Abstract
Legionella pneumophila, the causative agent of Legionnaires' disease, grows within macrophages and manipulates target cell signaling. Formation of a Legionella-containing replication vacuole requires the function of the bacterial type IV secretion system (Dot/Icm), which transfers protein substrates into the host cell cytoplasm. A global microarray analysis was used to examine the response of human macrophage-like U937 cells to low-dose infections with L. pneumophila. The most striking change in expression was the Dot/Icm-dependent up-regulation of antiapoptotic genes positively controlled by the transcriptional regulator nuclear factor κB (NF-κB). Consistent with this finding, L. pneumophila triggered nuclear localization of NF-κB in human and mouse macrophages in a Dot/Icm-dependent manner. The mechanism of activation at low-dose infections involved a signaling pathway that occurred independently of the Toll-like receptor adaptor MyD88 and the cytoplasmic sensor Nod1. In contrast, high multiplicity of infection conditions caused a host cell response that masked the unique Dot/Icm-dependent activation of NF-κB. Inhibition of NF-κB translocation into the nucleus resulted in premature host cell death and termination of bacterial replication. In the absence of one antiapoptotic protein, plasminogen activator inhibitor–2, host cell death increased in response to L. pneumophila infection, indicating that induction of antiapoptotic genes is critical for host cell survival.
Legionella pneumophila is the causative agent of Legionnaires's disease (1). The microorganism is a Gram-negative facultative intracellular bacterial pathogen that can grow in culture, as well as replicate within fresh water amoeba and mammalian cells (2). On internalization by macrophages or amoeba, L. pneumophila avoids fusion with the endocytic pathway and establishes a replication vacuole, which develops into a rough ER-like compartment (3–5). Legionella replicates within this compartment for up to 24 h, at which time the vacuole fills the host cell cytoplasm before lysis and liberation of intracellular bacteria.
Biogenesis of the L. pneumophila replication vacuole requires a functional Dot/Icm apparatus, which is similar to other type IV secretion systems that promote conjugative DNA transfer and translocation of proteins into target host cells (6). In the absence of Dot/Icm, the bacteria target into an endocytic compartment and fail to replicate (7, 8). Genetic and bioinformatics strategies have identified a large cadre of protein substrates that are translocated by this apparatus, and there appears to be considerable functional redundancy among these translocated proteins in regards to supporting L. pneumophila intracellular growth (9–14).
Although there has been extensive work on the L. pneumophila–encoded proteins required for growth within macrophages, it is still unclear what host cell factors regulate L. pneumophila growth. Several studies have identified common and specific host cell responses after contact with other bacterial pathogens, usually after challenging cells with bacteria at high multiplicities, to ensure the entire population of host cells contacts the pathogen (15–18). In the case of L. pneumophila, such a global analysis of gene expression patterns in the amoeba Dictyostelium discoideum has been performed (19). Most of the transcriptional changes occurred at 24 h after infection (hai), when L. pneumophila initiated replication in this study. Some of the major transcriptional changes reported include induction of heat shock proteins, tRNA synthetase genes, and repression of calcium-binding proteins involved in signal transduction (19). More directed analyses of cytokines and transporters produced by target host cells in response to L. pneumophila have been performed, and there is selective induction of TNF-α and IL-1α, IL-1β, and IL-6 in peritoneal A/J mouse macrophages (20). The transcriptional expression of another cytokine, IL-12, is variable and is probably dependent on specific experimental conditions (21, 22). In addition, one study identified an amino acid transporter, slc1a5, whose expression is required for L. pneumophila growth within human monocytes (23).
To understand the macrophage signaling pathways important for L. pneumophila growth, we investigated the global transcriptional response of host cells harboring a single bacterium. This highly sensitive approach allowed for maximum bacterial replication and minimized host cell cytotoxicity. We found that among the genes induced under these conditions were ones that encode antiapoptotic proteins. The antiapoptotic response appeared tightly linked to NF-κB activation, supporting a role for this regulator in L. pneumophila intracellular growth that involves the maintenance of host cell survival after challenge with low doses of bacteria.
RESULTS
Low-dose challenge with L. pneumophila and enrichment of infected macrophages
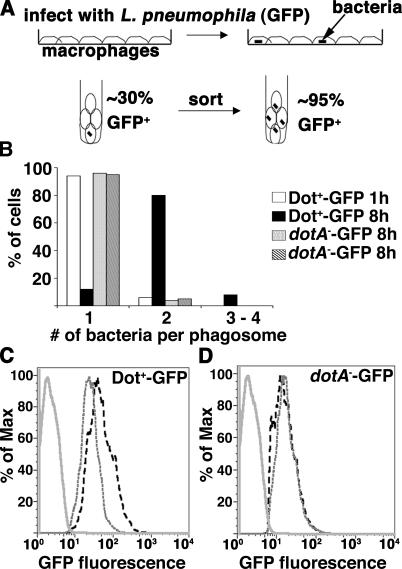
To identify host cell genes that may modulate intracellular growth of L. pneumophila, we compared the transcriptional profile of U937 cells incubated with either WT L. pneumophila (Dot+) or a dotA − mutant lacking a functional Dot/Icm translocator (24, 25). The isolation of a homogenously infected cell population was a prerequisite for this study. Unlike other pathogens, L. pneumophila causes rapid Dot/Icm-dependent cytotoxicity at the high multiplicities of infection (MOIs) (26) that are required to infect the majority of cells in the monolayer. To bypass cytotoxicity, all incubations were performed at MOI = 1 (one bacterium per macrophage), which results in a minority of the cultured cells having associated bacteria. To specifically analyze this population, U937 cells having associated bacteria were then sorted away from the vast excess of uninfected cells using the GFP marker (Fig. 1 A).
Figure 1.
Use of flow cytometry to isolate U937 cells associated with L. pneumophila. L. pneumophila strains Dot+-GFP and dotA −-GFP were introduced onto U937 macrophage monolayers at MOI = 1, allowed to incubate for 1 or 8 h, and infected cells were isolated by fluorescence sorting (Materials and methods). (A) Schematic diagram of sorting experiment. (B) One bacterial division occurs during the 8-h incubation. For each sorted GFP+ fraction, the cells were plated on coverslips, and the number of bacteria per phagosome was determined by fluorescence microscopy. (C and D) Examples of the fluorescence profile of sorted, uninfected macrophages (solid gray line); sorted, infected macrophages at 1 hai (gray dashed line); or sorted, infected macrophages at 8 hai (black dashed line) for Dot+-GFP (C) or dotA −-GFP (D), respectively. The percentage of max (% of max) indicates the number of cells relative to the peak fraction of cells.
Samples were collected 1 or 8 hai and sorted for the cell population harboring GFP bacteria, and total RNA was isolated from these sorted populations. These two time points were chosen because the Dot+ strain initiates biogenesis of the replication vacuole at 1 hai, with clear recruitment of ER-derived vesicles at this time point (5), and has completed one round of division by 8 hai (Fig. 1 B). In contrast, the type IV secretion–defective strain dotA −-GFP fails to establish an ER-like vacuole and to replicate. At each of these time points, bacterial multiplication was monitored by graphing the fluorescence profile of each sorted sample as a function of normalized cell number (% of max = percentage of cells relative to peak fraction of cells; Fig. 1, C and D). Between 1 and 8 hai, the peak fluorescence intensity of sorted GFP-positive cells increased from 40 to 80 fluorescent units (Fig. 1 C), corresponding to two bacteria per phagosome being present in 80% of the Dot+-infected cells (Fig. 1 B). In contrast, the peak fluorescence intensity of dotA −-GFP did not change over this time period (Fig. 1 D), which is consistent with a lack of replication.
To confirm that any changes in expression patterns were a result of a response to functions performed by the type IV translocation system, the mutant strain dimB − was included in this study. The dimB − mutant has an intact Dot/Icm translocator and shows proper formation of a replication vacuole but is defective for intracellular growth and stalls after one intracellular doubling (unpublished data). The peak fluorescence intensity of U937 cells infected with dimB − -GFP was identical to that observed for Dot+-GFP at 1 and 8 hai, as expected for a mutant that stalls after this time point (unpublished data).
