Abstract
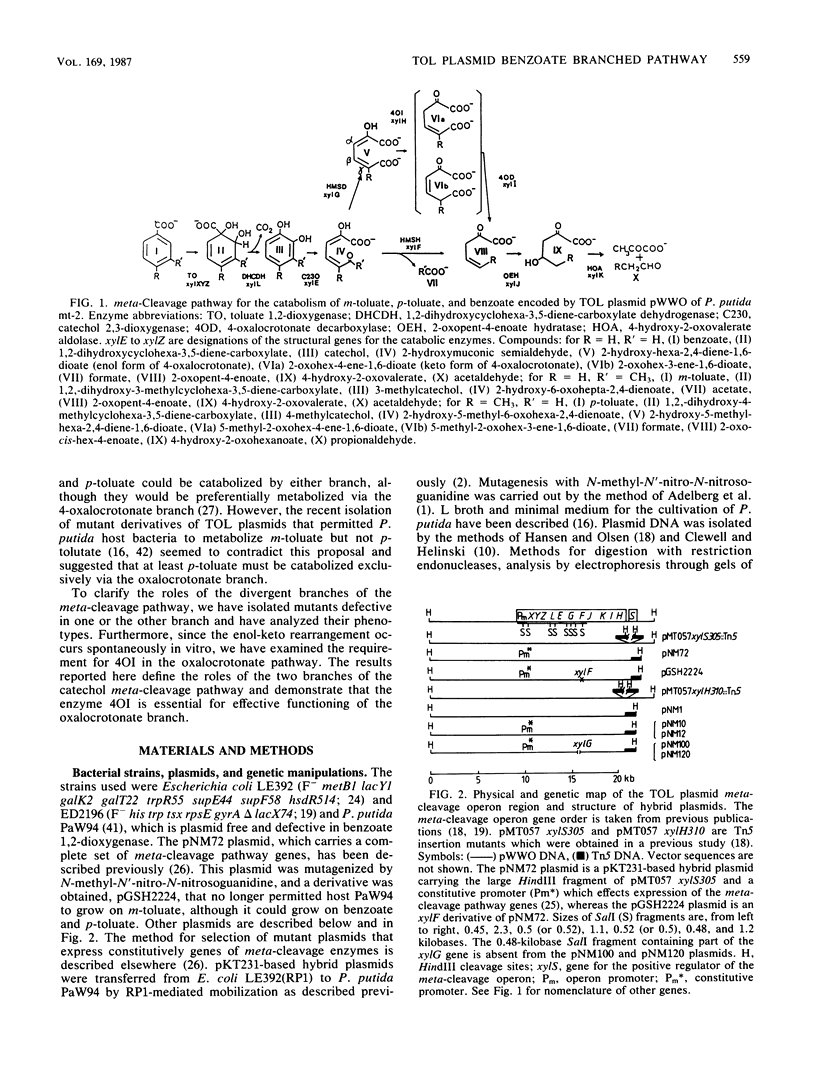
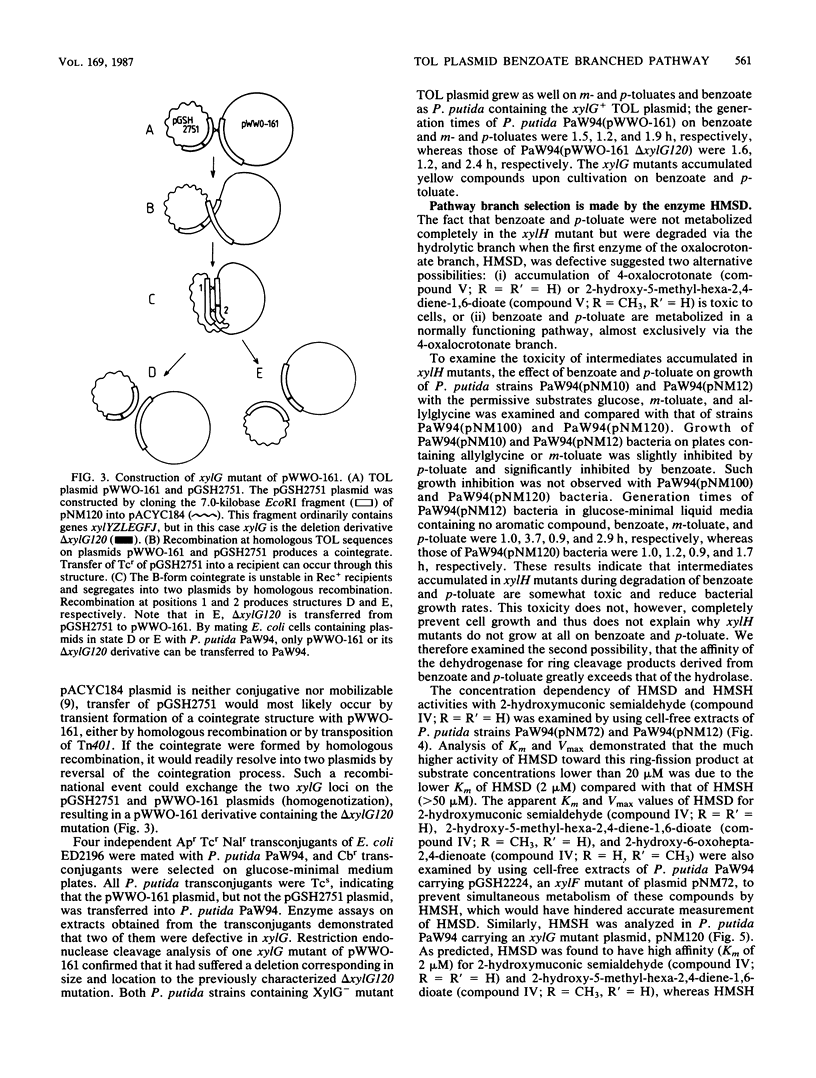
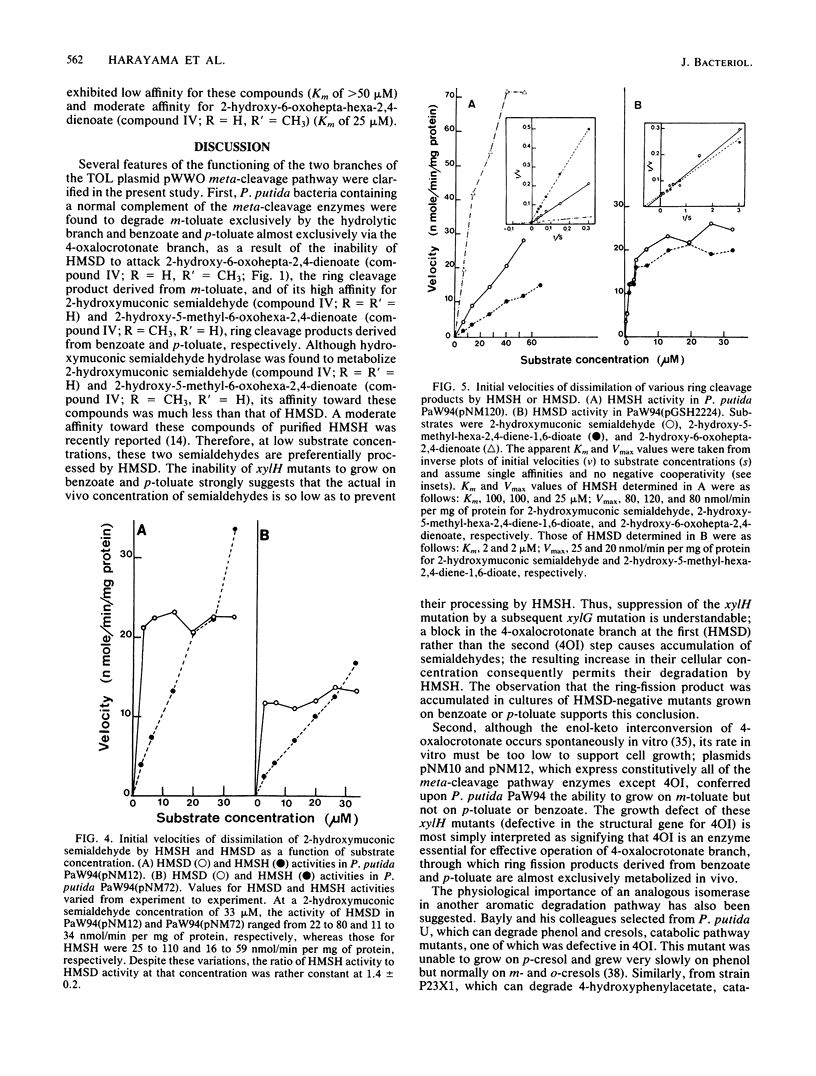
The TOL plasmid-specified meta-cleavage pathway for the oxidative catabolism of benzoate and toluates branches at the ring cleavage products of catechols and reconverges later at 2-oxopent-4-enoate or its corresponding substituted derivatives. The hydrolytic branch of the pathway involves the direct formation of 2-oxopent-4-enoate or its derivatives, whereas the oxalocrotonate branch involves three enzymatic steps effected by a dehydrogenase, an isomerase, and a decarboxylase, which produce the same compounds. Evidence is presented which shows that benzoate and p-toluate can, under certain circumstances, be catabolized by the hydrolytic branch. However, in a fully functional pathway, only m-toluate is dissimilated via this branch, and benzoate and p-toluate are catabolized almost exclusively by the oxalocrotonate branch. The biochemical basis of this selectivity was found to reside in the high affinity of the dehydrogenase for ring fission products derived from benzoate and p-toluate and its inability to attack the ring fission product derived from m-toluate. Although isomerization of 4-oxalocrotonate occurs spontaneously in vitro, enzymatic isomerization was found to be essential for effective functioning of this branch of the pathway in vivo.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Barbour M. G., Bayly R. C. Mutants defective in isomerase and decarboxylase activities of the 4-hydroxyphenylacetic acid meta-cleavage pathway in Pseudomonas putida. J Bacteriol. 1980 May;142(2):480–485. doi: 10.1128/jb.142.2.480-485.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R. C., Dagley S. Oxoenoic acids as metabolites in the bacterial degradation of catechols. Biochem J. 1969 Feb;111(3):303–307. doi: 10.1042/bj1110303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R. C., Wigmore G. J. Metabolism of phenol and cresols by mutants of Pseudomonas putida. J Bacteriol. 