profiles of cells expressing IL-22 and/or IL-17A on day 4 (Fig. 3 B). We did not observe any differences in CFSE between IL-22−IL-17A+ and IL-22−IL-17A− cells activated with TGF-β and IL-6 alone or when supplemented with TNF-α, IL-1β, or IL-23. This suggests that there is no correlation between IL-17A expression and proliferation. When we examined the CFSE profiles of IL-22+IL-17A− and IL-22+IL-17A+ cells activated with TGF-β and IL-6, we observed that these cells had proliferated less than IL-22− IL-17A− and IL-22−IL-17A+ cells. Similar findings were observed in cultures supplemented with IL-1β, TNF-α, or IL-12 (Fig. 3 B and not depicted). In contrast, IL-23 in the context of TGF-β and IL-6 enhanced the proliferation and expansion of IL-22+IL-17A− and IL-22+IL-17A+ cells (Fig. 3 B). These findings demonstrate that IL-23 drives the expansion of IL-22–producing cells in the Th17 lineage.
Figure 3.
IL-23 expands IL-22–producing cells. CFSE-labeled naive DO11 T cells were activated with OVAp, irradiated splenocytes, TGF-β, and IL-6. TNF-α, IL-1β, IL-23, or IL-12 was added to some cultures. Intracellular cytokine staining for IL-22 and IL-17A was performed on days 1–5. (A) Percentages of IL-22+ cells on day 1–5 and representative plots on days 2 and 4 are shown. (B) CFSE profiles on day 4 of cells separated into four populations: IL-22+IL-17A−, IL-22+IL-17A+, IL-22−IL-17A+, and IL-22−IL-17A−. Percentage of cells in each peak is shown. (C) Naive DO11 T cells were cultured with LPS-activated DCs, OVAp, and neutralizing antibodies to IL-23R, IL-12p40, or respective isotype controls. IL-22 concentrations were determined on day 5. Data are representative of at least two experiments.
To examine if endogenous IL-23 is necessary for optimal IL-22 expression, we activated naive DO11 T cells with LPS-treated DCs in the presence of anti–IL-23R to block IL-23 signaling or anti–IL-12p40 to neutralize both IL-12 and IL-23. Neutralization of IL-23R reduced IL-22 production by 62% (at 1 μg/ml OVAp) as compared with isotype control (Fig. 3 C). A similar reduction of IL-22 expression was observed with anti–IL-12p40 (64%), suggesting that IL-23, and not IL-12, is responsible for the majority of IL-22 production. Collectively, these data demonstrate that IL-23 is required for optimal expansion of IL-22–producing cells.
IL-22 is coexpressed with IL-17A and IL-17F in vivo
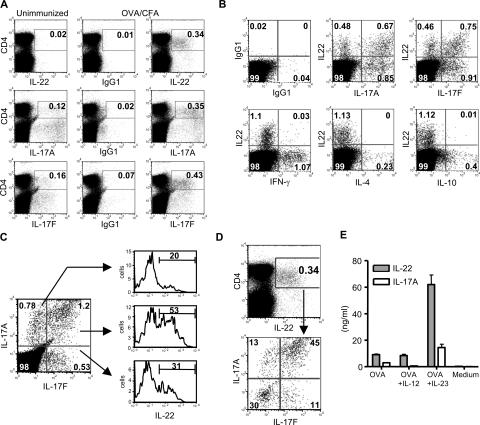
Our in vitro data demonstrate that IL-22 is coexpressed with IL-17A and IL-17F. To examine if this population exists in vivo, we performed intracellular cytokine staining on draining LN cells harvested from C57BL/6 mice that had been immunized with OVA/CFA 7 d earlier. Immunization with OVA/CFA increased the expansion of IL-22+ (0.34%), IL-17A+ (0.35%), and IL-17F+ (0.43%) cells as compared with unimmunized mice (Fig. 4 A). IL-22 was coexpressed with IL-17A (44% of IL-17A+ cells were IL-22+) and IL-17F (45% of IL-17F+ cells were IL-22+), but not with IFN-γ, IL-4, or IL-10 (Fig. 4 B). We also did not observe any coexpression of IL-17A or IL-17F with IFN-γ, IL-4, or IL-10 (not depicted). We did detect considerable, but not complete, coexpression between IL-17A and IL-17F (Fig. 4 C). These results demonstrate a heterogeneity of IL-17A and IL-17F expression within Th17 cells. We further examined IL-22 expression in different populations of IL-17A– and IL-17F–producing cells and observed the highest IL-22 expression in IL-17A+IL-17F+ cells (53%; Fig. 4 C). We also analyzed the expression of IL-17A and IL-17F in IL-22+ cells. The majority (70%) of IL-22+ cells expressed either IL-17A or IL-17F, with 45% of IL-22+ cells expressing both (Fig. 4 D). These in vivo expression profiles among IL-17A, IL-17F, and IL-22 are similar to the expression profiles generated in vitro with TGF-β, IL-6, IL-1β, TNF-α, and IL-23 (Fig. 2 D), suggesting that this in vitro condition is sufficient to replicate in vivo Th17 differentiation. Similar results were also observed on days 4 and 10 after immunization (not depicted). These data demonstrate that IL-22 is not coexpressed with IFN-γ, IL-4, and IL-10 in vivo, but rather with IL-17A and IL-17F.