Low-dose infection caused substantial changes in macrophage gene transcription
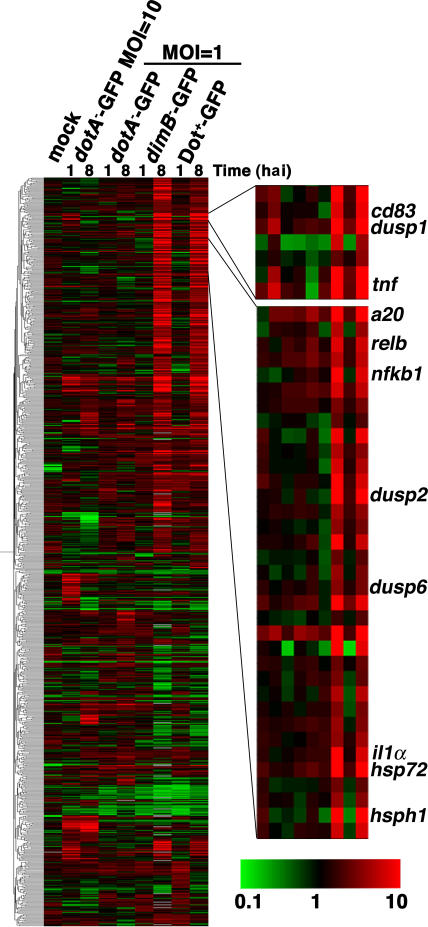
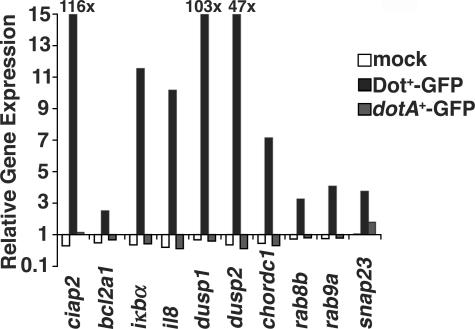
Total RNA from the enriched population of U937 cells harboring L. pneumophila was analyzed by probing oligonucleotide microarrays representing 21,173 human genes (see Materials and methods). Differentially expressed genes were identified and subjected to hierarchical clustering to reveal gene expression trends (see Materials and methods; Fig. 2; and Table S1, available at http://www.jem.org/cgi/content/full/jem.20060766/DC1). We observed that U937 cells responded almost identically to the Dot+-GFP (WT) and the replication-defective dimB − -GFP strains, with only a small subset of genes differentially expressed in these two infections (Table S2, available at http://www.jem.org/cgi/content/full/jem.20060766/DC1). In contrast, there were large differences in gene expression patterns between dotA −-GFP–infected and Dot+-GFP–infected U937 cells. We found that >200 genes were induced or repressed specifically by Dot+-GFP and not by dotA −-GFP at 1 or 8 hai (Table S3, available at http://www.jem.org/cgi/content/full/jem.20060766/DC1). Several differentially expressed genes that were up-regulated in response to infection with Dot+-GFP, but not dotA −-GFP, were verified by quantitative real-time PCR (qPCR; Fig. 3). qPCR revealed that the induction levels of some genes in response to Dot+-GFP, including dusp1, dusp2, and ciap2, appeared to be underestimated by the microarray analysis (Fig. 3), strengthening the significance of the induction that was observed in the microarray analysis.
Figure 2.
Microarray analysis of infected cells shows induction of host cell genes in response to functional Dot/Icm secretion system. Pearson hierarchical cluster analysis of genes differentially expressed relative to uninfected, unsorted reference cells. 799 genes are displayed. Shown are mock (uninfected cells, sorted/reference), dotA −-GFP (MOI = 10; unsorted dotA −-GFP/reference), dotA −-GFP (MOI = 1; sorted dotA −- GFP/reference), dimB −-GFP (sorted dimB −-GFP/reference), or Dot+-GFP (sorted Dot+-GFP/reference) at 1 and 8 hai. The same reference preparation of uninfected U937 cells was used for each comparison. Each condition is represented as the mean from three independent microarrays, each of which was performed on preparations of RNA from different infections. Genes were judged to be induced or repressed if they were twofold up- or down-regulated in comparison to unsorted, uninfected reference cells (P < 0.05 as determined by the t test). Green represents repressed genes (<1), black represents equally expressed genes (reference 1), and red represents induced genes (>1) on a log scale of 0.1–10. The complete list of differentially expressed genes from the hierarchical cluster is shown in Table S1. Microarray data has been deposited in the NCBI Gene Expression Omnibus under accession no. GSE5551.
Figure 3.
Induction of host genes after contact with Dot+-GFP predicted to be up-regulated. qPCR analysis was performed on infected U937 cells sorted 8 hai. Mock (sorted, uninfected); sorted, Dot+-GFP–infected; and sorted, dotA −-GFP–infected cells are shown. Relative gene expression represents the normalized value of the denoted sorted cDNA versus unsorted, uninfected cDNA.
Association of host cells with the Dot+ strain induces major changes in genes regulating immune response, heat shock, and vesicle trafficking
By 8 hai, Dot+-GFP induced transcription of a subset of the proinflammatory cytokines tnfα, il1α, and il1β, as well as the chemokine il8. Expression of il8 was verified by qPCR, and relative induction levels were similar to those found on the arrays (Fig. 3). As the dotA −-GFP strain did not seem to induce the expression of many inflammatory genes, we determined whether incubation of target cells with higher doses of the dotA − mutant could trigger induction of these genes. Microarray analysis of U937 cells infected with dotA −-GFP at MOI = 10 revealed Dot/Icm-independent induction of inflammatory genes such as il1β, tnfα, and il8 (Table I). Therefore, use of a low MOI is critical to observe Dot/Icm-dependent events in gene expression.
Table I.
Dot+ induces a subset of inflammatory, heat shock, and vesicle trafficking genes
|
|
|
MOI = 10
|
|
|
MOI = 1
|
|
|
||
|---|---|---|---|---|---|---|---|---|---|
| mock | 1 hai dotA − |
8 hai dotA − |
1 hai dotA − |
8 hai dotA − |
1 hai dimB − |
8 hai dimB − |
1 hai Dot+ |
8 hai Dot+ |
|
| Inflammatory genes | |||||||||
| il1α (NM_000575) | 1.3 | 0.9 | 1 | 1.2 | 1.4 | 1.4 | 10.1 | 1.1 | 7.6 |
| il1β (NM_ 000576) | 1.8 | 4.8 | 3.7 | 2.1 | 4.6 | 2.4 | 46.7 | 1.4 | 60.1 |
| il8 (NM_000584) | 1.3 | 8.1 | 8 | 2 | 3.5 | 1.8 | 25.3 | 1.4 | 17.2 |
| tnfα (NM_000594) | 0.8 | 4.4 | 1 | 1.2 | 0.5 | 1.4 | 33.4 | 2.4 | 80.4 |
| Heat shock genes | |||||||||
| hsdj2 (NM_005494) | 1.4 | 1.7 | 1 | 2.2 | 2 | 2.3 | 6.1 | 2 | 7.6 |
| hsp40 (NM_006145) | 1.3 | 1.3 | 1.7 | 1 | 1.2 | 1.1 | 11.1 | 1 | 9.7 |
| hsp72 (NM_005345) | 1.9 | 1.2 | 1.1 | 1.6 | 2.2 | 1.5 | 19.2 | 1.6 | 19.3 |
| hsp90 (AK056446) | 1.1 | 0.7 | 0.7 | 0.9 | 1.2 | 0.7 | 2.4 | 0.6 | 2.8 |
| hsph1 (NM_006644) | 1.8 | 0.8 | 0.9 | 0.8 | 1.2 | 0.6 | 12.3 | 0.5 | 12.1 |
| Vesicle trafficking genes | |||||||||
| rab9a (BC017265) | 1.6 | 1 | 1.3 | 1 | 0.9 | 1.2 | 5.4 | 1.6 | 4.2 |
| rab8b (AK001111) | 1.3 | 2.7 | 1.3 | 1.4 | 2.6 | 1.2 | 2.7 | 1.2 | 2.3 |
| arl4 (NM_005738) | 1.8 | 1 | 0.9 | 0.7 | 0.9 | 0.8 | 10.2 | 0.3 | 4.6 |
| snap23 (BC003686) | 1 | 0.9 | 1.7 | 1.5 | 1.6 | 0.9 | 1.3 | 1.3 | 2 |
| stx11 (NM_003764) | 1 | NA | 2.7 | 0.9 | 1.1 | 0.9 | 11.3 | 1.1 | 3.5 |
| stx6 (NM_005819) | 1.5 | 0.8 | 1.5 | 1.1 | 1.3 | 0.4 | 7 | 1.5 | 2.5 |
| trip11 (NM_004239) | 1.1 | 0.7 | 1 | 1.1 | 1.1 | 1.7 | 1.4 | 1.2 | 3.5 |
Genes in bold were considered to be induced or repressed if the expression was >2-fold higher or <0.5-fold lower in infected macrophage samples compared with the reference (P < 0.05, as determined by the Student's t test). GenBank/EMBL/DDBJ accession numbers are shown in parentheses. NA, not available.
We also observed induction of several heat shock genes (hsp40, hsdj2, hsph1, hsp72, and hsp90) in response to Dot+-GFP, but not dotA −-GFP (Table I). Two were DnaJ chaperone family members (hsp40 and hsdj2). In addition to their roles in protein folding, Hsp72, Hsp90, Hsp40, and Hdj2 have roles in interfering with apoptosis (27), which plays a critical role in supporting L. pneumophila replication, as will be shown below.
Multiple genes encoding proteins involved in membrane vesicle interactions were induced in response to the strain with an intact Dot/Icm translocator (Table I). Induction of expression of the snap23, rab8b, and rab9 genes was verified by qPCR to be dependent on an intact Dot/Icm translocator (Fig. 3), and the levels of induction were found to be similar to those observed on the arrays. This apparent selective induction of proteins associated with late secretory and endosomal system will be treated further in the Discussion.