1973 Mar;113(3):1112–1120. doi: 10.1128/jb.113.3.1112-1120.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R. C., di Berardino D. Purification and properties of 2-hydroxy-6-oxo-2,4-heptadienoate hydrolase from two strains of Pseudomonas putida. J Bacteriol. 1978 Apr;134(1):30–37. doi: 10.1128/jb.134.1.30-37.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain R. B., Farr D. R. Metabolism of arylsulphonates by micro-organisms. Biochem J. 1968 Feb;106(4):859–877. doi: 10.1042/bj1060859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall F. A., Sala-Trepat J. M., Williams P. A. The coexistence of two pathways for the metabolism of 2-hydroxymuconic semialdehyde in a naphthalene-grown pseudomonad. Biochem Biophys Res Commun. 1971 May 7;43(3):463–469. doi: 10.1016/0006-291x(71)90636-x. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinsworth W. L., Chapman P. J., Dagley S. Stereospecific enzymes in the degradation of aromatic compounds by pseudomonas putida. J Bacteriol. 1973 Feb;113(2):922–931. doi: 10.1128/jb.113.2.922-931.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAGLEY S., GIBSON D. T. THE BACTERIAL DEGRADATION OF CATECHOL. Biochem J. 1965 May;95:466–474. doi: 10.1042/bj0950466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist C. F., Hegeman G. D. Phenol and benzoate metabolism by Pseudomonas putida: regulation of tangential pathways. J Bacteriol. 1969 Nov;100(2):869–877. doi: 10.1128/jb.100.2.869-877.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Lehrbach P. R., Lurz R., Rueckert B., Bagdasarian M., Timmis K. N. Localization and functional analysis of transposon mutations in regulatory genes of the TOL catabolic pathway. J Bacteriol. 1983 May;154(2):676–685. doi: 10.1128/jb.154.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Lehrbach P. R., Timmis K. N. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J Bacteriol. 1984 Oct;160(1):251–255. doi: 10.1128/jb.160.1.251-255.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Timmis K. N. Genetic analysis of a relaxed substrate specificity aromatic ring dioxygenase, toluate 1,2-dioxygenase, encoded by TOL plasmid pWW0 of Pseudomonas putida. Mol Gen Genet. 1986 Feb;202(2):226–234. doi: 10.1007/BF00331641. [DOI] [PubMed] [Google Scholar]
- Harwood C. S., Ornston L. N. TOL plasmid can prevent induction of chemotactic responses to aromatic acids. J Bacteriol. 1984 Nov;160(2):797–800. doi: 10.1128/jb.160.2.797-800.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. J Bacteriol. 1981 Nov;148(2):413–418. doi: 10.1128/jb.148.2.413-418.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. F., Stanier R. Y. Dissimilation of aromatic compounds by Alcaligenes eutrophus. J Bacteriol. 1971 Aug;107(2):468–475. doi: 10.1128/jb.107.2.468-475.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach P. R., Zeyer J., Reineke W., Knackmuss H. J., Timmis K. N. Enzyme recruitment in vitro: use of cloned genes to extend the range of haloaromatics degraded by Pseudomonas sp. strain B13. J Bacteriol. 1984 Jun;158(3):1025–1032. doi: 10.1128/jb.158.3.1025-1032.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N., Lehrbach P. R., Reineke W., Timmis K. N. Transcription of the TOL plasmid toluate catabolic pathway operon of Pseudomonas putida is determined by a pair of co-ordinately and positively regulated overlapping promoters. EMBO J. 1984 Nov;3(11):2461–2466. doi: 10.1002/j.1460-2075.1984.tb02156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Duggleby C. J., Sala-Trepat J. M., Williams P. A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972 Jul 24;28(3):301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- NISHIZUKA Y., ICHIYAMA A., NAKAMURA S., HAYAISHI O. A new metabolic pathway of catechol. J Biol Chem. 1962 Jan;237:PC268–PC270. [PubMed] [Google Scholar]