Figure 4.
IL-22 is coexpressed with IL-17A and IL-17F in vivo. C57BL/6 mice were immunized with 100 μg OVA emulsified in CFA. 7 d after immunization, popliteal LN cells were harvested and the intracellular cytokine staining protocol was immediately performed. (A) LN cells were stained for CD4 and IL-22, IL-17A, and IL-17F. (B) IL-22 expression was analyzed in relation to IL-17A, IL-17F, IFN-γ, IL-4, and IL-10 in CD4+ T cells. (C) Expression of IL-22 in IL-17A+ and/or IL-17F+ populations was analyzed. (D) Expression of IL-17A and IL-17F in IL-22+ cells was determined. (E) LN cells were restimulated with 200 μg/ml OVA. IL-12 or IL-23 was added as indicated. IL-22 and IL-17A concentrations were examined on day 4. Data shown are representative of three independent experiments. Error bars are SD.
To examine if IL-12 or IL-23 stimulates IL-22 production from in vivo–primed T cells, LN cells were restimulated in the presence of exogenous IL-12 or IL-23. The addition of IL-23 enhanced the production of IL-22 by sevenfold compared with OVA alone, whereas IL-12 had no effect (Fig. 4 E). These data further demonstrate that IL-23, rather than IL-12, is the stimulus for enhancing IL-22 production.
IL-22, IL-17A, and IL-17F cooperatively induce antimicrobial peptides
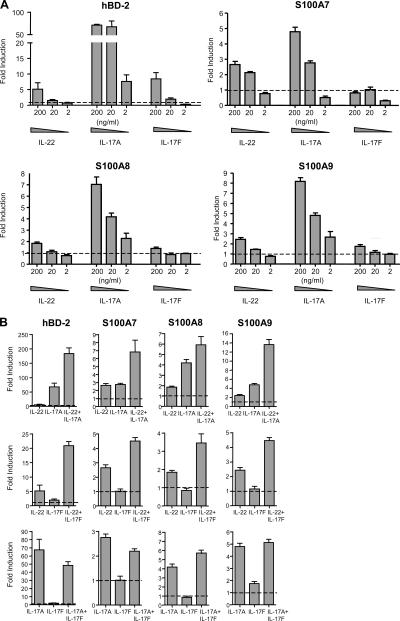
One function of IL-22 is to enhance the expression of antimicrobial peptides associated with host defense, including β-defensin 2 (hBD-2), S100A7, S100A8, and S100A9 (12, 15). To examine whether IL-17A, IL-17F, and IL-22 can act cooperatively to regulate these genes, we treated primary keratinocytes with individual or paired combinations of these cytokines. IL-17A enhanced transcript expression of all four antimicrobial peptides (5–70-fold induction at 200 ng/ml; Fig. 5 A). IL-22 also induced all four transcripts (two- to fivefold at 200 ng/ml), whereas IL-17F (200 ng/ml) induced hBD-2 by eightfold, S100A8 by 1.5-fold, and S100A9 by twofold, but did not up-regulate S100A7. We then cultured keratinocytes with paired cytokine combinations. Treatment with IL-22 (200 ng/ml) and IL-17A (20 ng/ml) led to a synergistic increase of hBD-2 (IL-22, fivefold; IL-17A, 70-fold; IL-22+IL-17A, 180-fold) and S100A9 (IL-22, twofold; IL-17A, fivefold; IL-22+IL-17A, 13-fold) and an additive increase of S100A7 and S100A8 expression (Fig. 5 B). Treatment with IL-22 (200 ng/ml) and IL-17F (20 ng/ml) also synergistically enhanced hBD-2 (IL-22, fivefold; IL-17F, twofold; IL-22+IL-17F, 20-fold). Even though S100A7, S100A8, and S100A9 were not up-regulated by IL-17F (20 ng/ml) alone, IL-17F plus IL-22 enhanced the expression of these three peptides by twofold over IL-22 