Activation of NF-κB pathway and manipulation of host cell survival
The most striking result from the analysis of low MOI challenge was that genes regulated by the mitogen-activated protein (MAP) kinase and NF-κB signaling pathways showed intense levels of response to Dot+-GFP but not dotA −-GFP infection (Table II). Although induction of the MAP kinase and NF-κB signaling pathways has been observed to be a general response to bacterial components at high MOI (17), in this case the response is specific to strains having an intact Dot/Icm translocator.
Table II.
MAP kinase and NF-κB cascade genes are induced by Dot+ infection
|
|
|
MOI = 10
|
|
|
MOI = 1
|
|
|
||
|---|---|---|---|---|---|---|---|---|---|
| mock | 1 hai dotA − |
8 hai dotA − |
1 hai dotA − |
8 hai dotA − |
1 hai dimB − |
8 hai dimB − |
1 hai Dot+ |
8 hai Dot+ |
|
| MAP kinase pathway | |||||||||
| dusp1 (NM_004417) | 1.9 | 5.2 | 1 | 1.5 | 1.8 | 1.5 | 25.7 | 2.6 | 13.7 |
| dusp2 (NM_0044 18) | 1 | 1.1 | 0.8 | 1.4 | 0.9 | 0.7 | 67.8 | 1.6 | 34.1 |
| dusp6 (NM_001946) | 1.3 | 1 | 1.2 | 1.1 | 1.7 | 0.9 | 3.4 | 1.2 | 3 |
| map2k3 (NM_002756) | 1.4 | 3.3 | 1.3 | 1.1 | 1.4 | 1 | 12 | 1.2 | 3.3 |
| NF-κB pathway genes | |||||||||
| nfkb1 (NM_003998) | 0.8 | 0.7 | 1 | 1.2 | 1.5 | 1.1 | 9.8 | 0.8 | 7.8 |
| nfkb2 (S76638) | 0.7 | 1.1 | 1.3 | 0.8 | 0.8 | 1 | 2.8 | 1 | 2.2 |
| rel (NM_002908) | 0.6 | 0.9 | 0.9 | 0.9 | 1.2 | 0.8 | 2.5 | 0.7 | 2.1 |
| relb (NM_006509) | 1.2 | 0.9 | 1.7 | 1.3 | 2.6 | 1.1 | 8 | 1.4 | 6.9 |
| Activators | |||||||||
| atp2c1 (AF189723) | 1.5 | 0.9 | 0.9 | 1.7 | 0.8 | 0.8 | 4.4 | 1.7 | 2.6 |
| bcl10 (NM_003921) | 1.4 | 1 | 1.6 | 1.2 | 1.6 | 0.7 | 2.1 | 1.8 | 2.3 |
| cl24751 (AF070530) | 0.8 | 0.9 | 1.2 | 1 | 1 | 0.9 | 1.9 | 1.1 | 2.2 |
| ikbkb (AF080158) | 1 | 1.1 | 0.6 | 1.3 | 1.5 | 1.9 | 1.9 | 0.8 | 2.3 |
| irak2 (NM_001570) | 0.5 | 2.8 | 1.7 | 1.3 | 1 | 0.8 | 6.9 | 0.9 | 4.6 |
| malt1 (NM_006785) | 1.5 | 1.1 | 1.1 | 1.3 | 1.5 | 2 | 4.8 | 1.8 | 2.8 |
| oprk1 (NM_000912) | 0.9 | 0.7 | 0.1 | 1.1 | 0.7 | 1.4 | 3.1 | 2.7 | 1.7 |
| tab2 (AL117407) | 1.3 | 2 | 0.9 | 0.9 | 0.8 | 1.1 | 4.9 | 1.6 | 2.3 |
| Inhibitors | |||||||||
| nfkbia (NM_020529) | 1.6 | 9.2 | 3.4 | 3.8 | 5.2 | 3.1 | 27.3 | 2.9 | 36 |
| nfkbie (NM_004556) | 1 | 1.6 | 1.8 | 1.1 | 1.6 | 1.3 | 7.1 | 1.2 | 3.7 |
|
tnfaip3 (A20)
(NM_006290) |
0.8 | 2.7 | 2.7 | 2.3 | 4.3 | 2 | 40.4 | 2.1 | 23.2 |
| Antiapoptotic | |||||||||
| bcl2a1 (NM_004049) | 1 | 1.2 | 1.3 | 1.5 | 1.5 | 1.6 | 2.7 | 1.2 | 2.6 |
| ciap1 (NM_001166) | 1.5 | 0.7 | 0.9 | 1.2 | 2.2 | 0.9 | 1.8 | 1.6 | 3.7 |
| ciap2 (AF070674) | 1 | 0.5 | 2.2 | 0.9 | 1.8 | 5.9 | 32.8 | 1.4 | 14.2 |
| ier3 (NM_052815) | 1 | 4.8 | 1.7 | 1.1 | 0.8 | 1.3 | 6.2 | 1.3 | 3.8 |
| pai-2 (NM_002575) | 1.5 | 4.4 | 2.2 | 0.8 | 2.7 | 0.8 | 7.4 | 0.7 | 7.8 |
Genes in bold were considered to be induced or repressed if the expression was >2-fold higher or <0.5-fold lower in infected macrophage samples compared with the reference (P < 0.05, as determined by the Student's t test). GenBank/EMBL/DDBJ accession numbers are shown in parentheses.
Dual specificity phosphatases 1, 2, and 6 (dusp1, 2, and 6), which down-modulate MAP kinase pathways, were among the most highly induced genes on arrays, and qPCR analysis showed even higher levels of up-regulation with 100-fold induction of dusp1 (Fig. 3). It would appear that induction of these specific Dusp proteins allows fine tuning and potential inactivation of MAP kinase pathways in response to L. pneumophila infection. Dusps also play a role in limiting c-Jun N-terminal kinase activation, thereby preventing host cell death (28). Even more striking was the induction of a large number of genes upstream and downstream of the NF-κB pathway (Table II). Genes that encode NF-κB activators, such as irak2, tab2, and malt1, were induced by 8 hai (29, 30). Also induced were genes that encode factors involved in down-regulating NF-κB signaling, tnfaip3 (A20) and iκbα (31), and those encoding antiapoptotic proteins under transcriptional control of NF-κB subunit p65, such as ciap1, ciap2, pai-2, and bcl2a1 (32–34). These latter proteins inhibit apoptosis by either binding directly to caspases (Ciaps) (35) or blocking release of cytochrome c from mitochondria (Blc2a1) (36).
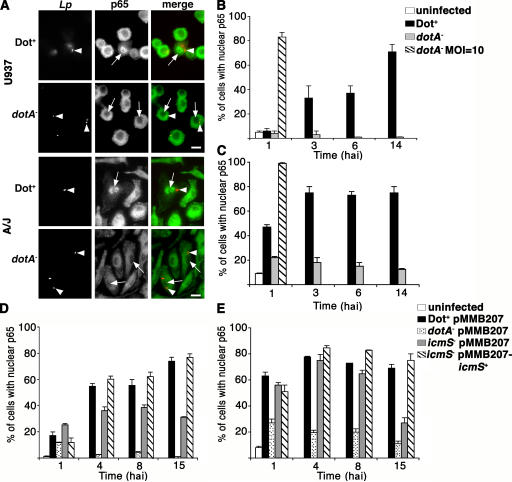
The NF-κB family includes five transcription factors: p65 (RelA), p50, p52, Relb, and c-Rel. The subunits form homo- and heterodimers, and the canonical form is p50-p65. Translocation of these NF-κB dimers from the cytoplasm to the nucleus is necessary for activation of this signaling pathway. Because p65 is known to regulate antiapoptotic gene expression (32, 34), we tested whether nuclear translocation of NF-κB p65 occurs in response to L. pneumophila. U937 cells were infected at MOI = 1 with Dot+ or dotA − strains lacking the GFP plasmid, and the cells harboring bacteria were analyzed by immunofluorescence microscopy for localization of p65. The NF-κB subunit p65 was detected in the nucleus at 6 hai in individual cells associated with Dot+, whereas there was no substantial nuclear p65 staining in cells harboring dotA − at MOI = 1 (Fig. 4 A, top). A time course was conducted to determine the kinetics of NF-κB p65 activation in response to infection and the persistence of the protein in the nucleus (Fig. 4 B). p65 translocation was first observed at 3 hai in U937 cells infected with Dot+. Between 6 and 14 hai, ∼50% of Dot+-containing cells had nuclear p65 staining. In contrast, the Dot/Icm defective strain dotA − did not notably activate NF-κB p65 at any time point tested. Only when dotA − was added at MOI = 10 was NF-κB translocation observed by this assay (Fig. 4 B). This is consistent with the microarray analysis in which we did not observe activation of the NF-κB cascade at MOI = 1, but we did observe a subset of NF-κB genes induced with dotA −-GFP at MOI = 10 (Table II).
Figure 4.