- Nakazawa T., Yokota T. Benzoate metabolism in Pseudomonas putida(arvilla) mt-2: demonstration of two benzoate pathways. J Bacteriol. 1973 Jul;115(1):262–267. doi: 10.1128/jb.115.1.262-267.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. IV. Regulation. J Biol Chem. 1966 Aug 25;241(16):3800–3810. [PubMed] [Google Scholar]
- Reineke W., Jeenes D. J., Williams P. A., Knackmuss H. J. TOL plasmid pWW0 in constructed halobenzoate-degrading Pseudomonas strains: prevention of meta pathway. J Bacteriol. 1982 Apr;150(1):195–201. doi: 10.1128/jb.150.1.195-201.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Murray K., Williams P. A. The metabolic divergence in the meta cleavage of catechols by Pseudomonas putida NCIB 10015. Physiological significance and evolutionary implications. Eur J Biochem. 1972 Jul 24;28(3):347–356. doi: 10.1111/j.1432-1033.1972.tb01920.x. [DOI] [PubMed] [Google Scholar]
- Sparnins V. L., Chapman P. J., Dagley S. Bacterial degradation of 4-hydroxyphenylacetic acid and homoprotocatechuic acid. J Bacteriol. 1974 Oct;120(1):159–167. doi: 10.1128/jb.120.1.159-167.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigmore G. J., Bayly R. C., Di Berardino D. Pseudomonas putida mutants defective in the metabolism of the products of meta fission of catechol and its methyl analogues. J Bacteriol. 1974 Oct;120(1):31–37. doi: 10.1128/jb.120.1.31-37.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. A., Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974 Oct;120(1):416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. L., Dunn N. W. Transmissible plasmid coding for the degradation of benzoate and m-toluate in Pseudomonas arvilla mt-2. Genet Res. 1974 Apr;23(2):227–232. doi: 10.1017/s0016672300014853. [DOI] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]