alone. These data demonstrate that IL-22 can act cooperatively, either synergistically or additively, with IL-17A or IL-17F. Keratinocytes treated with a combination of IL-17A and IL-17F enhanced S100A8, but did not further enhance expression of hBD-2, S100A7, or S100A9. The combination of IL-17A and IL-17F resulted in less induction of these genes than the combination of IL-22 with IL-17A or IL-17F. Expression of receptors for IL-22 (IL-22R1) or IL-17 (IL-17RA) were not altered by IL-22, IL-17A, or IL-17F (not depicted), suggesting that these effects are not related to changes in receptor expression. These data demonstrate that IL-22 in combination with IL-17A or IL-17F cooperatively enhances the expression of antimicrobial peptides.
Figure 5.
IL-22 in combination with IL-17A or IL-17F cooperatively enhances the expression of antimicrobial peptides. Primary human keratinocytes were stimulated with IL-22, IL-17A, or IL-17F individually (A) or with pairwise combinations (B) of 200 ng/ml IL-22, 20 ng/ml IL-17A, and 20 ng/ml IL-17F for 44 h. Relative amounts of hBD-2, S100A7, S100A8, and S100A9 transcript were determined by quantitative PCR and normalized to GAPDH. Fold induction was calculated relative to untreated keratinocytes (dashed line). Data are representative of at least two experiments performed on each of three separate donors. Error bars are SD.
IL-22 was initially characterized as a Th1 cytokine because IL-12 treatment of T cells up-regulates IL-22 mRNA (17). Here, we show that IL-22 protein is expressed in the Th17 lineage at substantially higher amounts than Th1 or Th2, revealing a new effector cytokine secreted by Th17 cells. Despite IL-22 being a Th17 cytokine, the IL-22 locus is located ∼90 kb in proximity to IFN-γ. The distinct expression pattern of IL-22 and IFN-γ suggests cis-regulatory elements exist near these loci that regulate the differentiation of Th1 versus Th17 cells. Further analysis of these loci may improve our understanding of the elements that control T cell differentiation.
Our data also define a new function for IL-23: the induction of IL-22 from T cells. Although IL-17A is a critical cytokine induced by IL-23, certain data suggests that IL-17A may not account for all the downstream effects of IL-23. For example, IL-23p19–deficient mice are completely resistant to disease in collagen-induced arthritis, whereas IL-17A–deficient mice remain susceptible, albeit with a significantly reduced incidence and severity (9, 10). Also, IL-23p19–deficient mice are very susceptible to Citrobacter rodentium infection despite maintaining wild-type expression of IL-17A (7). IL-22 may play a role in these in vivo model systems.