NF-κB p65 translocation is dependent on functional type IV secretion system. (A) U937 macrophages (top) or A/J BM macrophages (bottom) were incubated with Dot+ or dotA − at MOI = 1 for 6 h. NF-κB p65 localization and L. pneumophila (Lp) were visualized by immunofluorescence microscopy after probing with anti–L. pneumophila (red) or anti-p65 (green). Arrowheads point to bacteria, and arrows point to macrophages with associated bacteria. Bar, 10 μm. Time course of NF-κB p65 translocation in infected U937 cells (B and D) or A/J BM macrophages (C and E). Each graph shows the percentage of cells with nuclear p65 staining. (B and C) The following incubations were analyzed: uninfected cells, cells harboring Dot+, cells harboring dotA −, and cells harboring dotA − introduced at MOI = 10. (D and E) The following incubations were analyzed: uninfected cells, cells harboring Dot+ pMMB207, cells harboring dotA − pMMB207, cells harboring icmS − pMMB207, and cells harboring icmS −/pMMB207icmS +. Means + SE from three coverslips from a representative experiment are displayed. A total of 100 infected cells were counted per coverslip.
To further analyze the response of NF-κB to L. pneumophila in primary macrophages, p65 translocation was evaluated in BM macrophages from A/J mice (see Materials and methods). In general, the response of these mouse macrophages was more rapid than that observed in U937 cells, with 50% and 80% of the cells harboring Dot+ showing NF-κB nuclear staining by 1 and 14 hai, respectively (Fig. 4, A and C). In contrast to U937 cells, we observed some NF-κB translocation in BM macrophages harboring dotA − at MOI = 1 (Fig. 4 C). Consistent with what we observed in U937 cells, increasing the bacterial dose to MOI = 10 resulted in NF-κB translocation in nearly 100% of dotA −-infected macrophages shortly after infection (Fig. 4 C). Therefore, dependence of NF-κB translocation on a functional type IV secretion system required low MOI conditions, indicating that there are mechanistic differences in the pathways promoted by these two strains that lead to the NF-κB response.
As is true of mutations in most dot/icm genes, lesions in dotA prevent function of the type IV complex. The absence of the IcmS chaperone, however, allows Dot/Icm function but causes defective translocation of a subset of substrates (13, 37). To determine if these substrates were required for NF-κB translocation, p65 nuclear localization was analyzed during an icmS − infection. Nuclear localization of p65 was partly dependent on icmS in U937 cells at early time points (Fig. 4 D), with NF-κB translocation observed in only 30–40% of icmS −-infected cells at 4 hai (Fig. 4 D). Any defect in activation could be complemented by icmS located in trans on a plasmid. In contrast, after contact of the icmS − mutant with A/J BM macrophages, there was very little defect in NF-κB translocation during the first 8 h of incubation, although by 15 hai there were clearly lowered levels of nuclear NF-κB (Fig. 4 E). This late decrease is likely caused by the restriction of icmS − growth and consequent degradation of bacteria rather than the loss of specific effectors of the NF-κB response. Therefore, most of the p65 nuclear localization observed was either a result of Dot/Icm substrates that do not require IcmS for their deposition into host cells or because of direct interaction of the type IV secretion system with target cells.
NF-κB translocation is necessary to promote host cell survival after L. pneumophila infection
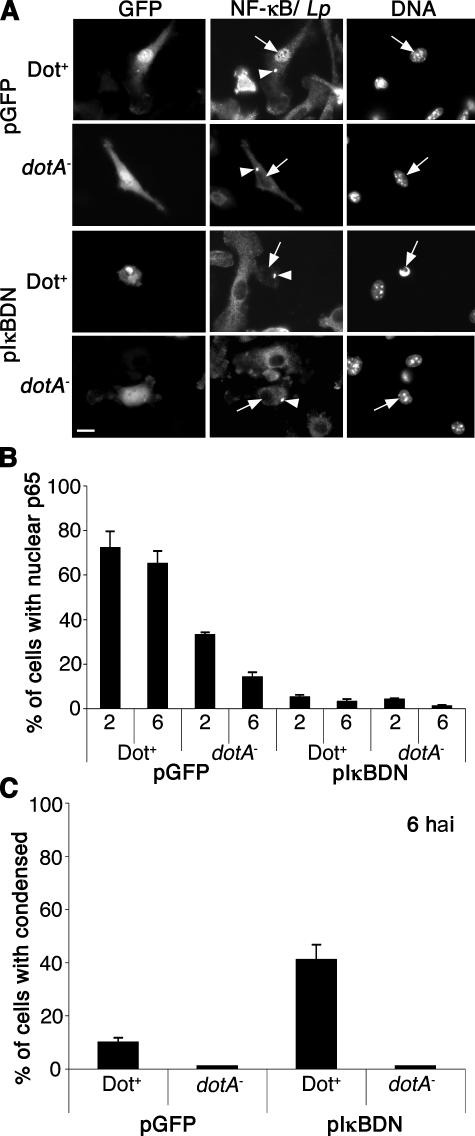
NF-κB nuclear translocation may be required for host cell survival during L. pneumophila infection to maintain a niche for L. pneumophila replication. To address this possibility, the overexpression of the dominant-negative form of IκB, IκBDN, was used to inhibit NF-κB translocation (38). Specifically, BM macrophages from A/J mice were transduced with a retrovirus expressing either GFP alone or IκBDN-IRES-GFP followed by infection with Dot+ or dotA − at MOI = 1. At 2 hai, nuclear localization of NF-κB p65 was detected in ∼70% of the GFP-producing cells harboring Dot+ bacteria, whereas NF-κB translocation was almost completely inhibited in infected macrophages expressing IκBDN (Figs. 5, A and B). A microscopic assay was then used to examine the consequence of NF-κB inhibition on cell survival after L. pneumophila infection. A hallmark of apoptotic cell death is the condensation of the cell nucleus, and cell death was quantitated in infected cells using Hoechst dye to analyze nuclear morphology (Fig. 5 A). By 6 hai, ∼45% of pIκBDN-producing macrophages harboring Dot+ had condensed nuclei, which is indicative of cell death (Fig. 5, A and C), whereas there was only 10% cell death in GFP-producing macrophages harboring Dot+. In contrast, no more than 5% of macrophages harboring the dotA − showed evidence of cell death in either GFP- or IκBDN- producing cells (Fig. 5 C). This is consistent with model that NF-κB translocation is necessary for host cell survival after L. pneumophila infection.
Figure 5.
NF-κB activation is required for host cell survival after L. pneumophila infection. A/J BM macrophages were transduced with retroviruses expressing either GFP or the dominant negative IκBDN-IRES-GFP construct (reference 38). Transduced A/J macrophages were then incubated with the Dot+ or dotA − strains at MOI = 1, and NF-κB p65 translocation and bacteria were visualized by immunofluorescence microscopy at 2 and 6 hai. (A) Images are examples of transduced macrophages harboring bacteria at 6 hai. NF-κB p65 and L. pnenumophila (Lp) were stained with same flour. Bacteria are marked with arrowheads, and cell nuclei are marked with arrows. Nuclear morphology was observed by Hoechst DNA staining. Bar, 10 μm. (B) NF-κB p65 translocation and (C) cell death (condensed nuclei) were observed by immunofluorescence microscopy and quantitated. Means + SE from three independent experiments are explained. A total of 100 cells harboring bacteria were counted per experiment.
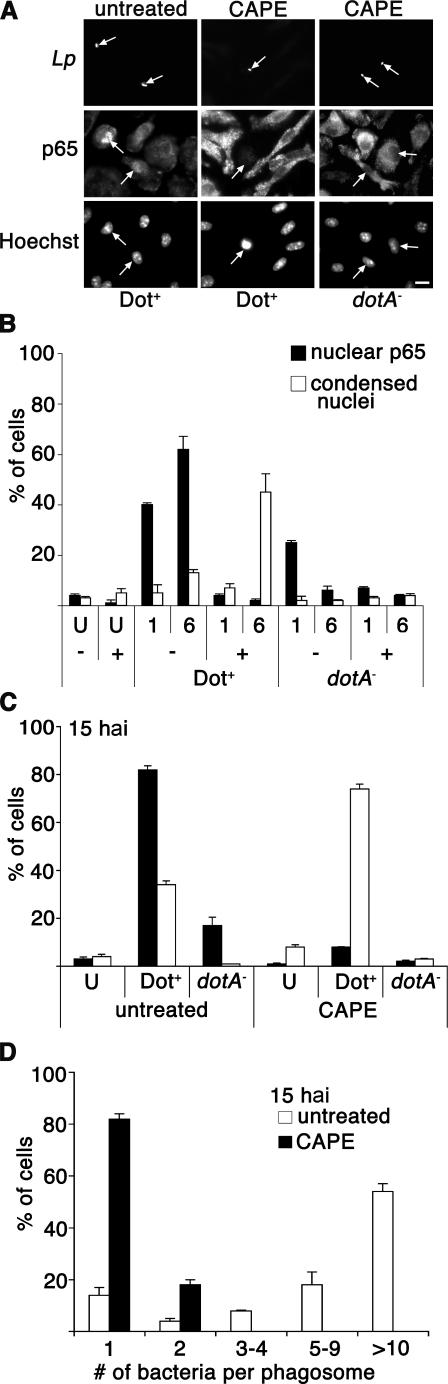
To investigate whether lack of NF-κB translocation inhibits intracellular growth of L. pneumophila, cultures were treated with caffeic acid phenethyl ester (CAPE), which is known to block translocation of NF-κB (39). This assay is not only an independent measure of the consequences of NF-κB inhibition, but it also allowed us to evaluate a larger number of infected cells in the monolayer than we could with IκBDN transduction assay. A/J BM macrophages were pretreated with CAPE at 10 μg/ml for 2 h and incubated with Dot+ or dotA − as described above, and NF-κB translocation, cell death, and bacterial replication were determined. Translocation of p65 was efficiently inhibited by CAPE in Dot+-infected macrophages at both 1 and 6 hai (Fig. 6, A and B). Similar to what was observed in the IκBDN-expressing macrophages, 50% of the CAPE-treated macrophages harboring the Dot+ strain had condensed nuclei by 6 hai (Fig. 6, A and B), and by 15 hai 80% of the cells with associated bacteria had condensed nuclei-based Hoechst staining (Fig. 6 C). In contrast, the nuclear morphology of neighboring uninfected cells or cells harboring the dotA − strain were normal (Fig. 6, B and C), with punctate nucleoli visible (Fig. 6 A). Furthermore, the CAPE-treated macrophages undergoing apoptosis were not permissive for growth of the Dot+ L. pneumophila strain. Replication of the Dot+ strain was inhibited in CAPE-treated macrophages, as almost all of the cells at 15 hai contained 1 bacterium per phagosome, whereas 60% of the untreated cells had >10 bacteria per phagosome (Fig. 6 D).
Figure 6.
NF-κB translocation is necessary for host cell survival and bacterial replication. A/J BM macrophages were pretreated for 2 h in the presence or absence of CAPE at 10 μg/ml (reference 39). Cells were incubated with the Dot+ or dotA − strains at MOI = 1, and NF-κB p65 translocation and bacteria were visualized by immunofluorescence microscopy at 1 and 6 hai. Nuclear morphology was observed by Hoechst DNA staining. (A) Examples of p65 staining and nuclear morphology at 6 hai. Bar, 10 μm. (B) NF-κB p65 translocation and cell death (condensed nuclei) were quantitated at 1 or 6 hai. (C) Percentage of condensed nuclei at 15 hai. (D) Bacterial replication at 15 hai. The percentage of cells having phagosomes bearing the denoted number of bacteria is shown. The only cells counted were those harboring bacteria. Means + SE of three coverslips from a representative experiment are given. U, uninfected cells; −, untreated cells; +, cells treated with CAPE.
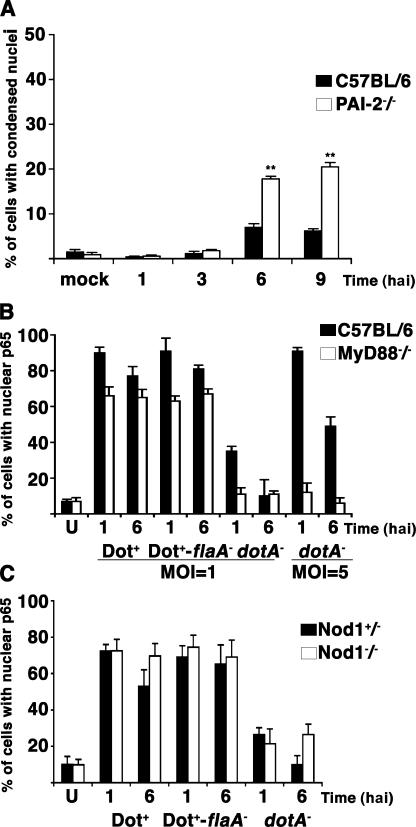
Many of the NF-κB–regulated antiapoptotic proteins identified in the arrays could contribute to protecting from host cell death after L. pneumophila infection (Table II). To address whether a single antiapoptotic protein among this group contributed to host cell survival, we used BM macrophages derived from a plasminogen activator inhibitor–2 (PAI-2)–deficient C57BL/6 mouse, which has been shown to be more susceptible to LPS-stimulated cell death (33). To determine if this protein contributes to host cell survival in response to L. pneumophila, macrophages were isolated from the C57BL/6 pai-2 −/− mouse strain and challenged with L. pneumophila flaA −, which has been shown to grow in the normally restrictive C57BL/6 background (40, 41). By 6 hai, we observed a 2.5-fold increase in cell death in PAI-2–deficient macrophages compared with the C57BL/6 control (P < 0.0005, determined by the Student's t test; Fig. 7 A). Therefore, NF-κB–regulated antiapoptotic genes directly contribute to host cell survival.
Figure 7.
PAI-2–dependent and MyD88/Nod1-independent effects associated with L. pneumophila control of NF-κB translocation. (A) BM macrophages derived from C57BL/6 or C57BL/6 pai-2 −/− mice were incubated with L. pneumophila Dot+-flaA − at MOI = 1 for the indicated times. Nuclear morphology was observed by Hoechst DNA staining, and the percentage of infected cells with condensed nuclei was quantitated. **, P < 0.0005 as determined by the Student's t test. (B) BM macrophages from C57BL/6 and C57BL/6 myd88 −/− mice were incubated with the Dot+, Dot+-flaA −, or dotA − strains at MOI = 1 or 5 for the noted times. The percentage of infected cells with NF-κB p65 staining in the nucleus was quantitated by immunofluorescence microscopy. (C) BM macrophages from C57BL/6 nod1 +/− heterozygous or C57BL/6 nod1 −/− homozygous mice were incubated with the Dot+, Dot+-flaA −, or dotA − strains at MOI = 1 for the noted times, and nuclear p65 was determined as in B. Means + SE from three independent experiments are shown.
Dot+ NF-κB activation is independent of MyD88 and Nod1
Toll-like receptors (TLRs) have been shown to activate NF-κB transcription factors in response to bacterial macromolecules. In several experiments, engagement of a variety of TLRs results in signaling that is mediated at least in part by the adaptor molecule MyD88 (42). To determine whether the observed NF-κB activation is dependent on MyD88 signaling, BM macrophages from MyD88-deficient and C57BL/6 control mice were used for L. pneumophila infections. Consistent with previous experiments, NF-κB translocation was readily observed in macrophages infected by dotA − at MOI = 5 but only marginally at MOI = 1 in the control C57BL/6 BM macrophages (Fig. 7 B, shaded bars). The observed NF-κB translocation by this dotA − strain required the presence of MyD88 (Fig. 7 B, open bars). In contrast, the type IV secretion–competent strains Dot+ and Dot+-flaA − were still able to activate NF-κB in macrophages deficient for MyD88 (Fig. 7 B). Thus, L. pneumophila appeared to trigger two pathways resulting in NF-κB translocation; one bypasses MyD88 and requires the Dot/Icm system, whereas the other requires MyD88 and can be detected in the absence of the type IV secretion system when using elevated MOIs.
A pathway exists for NF-κB activation via the cytosolic sensor Nod, which recognizes intracellularly localized peptidoglycan products (43). There is evidence that peptidoglycan products are delivered across the plasma membrane by the Helicobacter pylori type IV secretion system, promoting Nod1-dependent NF-κB activation, so it is possible that Dot/Icm uses a similar strategy to promote translocation of p65 (44). To determine if activation of NF-κB by the Dot/Icm system is dependent on Nod1, we compared Nod1-deficient C57BL/6 BM macrophages with a heterozygous littermate control. L. pneumophila incubation with macrophages derived from the two mouse strains showed no considerable difference in p65 translocation (Fig. 7 C), indicating that NF-κB activation in this system was independent of Nod1.
DISCUSSION
In this paper we analyzed the gene expression patterns of host cells harboring a single bacterium after incubation with different L. pneumophila strains. This strategy allowed us to identify >200 genes that were differentially expressed in response to events promoted by the L. pneumophila Dot/Icm secretion system. Most striking among the panel of differentially expressed genes was the induction of antiapoptotic genes, many of which are positively regulated by transcription factor NF-κB p65 (32, 34).
It has been reported that L. pneumophila strain AA100 can induce apoptosis in U937 cells at high doses of bacteria (45). We found that the L. pneumophila Philadelphia 1 strain only causes small increases in cell death at low MOIs and instead stimulates an antiapoptotic response (46). Inducing apoptosis with staurosporine or TNF-α has been shown to be restrictive for L. pneumophila replication (46). Thus, interference with cell death is critical for optimal growth of the L. pneumophila and, based on our experiments, is dependent on NF-κB p65 translocation, at least in mouse BM macrophages. The importance of this response was emphasized by the fact that blocking NF-κB translocation by CAPE or expression of dominant-negative IκB resulted in premature cell death in BM macrophages incubated with the Dot+ strain. Both the activation of the NF-κB cell survival pathway and stimulation of cell death in the absence of NF-κB translocation were dependent on a functional type IV secretion system, as the dotA − strain did not induce cell death under any condition used in this study.
Several intracellular pathogens have been shown to activate the NF-κB–antiapoptotic pathway. For instance, host cell survival after infection with parasites such as Toxoplasma gondii (47) or bacteria such as Rickettsia ricketsii (48) is associated with NF-κB translocation. NF-κB can be activated by engagement of TLRs at the host cell surface in response to microbial molecules that are present on both pathogens and nonpathogens (43, 49). TLR2 and TLR5, in fact, are engaged by surface-exposed L. pneumophila LPS and flagellum, respectively (50, 51), but at low bacterial doses it seems unlikely that these common TLR ligands activate NF-κB translocation. Under conditions in which translocation of NF-κB was dependent on the Dot/Icm system, we observed nuclear localization of this protein in both the absence of MyD88 and with strains lacking the flagellin protein, arguing against activation occurring via TLR2 or TLR5. The dependence on the type IV system for nuclear localization of NF-κB, however, could be bypassed by challenging macrophages with the dotA − strain at high MOIs. Under this condition, elevated MOIs allowed signaling through the adaptor MyD88 to promote nuclear localization of NF-κB.