The coexpression of IL-22 with IL-17A and IL-17F from T cells and the presence of their receptors on fibroblast and epithelial cells suggest that these cytokines may act together to regulate local tissue inflammation. One mechanism is by the induction of cytokine and chemokine expression. Similar to IL-17A and IL-17F, IL-22 induces IL-6, CXCL8, and MCP-1 expression by fibroblast cells (1, 13, 14). On colonic myofibroblasts, IL-22 and IL-17A additively enhance expression of CXCL8, IL-6, and LIF, as well as activation of transcription factors, further demonstrating a cooperative signaling relationship between these cytokines (14). IL-22, in contrast to IL-17A, did not enhance mRNA expression for a variety of chemokines from primary keratinocytes (not depicted), suggesting that the functions of IL-22 and IL-17A are cell type dependent. We have demonstrated that IL-22 can function in synergy with IL-17A or IL-17F to enhance keratinocyte expression of antimicrobial peptides. These peptides are known to have potent activity against bacteria such as K. pneumoniae, supporting the idea that Th17 cells may have evolved to combat extracellular pathogens (11, 18). Although these data point to a function for IL-22 in inflammation, the essential role of IL-22 and its interplay with IL-17A and IL-17F in vivo are not well understood. The coexpression of IL-22 with IL-17A and IL-17F adds a new layer of complexity and prompts new directions for future investigation on the development and functions of the Th17 lineage.
MATERIALS AND METHODS
Antibodies and recombinant cytokines.
Murine IL-4, IL-6, IL-12, IL-23, TNF-α, and GM-CSF and human IL-17A were purchased from R&D Systems. Murine IL-2 and human TGF-β were purchased from Sigma-Aldrich. Murine IL-1β was obtained from Bender MedSystems. IL-22 and IL-17F cytokines were prepared by previously described methods (19). Antibodies to IFN-γ (XMG1.2), IL-4 (BVD4-1D11), IL-17A (TC11-18H10), IL-10 (JES5-16E3), and CD4 (RM4-5) were purchased from BD Biosciences. Anti–IL-12p40 (C17.8) and anti–IL-23R (258010) were obtained from R&D Systems. Anti-DO11 antibody (KJ126) was purchased from Caltag Laboratories. Antibodies to murine IL-22 (Ab-01, Ab-02, and Ab-03) and murine IL-17F (RK015-1) were generated by methods as described previously (19).
Mice.
BALB/cByJ, C57BL/6, and C.Cg-Tg(DO11.10)10Dlo TCR transgenic mice were purchased from The Jackson Laboratory. All mice were used between 6 to 10 wk of age. Mice were immunized subcutaneously in the flanks with 100 μg OVA (Sigma-Aldrich) emulsified in CFA (Sigma-Aldrich). 7 d later, inguinal LNs were harvested. All mice were maintained in strict accordance with Wyeth Research Institutional Animal Care and Use Committee regulations.
In vitro T cell activation.
Naive (CD62LhiCD4+) T cells were purified from the spleens of DO11 mice by CD4 negative selection followed by CD62L positive selection according to the manufacturer's directions (Miltenyi Biotec). In a 24-well plate, 2 × 105 DO11 T cells were cultured with 4 × 106 irradiated BALB/cByJ splenocytes (3,300 rad) and 1 μg/ml OVA323–339 peptide (OVAp; New England Peptide). Recombinant cytokines and neutralizing antibodies were used at the following concentrations: 10 ng/ml IL-12, 10 ng/ml IL-4, 1 ng/ml TGF-β, 20 ng/ml IL-6, 10 ng/ml IL-1β, 10 ng/ml TNF-α, 10 ng/ml IL-23, 10 μg/ml anti–IFN-γ, and 10 μg/ml anti–IL-4. In some cases, cells were labeled with CFSE (Invitrogen) according to the manufacturer's directions. For restimulation cultures, cells were harvested on day 7 of primary stimulation, washed extensively, and rested overnight. 2 × 105 DO11 T cells were restimulated with 4 × 106 irradiated splenocytes, 5 ng/ml IL-2, and various conditions as indicated. Conditioned media was collected on day 5 for optimal cytokine accumulation. For generation of DCs, BM cells were cultured with 10 ng/ml GM-CSF and 1 ng/ml IL-4 for 7 d. After purification by CD11c positive selection (Miltenyi Biotec), DCs were matured for 24 h with 1 μg/ml LPS (Escherichia Coli serotype 0111-B4; Sigma-Aldrich). DCs were then washed, and 104 DCs were cultured with 2 × 104 purified naive DO11 T cells, OVAp, and 10 μg/ml anti–IL-12p40, anti–IL-23R, or relevant isotype controls in a 96-well plate. For ex vivo restimulations, 4 × 105 inguinal LN cells were restimulated with 200 μg/ml OVA alone or with 10 ng/ml exogenous IL-12 or IL-23. All lymphocyte cultures were grown in RPMI 1640 supplemented with 10% FBS, 2 mM L-glutamine, 5 mM Hepes, 100 U/ml Pen-Strep, and 2.5 μM β-mercaptoethanol.