One route for bypass of the MyD88 adaptor occurs via the intracellular signaling molecules Nod1 and Nod2, which are involved in recognition of bacterial peptidoglycan (43). It has been suggested that the extracellular pathogen H. pylori activates NF-κB via Nod1 by translocating peptidoglycan into the host cell through its type IV secretion system (44). In contrast, we observed that NF-κB translocation after L. pneumophila challenge occurred independently of Nod1, which is consistent with a model in which protein substrates of the Dot/Icm system stimulate this event and promote host cell survival. In the absence of Nod1, however, Nod2 may still sense peptidoglycan released through L. pneumophila's type IV secretion system, so this experiment does not totally eliminate the possibility that a substrate other than protein is the inducer of NF-κB translocation. If a protein is an inducer, then it must be transferred in a process that is largely independent of the L. pneumophila IcmS/IcmW chaperone, which is involved in the egress of a subset of the type IV secretion substrates (13). Alternatively, the observed nuclear localization of p65 could be a direct result of contact of the Dot/Icm apparatus with the host cell.
In addition to activating antiapoptotic pathways, the NF-κB family of transcription factors is known to control many proinflammatory processes. We observed selective strong induction of proinflammatory cytokines TNF-α, IL-1α, and IL-1β, which is consistent with previous observations (20), as well as the induction of the chemokine IL-8. These inflammatory genes all are activated in part by NF-κB (52). In general, however, there was a lack of response of type I IFN–regulated genes. It has been recently reported that IL-6 and IFN-β are highly induced by opsonized L. pneumophila in restrictive C57BL/6 MyD88/Trif double knockout macrophages (53). The discrepancy in these results may be caused by the fact that our analysis used conditions that favored intracellular growth rather than restriction. Furthermore, the host and bacterial strains that were analyzed in our experiments were different from those used in this previous study (53). When we directly assayed for secretion of IL-6 using BM macrophages from A/J mice and the Dot+ bacterial strain used in our experiments, IL-6 production was not apparent until 13 hai, a time at which the intracellular replication cycle is close to being completed and bacteria liberated from host cells begin to appear in the culture (unpublished data).
Heat shock genes were induced in host cells at 8 hai in response to a strain encoding an intact Dot/Icm translocator. Heat shock genes were also shown to be induced in Dictyostelium discoideum in response to L. pneumophila infection (19) and may be playing similar roles in both host cell types. L. pneumophila effector proteins are deposited across the translocation channel, presumably in either an unfolded or partially folded form, which may induce the heat shock response and potentially facilitate the folding of translocated effectors. In addition, Hsps are known to play a role in preventing apoptosis (27), and their induction may support L. pneumophila replication in a manner similar to that proposed for NF-κB.
The L. pneumophila replication vacuole intercepts vesicles derived from the ER and destined for the cis-Golgi, yet the Golgi remains intact throughout the intracellular replication cycle (54). To compensate for perturbation caused by the formation of the replication vacuole, it might have been predicted that the cell would up-regulate components involved in ER to Golgi traffic, but we found no evidence for this occurring at the transcriptional level. Instead, there was induction of a distinct subset of genes encoding proteins that appeared to participate in traffic between the trans-Golgi network and either the plasma membrane or various endocytic compartments (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20060766/DC1). These may be involved in maintaining Golgi integrity via a retrograde pathway. Alternatively, these proteins may facilitate cytokine secretion, as the soluble NSF attachment protein syntaxin 6 (stx6) is involved in the secretion of TNF-α (55), and both stx6 and tnfα were found to be induced after L. pneumophila infection (Table I).
In conclusion, we have performed a global screen for host cell genes differentially expressed during L. pneumophila infection at a low infectious dose, with unexpected findings. We have shown that NF-κB translocation is stimulated by the L. pneumophila virulence system and is required to support bacterial intracellular growth within mouse macrophages. Therefore, the NF-κB signaling pathway is likely to be a major target for manipulation by the L. pneumophila type IV secretion system. Continued research on this signaling will help to uncover the mechanism used by L. pneumophila to regulate this pathway and control intracellular growth.
MATERIALS AND METHODS
Bacterial strains and media.
L. pneumophila philadelphia 1 strains Lp02 (thyA, referred to as Dot+) and Lp03 (dotA−, referred to as dotA−) are derivatives of Lp01 (hsdR rpsL) (56). The dimB− mutant (Lp02 dimB) has the kanamycin resistance transposon miniTn10(kan) in the open reading frame Lpg2815. The icmS mutant was generated as previously described (37), and the icmS complementing plasmid pMMB207icmS+ (pCR43) was provided by Craig Roy (Yale University School of Medicine, New Haven, CT) (37). Lp02-flaA− (referred to as Dot+-flaA−) was provided by Tao Ren and William Dietrich (Harvard Medical School, Boston, MA). L. pneumophila strains carrying GFP on an isopropyl-β-D-thiogalactopyranoside (IPTG)–inducible plasmid have been described previously (57). L. pneumophila strains were maintained on buffered charcoal yeast extract solid medium and ACES-buffered yeast extract broth culture media (58–60). GFP strains were maintained on medium containing 100 μg thymidine/ml and 5 μg chloramphenicol/ml and grown in liquid culture containing 100 μg/ml thymidine and 1 mM IPTG.
Cell culture.
U937 cells (American Type Culture Collection) were grown in RPMI 1640 (Irvine) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone) and 1 mM glutamine (Invitrogen). 5 × 107 U937 cells were differentiated using 10 ng/ml 12-tetradecanoyl phorbol 13-acetate (TPA) for 48 h in a T175 tissue culture flask, after which cells were washed, replated with fresh media in the absence of TPA, and used for incubations with L. pneumophila ∼16 h later. A/J and C57BL/6 BM-derived macrophages were isolated from the femurs of 6–8-wk-old female mice and prepared by standard procedures (58). C57BL/6 myd88 −/− femurs were provided by Sarah Stanley in the laboratory of Jeffrey Cox (University of California, San Francisco, San Francisco, CA), and permission to use these mice in our study was granted by Shizuo Akira (Osaka University, Osaka, Japan). Femurs from C57BL/6 pai2 −/− mice were provided by Randal Westrick in the laboratory of David Ginsburg (University of Michigan Medical School, Ann Arbor, MI). Femurs from nod1 +/− and nod1 −/− littermates were provided by Mary O'Riordan (University of Michigan Medical School, Ann Arbor, MI). Macrophages were isolated from femurs and differentiated for 7 d, collected, and frozen for use in multiple experiments in media containing 20% serum, 10% DMSO. Macrophages were replated in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum and 1 mM glutamine for use.
Flow cytometry sorting.
5 × 107 TPA-differentiated U937 cells were replated in 10 cm tissue culture dishes in RPMI medium containing 10% FBS, 200 μg/ml thymidine, and 1 mM IPTG. After ∼16 h of incubation at 37°C, the U937 cells were infected with L. pneumophila grown in AYE broth medium to a post-exponential phase (A600, OD 3.5–4), as judged by the presence of motility in at least 50% of the bacteria observed in 40× fields (61). Individual plates were infected with Dot+-GFP (Lp02), dimB −-GFP, or dotA − -GFP adjusted to MOI = 1 bacterium/macrophage, assuming that 109 bacteria is equivalent to A600 = 1. Bacteria were then pelleted onto the monolayers by a 5-min centrifugation at 1,000 rpm in a tabletop centrifuge fitted with tissue culture plate carriers to synchronize infections, incubated at 37°C, 5% C02 for 1 h, and then washed three times before adding fresh medium. 1 or 8 h after centrifugation, uninfected (mock) or L. pneumophila-infected U937 cells were lifted by washing the monolayer once with PBS + 0.1 mM EDTA and then treating with 0.05% Trypsin-EDTA solution (Invitrogen) for 1 min. Complete RPMI/10% FBS medium was added back immediately, and cells were collected by centrifugation at 1,000 RPM. U937 cells were then resuspended in <1 ml of media to concentrate the suspension. Cells were sorted at Tufts Laser Cytometry facility (http://www.tufts.edu/med/research/TLC.html) using a FACS (MoFlo; DakoCytomation), and appropriate gates were collected for further analysis. For each sample, 1–2 × 106 GFP-positive cells and mock uninfected cells were sorted.
Microarray analysis.
QIAGEN oligonucleotide microarrays (Human v2.1.2) were printed by and purchased from Tufts University Expression Analysis Core facility (TEAC; http://www.tufts.edu/med/teac/index.html). According to TEAC-provided protocols, 10–20 μg of total RNA was isolated from U937 cells using an RNAeasy kit (QIAGEN) according to the manufacturer's instructions. Total RNA was reverse transcribed with superscript II RT (Invitrogen) to incorporate the modified nucleotide 5-(3-aminoallyl)-dUTP (Ambion) into cDNA. The resulting reactions were purified on GFX columns (Cyscribe; GE Healthcare) and coupled with either Cy3 or Cy5 normal human serum ester dye (GE Healthcare) in 50 mM NaHC03 (pH 9). As a reference, cDNA isolated from uninfected, unsorted U937 cells were always labeled with Cy3, and the sorted sample or high MOI sample was always labeled with Cy5. CyDye-labeled cDNA was purified on the Cyscribe GFX column, and incorporation of CyDye was quantitated spectrophotometrically to normalize each sample set. Reference Cy3- and Cy5-labeled cDNAs were hybridized in 1× GE Healthcare hybridization buffer and 10% formamide in a Corning hybridization chamber for 48 h at 42°C. Glass slides were washed with increasing stringency in 1 × SSC, 0.2% SDS, followed by 0.1 × SSC, 0.2% SDS, and 0.1 × SSC buffers. The glass microarrays were spun dry and scanned immediately (ScanArray 4000; Packard Instrument Co.). Microarray accession data are provided in Tables I and II and Table S1.
Microarray data analysis.
Hybridization levels were measured using analysis software (Imagene; Biodiscovery). Exported raw gene expression data was imported into a microarray program (Genespring; Agilent Technologies) for further analysis. The mean of expression levels for individual genes from three independent microarrays performed on cells isolated from three independent infections were used to identify differentially expressed genes. Genes were considered to be induced or repressed when the ratio of sorted sample/reference (unsorted and uninfected) was >2 or <0.5 on a log scale of normalized intensities (lowest intensity dependent on normalization) and generated a p-value < 0.05 by the Student's t test. The one-way analysis of variance test was used to select differentially expressed genes comparing U937 cells incubated with Dot+-GFP, dimB −-GFP, dotA −-GFP (all at MOI = 1), dotA −-GFP (at MOI = 10), or mock infections. Hierarchical clustering to identify gene expression trends was performed using Pearson correlation.
qPCR.
1 μg of total RNA from sorted cell samples and references was first reverse transcribed using superscript II RT (Invitrogen) in a total reaction volume of 60 μl containing DNase/Rnase-free water and a buffer suggested by the manufacturer. 1 μl of the resulting cDNA reaction mix was then used for qPCR analysis. qPCR was then performed using the GenAmp5700 system with the SYBR green PCR reagent (Applied Biosystems). Primer sequences are available on request. For each quantification, three reactions were performed in parallel, results were normalized based on a β-actin mRNA control, and mean values were calculated. Real-time PCR reactions were performed in duplicate, and all results were found to be highly reproducible.
Retroviral transduction.
pCLXSN-IκBDN-IRES-GFP and pCLXSN-GFP vectors were provided by James B. Bliska (State University of New York at Stony Brook, Stony Brook, NY). Retroviral transductions were performed as previously described (38, 62), with the following modifications. A/J BM macrophages grown for 7–8 d were used for transductions. Macrophages were cultured on glass coverslips in fresh retroviral 293T supernatant for 48 h using 24-well plates. The transduced macrophages were than washed with fresh media containing 200 μg/ml thymidine and incubated with bacteria 30 min later.
NF-κB translocation assay.
For immunofluorescence analyses, mammalian cells (either U937 cells or BM macrophages) were plated at a density of 2–3 × 105 cells per well with 200 μg/ ml thymidine on coverslips placed in 24-well dishes. For incubations with host cells, Dot+, dotA −, or Dot+-flaA were grown to the post-exponential phase, and cell monolayers were infected as previously described (10). Cells were allowed to continue to incubate in fresh RPMI/FBS medium for the times noted in the figures and were then fixed with 3.7% paraformaldehyde in PBS for 20 min at room temperature. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 10–20 min at room temperature, blocked in PBS containing 4% goat serum (Invitrogen) for 30 min, stained with primary rat anti–L. pneumophila serum (1:10,000) and rabbit anti-p65 (1:1,000; Santa Cruz Biotechnology, Inc.) in PBS containing 4% goat serum overnight at 4°C. Appropriate secondary antibodies were used, either goat anti–rat IgG conjugated to Texas red (Invitrogen), donkey anti–rabbit IgG conjugated to FITC (Jackson ImmunoResearch Laboratories), and/or goat anti–rabbit IgG conjugated to AlexaFluor 594 (Invitrogen). Nuclei were stained using Hoechst DNA dye (1:10,000; Invitrogen). Microscope images were taken using a 100× 1.4 NA lens on a microscope (IM200; Carl Zeiss MicroImaging, Inc.). Contrast and brightness of individual channels were adjusted linearly in Photoshop (Adobe).
Online supplemental material.
Table S1 contains the complete list of differentially expressed genes from the hierarchical cluster shown in Fig. 2. Table S2 lists the human genes that were induced or repressed in response to the dimB − -GFP strain and not other strains. Table S3 lists the human genes that were induced or repressed in response to Dot+-GFP but not by dotA − -GFP at MOI = 1. Fig. S1 is a diagram of the vesicle trafficking pathways affected by the genes induced in response to Dot+-GFP at 8 hai. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20060766/DC1.
Supplemental Material
Acknowledgments
We would like to thank Molly Bergman, Marion Dorer, Matthias Machner, Vicki Stone, and Joyce Yang for reviews of this manuscript. We thank Shizuo Akira, Tao Ren, Craig Roy, James Bliska, and Sarah Stanley for gifts of mouse and bacterial strains. We also thank Randal Westrick for providing BM from pai2 −/− mice and Mary O'Riordan for bone marrow from nod1 −/− mice.
Ralph R. Isberg is an investigator of the Howard Hughes Medical Institute (HHMI), and this work was supported by HHMI and Training Grant T32AI1007422.
The authors have no conflicting financial interest.
Abbreviations used: CAPE, caffeic acid phenethyl ester; Dot+, WT Legionella pneumophila; hai, h after infection; MAP, mitogen-activated protein; MOI, multiplicity of infection; PAI-2, plasminogen activator inhibitor–2; qPCR, quantitative real-time PCR; TLR, Toll-like receptor.
References
- 1.Fraser, D.W., T.R. Tsai, W. Orenstein, W.E. Parkin, H.J. Beecham, R.G. Sharrar, J. Harris, G.F. Mallison, S.M. Martin, J.E. McDade, et al. 1977. Legionnaires' disease: description of an epidemic of pneumonia. N. Engl. J. Med. 297:1189–1197. [DOI] [PubMed] [Google Scholar]
- 2.Fields, B.S. 1996. The molecular ecology of Legionellae. Trends Microbiol. 4:286–290. [DOI] [PubMed] [Google Scholar]
- 3.Horwitz, M.A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horwitz, M.A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tilney, L.G., O.S. Harb, P.S. Connelly, C.G. Robinson, and C.R. Roy. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 114:4637–4650. [DOI] [PubMed] [Google Scholar]
- 6.Christie, P.J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segal, G., M. Purcell, and H.A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA. 95:1669–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogel, J.P., H.L. Andrews, S.K. Wong, and R.R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 279:873–876. [DOI] [PubMed] [Google Scholar]
- 9.Campodonico, E.M., L. Chesnel, and C.R. Roy. 2005. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 56:918–933. [DOI] [PubMed] [Google Scholar]
- 10.Conover, G.M., I. Derre, J.P. Vogel, and R.R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305–321. [DOI] [PubMed] [Google Scholar]
- 11.Luo, Z.Q., and R.R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA. 101:841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai, H., J.C. Kagan, X. Zhu, R.A. Kahn, and C.R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 295:679–682. [DOI] [PubMed] [Google Scholar]
- 13.Ninio, S., D.M. Zuckman-Cholon, E.D. Cambronne, and C.R. Roy. 2005. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol. Microbiol. 55:912–926. [DOI] [PubMed] [Google Scholar]
- 14.Shohdy, N., J.A. Efe, S.D. Emr, and H.A. Shuman. 2005. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc. Natl. Acad. Sci. USA. 102:4866–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenberger, C.M., M.G. Scott, M.R. Gold, R.E. Hancock, and B.B. Finlay. 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 164:5894–5904. [DOI] [PubMed] [Google Scholar]
- 16.McCaffrey, R.L., P. Fawcett, M. O'Riordan, K.D. Lee, E.A. Havell, P.O. Brown, and D.A. Portnoy. 2004. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc. Natl. Acad. Sci. USA. 101:11386–11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenner, R.G., and R.A. Young. 2005. Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol. 3:281–294. [DOI] [PubMed] [Google Scholar]