Quantitation of cytokine transcripts.
Naive DO11 T cells were differentiated to Th1, Th2, and Th17 as described above. On day 7, CD4+ T cells were purified by positive selection (Miltenyi Biotec) and rested overnight. Cells were then restimulated with 50 ng/ml PMA (Sigma-Aldrich), 1 μg/ml ionomycin (Sigma-Aldrich), and with the following conditions: Th1 cells (IL-12, anti–IL-4), Th2 cells (IL-4, anti–IFN-γ), or Th17 (IL-23, anti–IFN-γ, anti–IL-4) for 6 h before RNA was isolated. Quantitative PCR for cytokine transcripts was performed using SYBR Green Platinum Taq (Invitrogen) and prequalified primers (QIAGEN). The ΔΔCt method was used to normalize transcript to HPRT and to calculate fold induction relative to purified, unactivated naive DO11 T cells.
Intracellular cytokine staining.
Cells were restimulated with 50 ng/ml PMA, 1 μg/ml ionomycin, and GolgiPlug (BD Biosciences) for 6 h. Cells were first stained for surface antigens and then treated with Cytofix/Cytoperm (BD Biosciences) according to the manufacturer's directions. Intracellular cytokine staining was performed using PE-conjugated antibodies to IFN-γ, IL-4, IL-10, and IL-17A as described above. Anti–IL-22 (Ab-02) was labeled with Alexa 647 (Invitrogen) and anti–IL-17F (RK015-1) was labeled with FITC (Pierce Biotechnologies) according to the manufacturer's directions.
Quantitation of cytokine concentrations.
Antibody pairs (coating, detection) were used to detect IFN-γ (AN-18, R4-6A2; eBioscience), IL-17A (MAB721, BAF421; R&D Systems), and IL-22 (Ab-01, biotinylated Ab-03) by standard sandwich ELISA.
Keratinocyte cultures.
Primary human keratinocytes (ScienCell) were cultured in keratinocyte medium (ScienCell) on human fibronectin-coated plates (BD Biosciences). Cells were passaged at 80% confluency and all experiments were done between passages 2 and 4. For evaluation of cytokine effects, 15,000 cells were seeded into a 24-well plate and allowed to adhere for 48 h. Cells were then treated with human IL-22, IL-17A, and IL-17F for 44 h. RNA was purified and quantitative PCR was performed using Taqman Real-Time PCR and prequalified primer probes (Applied Biosystems). Fold induction was calculated as described above.
Online supplemental material.
Fig. S1 shows the mRNA expression of non–T cell–derived cytokines by Th1, Th2, and Th17 differentiated cells. Fig. S2 shows the effects of IL-22 on naive and differentiated T cells. Fig. S3 shows the role of endogenous TGF-β on IL-22 expression induced by IL-6, IL-1β, and TNF-α. Figs. S1–S3 are available at http://www.jem.org/cgi/content/full/jem.20061308/DC1.
Supplemental Material
Acknowledgments
We thank R. Zollner, J. Wright, M. Whitters, S. Olland, G. Yan, D. Taylor, and K. Lam for preparation of cytokines and antibodies, and J. Douhan and P. Thakker for reading the manuscript.
The authors have no conflicting financial interests.
References
- 1.Kolls, J.K., and A. Linden. 2004. Interleukin-17 family members and inflammation. Immunity. 21:467–476. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal, S., N. Ghilardi, M.H. Xie, F.J. de Sauvage, and A.L. Gurney. 2003. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J. Biol. Chem. 278:1910–1914. [DOI] [PubMed] [Google Scholar]
- 3.Langrish, C.L., Y. Chen, W.M. Blumenschein, J. Mattson, B. Basham, J.D. Sedgwick, T. McClanahan, R.A. Kastelein, and D.J. Cua. 2005. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 201:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrington, L.E., R.D. Hatton, P.R. Mangan, H. Turner, T.L. Murphy, K.M. Murphy, and C.T. Weaver. 2005. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 6:1123–1132. [DOI] [PubMed] [Google Scholar]
- 5.Park, H., Z. Li, X.O. Yang, S.H. Chang, R. Nurieva, Y.H. Wang, Y. Wang, L. Hood, Z. Zhu, Q. Tian, and C. Dong. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6:1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veldhoen, M., R.J. Hocking, C.J. Atkins, R.M. Locksley, and B. Stockinger. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. [DOI] [PubMed] [Google Scholar]
- 7.Mangan, P.R., L.E. Harrington, D.B. O'Quinn, W.S. Helms, D.C. Bullard, C.O. Elson, R.D. Hatton, S.M. Wahl, T.R. Schoeb, and C.T. Weaver. 2006. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 441:231–234. [DOI] [PubMed] [Google Scholar]
- 8.Bettelli, E., Y. Carrier, W. Gao, T. Korn, T.B. Strom, M. Oukka, H.L. Weiner, and V.K. Kuchroo. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. [DOI] [PubMed] [Google Scholar]
- 9.Nakae, S., A. Nambu, K. Sudo, and Y. Iwakura. 2003. Suppression of immune induction of collagen-induced arthritis in IL-17-deficient mice. J. Immunol. 171:6173–6177. [DOI] [PubMed] [Google Scholar]
- 10.Murphy, C.A., C.L. Langrish, Y. Chen, W. Blumenschein, T. McClanahan, R.A. Kastelein, J.D. Sedgwick, and D.J. Cua. 2003. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J. Exp. Med. 198:1951–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Happel, K.I., P.J. Dubin, M. Zheng, N. Ghilardi, C. Lockhart, L.J. Quinton, A.R. Odden, J.E. Shellito, G.J. Bagby, S. Nelson, and J.K. Kolls. 2005. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 202:761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolk, K., S. Kunz, E. Witte, M. Friedrich, K. Asadullah, and R. Sabat. 2004. IL-22 increases the innate immunity of tissues. Immunity. 21:241–254. [DOI] [PubMed] [Google Scholar]
- 13.Ikeuchi, H., T. Kuroiwa, N. Hiramatsu, Y. Kaneko, K. Hiromura, K. Ueki, and Y. Nojima. 2005. Expression of interleukin-22 in rheumatoid arthritis: potential role as a proinflammatory cytokine. Arthritis Rheum. 52:1037–1046. [DOI] [PubMed] [Google Scholar]
- 14.Andoh, A., Z. Zhang, O. Inatomi, S. Fujino, Y. Deguchi, Y. Araki, T. Tsujikawa, K. Kitoh, S. Kim-Mitsuyama, A. Takayanagi, et al. 2005. Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterology. 129:969–984. [DOI] [PubMed] [Google Scholar]
- 15.Boniface, K., F.X. Bernard, M. Garcia, A.L. Gurney, J.C. Lecron, and F. Morel. 2005. IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J. Immunol. 174:3695–3702. [DOI] [PubMed] [Google Scholar]
- 16.Renauld, J.C. 2003. Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat. Rev. Immunol. 3:667–676. [DOI] [PubMed] [Google Scholar]
- 17.Wolk, K., S. Kunz, K. Asadullah, and R. Sabat. 2002. Cutting edge: immune cells as sources and targets of the IL-10 family members? J. Immunol. 168:5397–5402. [DOI] [PubMed] [Google Scholar]
- 18.Iwakura, Y., and H. Ishigame. 2006. The IL-23/IL-17 axis in inflammation. J. Clin. Invest. 116:1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, J., K.N. Tomkinson, X.Y. Tan, P. Wu, G. Yan, V. Spaulding, B. Deng, B. Annis-Freeman, K. Heveron, R. Zollner, et al. 2004. Temporal associations between interleukin 22 and the extracellular domains of IL-22R and IL-10R2. Int. Immunopharmacol. 4:693–708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.