- 18.Guillemin, K., N.R. Salama, L.S. Tompkins, and S. Falkow. 2002. Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA. 99:15136–15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farbrother, P., C. Wagner, J. Na, B. Tunggai, T. Morio, H. Urushihara, Y. Tanaka, M. Schleicher, M. Steinert, and L. Eichinger. 2005. Dictyostelium transcriptional host cell response upon infection with Legionella. Cell. Microbiol. 8:438–456. [DOI] [PubMed] [Google Scholar]
- 20.McHugh, S.L., Y. Yamamoto, T.W. Klein, and H. Friedman. 2000. Murine macrophages differentially produce proinflammatory cytokines after infection with virulent vs. avirulent Legionella pneumophila. J. Leukoc. Biol. 67:863–868. [DOI] [PubMed] [Google Scholar]
- 21.Matsunaga, K., T.W. Klein, C. Newton, H. Friedman, and Y. Yamamoto. 2001. Legionella pneumophila suppresses interleukin-12 production by macrophages. Infect. Immun. 69:1929–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neild, A.L., and C.R. Roy. 2003. Legionella reveal dendritic cell functions that facilitate selection of antigens for MHC class II presentation. Immunity. 18:813–823. [DOI] [PubMed] [Google Scholar]
- 23.Wieland, H., S. Ullrich, F. Lang, and B. Neumeister. 2005. Intracellular multiplication of Legionella pneumophila depends on host cell amino acid transporter SLC1A5. Mol. Microbiol. 55:1528–1537. [DOI] [PubMed] [Google Scholar]
- 24.Roy, C.R., K.H. Berger, and R.R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28:663–674. [DOI] [PubMed] [Google Scholar]
- 25.Roy, C.R., and R.R. Isberg. 1997. Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infect. Immun. 65:571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirby, J.E., J.P. Vogel, H.L. Andrews, and R.R. Isberg. 1998. Evidence for pore-forming ability by Legionella pneumophila. Mol. Microbiol. 27:323–336. [DOI] [PubMed] [Google Scholar]
- 27.Beere, H.M. 2004. “The stress of dying”: the role of heat shock proteins in the regulation of apoptosis. J. Cell Sci. 117:2641–2651. [DOI] [PubMed] [Google Scholar]
- 28.Kamata, H., S. Honda, S. Maeda, L. Chang, H. Hirata, and M. Karin. 2005. Reactive oxygen species promote TNFα-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 120:649–661. [DOI] [PubMed] [Google Scholar]
- 29.Matsuda, A., Y. Suzuki, G. Honda, S. Muramatsu, O. Matsuzaki, Y. Nagano, T. Doi, K. Shimotohno, T. Harada, E. Nishida, et al. 2003. Large-scale identification and characterization of human genes that activate NF-κB and MAPK signaling pathways. Oncogene. 22:3307–3318. [DOI] [PubMed] [Google Scholar]
- 30.Muzio, M., J. Ni, P. Feng, and V.M. Dixit. 1997. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 278:1612–1615. [DOI] [PubMed] [Google Scholar]
- 31.Heyninck, K., and R. Beyaert. 2005. A20 inhibits NF-κB activation by dual ubiquitin-editing functions. Trends Biochem. Sci. 30:1–4. [DOI] [PubMed] [Google Scholar]
- 32.Wang, C.Y., M.W. Mayo, R.G. Korneluk, D.V. Goeddel, and A.S. Baldwin Jr. 1998. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 281:1680–1683. [DOI] [PubMed] [Google Scholar]
- 33.Park, J.M., F.R. Greten, A. Wong, R.J. Westrick, J.S. Arthur, K. Otsu, A. Hoffmann, M. Montminy, and M. Karin. 2005. Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis–CREB and NF-κB as key regulators. Immunity. 23:319–329. [DOI] [PubMed] [Google Scholar]
- 34.Zong, W.X., L.C. Edelstein, C. Chen, J. Bash, and C. Gelinas. 1999. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNFα-induced apoptosis. Genes Dev. 13:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deveraux, Q.L., and J.C. Reed. 1999. IAP family proteins–suppressors of apoptosis. Genes Dev. 13:239–252. [DOI] [PubMed] [Google Scholar]
- 36.Werner, A.B., E. de Vries, S.W. Tait, I. Bontjer, and J. Borst. 2002. Bcl-2 family member Bfl-1/A1 sequesters truncated bid to inhibit is collaboration with pro-apoptotic Bak or Bax. J. Biol. Chem. 277:22781–22788. [DOI] [PubMed] [Google Scholar]
- 37.Coers, J., J.C. Kagan, M. Matthews, H. Nagai, D.M. Zuckman, and C.R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38:719–736. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, Y., A.T. Ting, K.B. Marcu, and J.B. Bliska. 2005. Inhibition of MAPK and NF-κB pathways is necessary for rapid apoptosis in macrophages infected with Yersinia. J. Immunol. 174:7939–7949. [DOI] [PubMed] [Google Scholar]
- 39.Natarajan, K., S. Singh, T.R. Burke Jr., D. Grunberger, and B.B. Aggarwal. 1996. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-κB. Proc. Natl. Acad. Sci. USA. 93:9090–9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molofsky, A.B., B.G. Byrne, N.N. Whitfield, C.A. Madigan, E.T. Fuse, K. Tateda, and M.S. Swanson. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203:1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren, T., D.S. Zamboni, C.R. Roy, W.F. Dietrich, and R.E. Vance. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai, T., and S. Akira. 2006. TLR signaling. Cell Death Differ. 13:816–825. [DOI] [PubMed] [Google Scholar]
- 43.Philpott, D.J., and S.E. Girardin. 2004. The role of Toll-like receptors and Nod proteins in bacterial infection. Mol. Immunol. 41:1099–1108. [DOI] [PubMed] [Google Scholar]
- 44.Viala, J., C. Chaput, I.G. Boneca, A. Cardona, S.E. Girardin, A.P. Moran, R. Athman, S. Memet, M.R. Huerre, A.J. Coyle, et al. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5:1166–1174. [DOI] [PubMed] [Google Scholar]
- 45.Gao, L.Y., and Y. Abu Kwaik. 1999. Apoptosis in macrophages and alveolar epithelial cells during early stages of infection by Legionella pneumophila and its role in cytopathogenicity. Infect. Immun. 67:862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abu-Zant, A., M. Santic, M. Molmeret, S. Jones, J. Helbig, and Y. Abu Kwaik. 2005. Incomplete activation of macrophage apoptosis during intracellular replication of Legionella pneumophila. Infect. Immun. 73:5339–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molestina, R.E., T.M. Payne, I. Coppens, and A.P. Sinai. 2003. Activation of NF-κB by Toxoplasma gondii correlates with increased expression of antiapoptotic genes and localization of phosphorylated IκB to the parasitophorous vacuole membrane. J. Cell Sci. 116:4359–4371. [DOI] [PubMed] [Google Scholar]
- 48.Clifton, D.R., R.A. Goss, S.K. Sahni, D. van Antwerp, R.B. Baggs, V.J. Marder, D.J. Silverman, and L.A. Sporn. 1998. NF-κB-dependent inhibition of apoptosis is essential for host cell survival during Rickettsia rickettsii infection. Proc. Natl. Acad. Sci. USA. 95:4646–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naumann, M. 2000. NF-κB activation and innate immune response in microbial pathogen infection. Biochem. Pharmacol. 60:1109–1114. [DOI] [PubMed] [Google Scholar]
- 50.Girard, R., T. Pedron, S. Uematsu, V. Balloy, M. Chignard, S. Akira, and R. Chaby. 2003. Lipopolysaccharides from Legionella and Rhizobium stimulate mouse bone marrow granulocytes via Toll-like receptor 2. J. Cell Sci. 116:293–302. [DOI] [PubMed] [Google Scholar]
- 51.Hawn, T.R., A. Verbon, K.D. Lettinga, L.P. Zhao, S.S. Li, R.J. Laws, S.J. Skerrett, B. Beutler, L. Schroeder, A. Nachman, et al. 2003. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to Legionnaires' disease. J. Exp. Med. 198:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blackwell, T.S., and J.W. Christman. 1997. The role of NF-κB in cytokine gene regulation. Am. J. Respir. Cell Mol. Biol. 17:3–9. [DOI] [PubMed] [Google Scholar]
- 53.Stetson, D.B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 24:93–103. [DOI] [PubMed] [Google Scholar]
- 54.Derre, I., and R.R. Isberg. 2005. LidA, a translocated substrate of the Legionella pneumophila type IV secretion system, interferes with the early secretory pathway. Infect. Immun. 73:4370–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murray, R.Z., F.G. Wylie, T. Khromykh, D.A. Hume, and J.L. Stow. 2005. Syntaxin 6 and Vti1b form a novel SNARE complex, which is up-regulated in activated macrophages to facilitate exocytosis of tumor necrosis factor-alpha. J. Biol. Chem. 280:10478–10483. [DOI] [PubMed] [Google Scholar]
- 56.Berger, K.H., and R.R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7–19. [DOI] [PubMed] [Google Scholar]
- 57.Solomon, J.M., A. Rupper, J.A. Cardelli, and R.R. Isberg. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68:2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swanson, M.S., and R.R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gabay, J.E., M. Blake, W.D. Niles, and M.A. Horwitz. 1985. Purification of Legionella pneumophila major outer membrane protein and demonstration that it is a porin. J. Bacteriol. 162:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feeley, J.C., R.J. Gibson, G.W. Gorman, N.C. Langford, J.K. Rasheed, D.C. Mackel, and W.B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Byrne, B., and M.S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, Y., and J.B. Bliska. 2003. Role of Toll-like receptor signaling in the apoptotic response of macrophages to Yersinia infection. Infect. Immun. 71:1513–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.