Abstract
Transcriptional control of gene expression in double-positive (DP) thymocytes remains poorly understood. We show that the transcription factor BCL11B plays a critical role in DP thymocytes by controlling positive selection of both CD4 and CD8 lineages. BCL11B-deficient DP thymocytes rearrange T cell receptor (TCR) α; however, they display impaired proximal TCR signaling and attenuated extracellular signal-regulated kinase phosphorylation and calcium flux, which are all required for initiation of positive selection. Further, provision of transgenic TCRs did not improve positive selection of BCL11B-deficient DP thymocytes. BCL11B-deficient DP thymocytes have altered expression of genes with a role in positive selection, TCR signaling, and other signaling pathways intersecting the TCR, which may account for the defect. BCL11B-deficient DP thymocytes also presented increased susceptibility to spontaneous apoptosis associated with high levels of cleaved caspase-3 and an altered balance of proapoptotic/prosurvival factors. This latter susceptibility was manifested even in the absence of TCR signaling and was only partially rescued by provision of the BCL2 transgene, indicating that control of DP thymocyte survival by BCL11B is nonredundant and, at least in part, independent of BCL2 prosurvival factors.
Double-positive (DP) thymocytes express TCRβ on their surface and actively rearrange the TCRα locus. After TCRα rearrangement and expression of surface heterodimeric TCRα/β, DP thymocytes have three fates, depending on the avidity of the TCR∷self peptide-MHC interaction. Cells that undergo a weak and transient interaction with self peptide-MHC will be positively selected, survive, and mature into CD4 or CD8 single-positive (SP) T cells, whereas a strong and prolonged interaction will result in elimination by negative selection. A total lack of signal results in death by neglect (for review see references [1, 2]). During positive selection, there are complex changes in gene expression, including the coreceptor genes CD4 and CD8 (3), CD69 (4), CD5 (5, 6), and TCRβ (1). Signaled DP thymocytes also induce expression of BCL2 and IL7Rα, which are both required for survival of mature T lymphocytes (7, 8). Several transcription factors found to be either direct or immediate-early targets of TCR signaling, such as SAP1 (ELK4) (9), EGR1 (10), Id3 (11), and NFAT4 (12) were shown to control positive selection of both CD4 and CD8 lineages. Runx1 was also recently shown to alter selection of both lineages (13). Other transcription factors act later and have a differential role in positive selection of CD4 or CD8 lineages and commitment, such as Thpok/cKrox for the CD4 lineage (14, 15), Runx3 (16), and Tox for the CD8 lineage (17, 18). Transcriptional control of the DP stage of T cell development still remains poorly defined.
BCL11B is a C2H2 zinc finger protein found to act both as a transcriptional repressor and activator (19–22). Germline removal of BCL11B alters early stages of T cell development, with a block at the DN3 stage, and causes enhanced susceptibility to apoptosis (23). The role of BCL11B at the DP stage of T cell development and beyond this stage is unknown. We provide evidence herein that BCL11B plays an essential and unique role in controlling early events of positive selection and survival of DP thymocytes.
RESULTS
Thymic and peripheral cellularity is altered in BCL11BF/FCD4cre mice, and CD4 and CD8 SP thymocytes and peripheral T lymphocytes are abnormal
To study the role of BCL11B beyond the DN3 stage of T cell development, we used the “floxed” BCL11B mice (BCL11BF/F) in which the exon 3, covering most of the BCL11B open reading frame (24), was flanked by LoxP sites (unpublished data). Removal of BCL11B was achieved by crossing the BCL11BF/F mice with the CD4cre transgenic mice, expressing the Cre recombinase after β selection (25). Cre-mediated removal of BCL11B was confirmed by quantitative (q) RT-PCR, immunoblot, and flow cytometry analysis. BCL11B was removed in DP thymocytes, as well as in the small numbers of CD4 and CD8 SP thymocytes, but was still present in DN thymocytes (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20070863/DC1). The antibodies used for detection of BCL11B correspond to an epitope in exon 2, aa 26–44 (21, 22), outside of the removed exon.
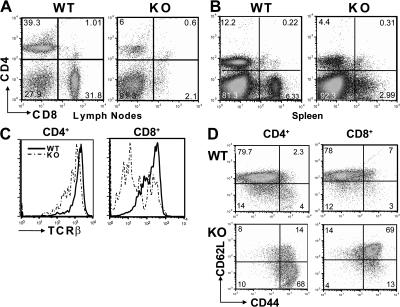
Thymi of 5–6 wk old BCL11BF/FCD4cre mice were reduced in size (Fig. 1), and thymic cellularity was reduced by 35%; however, DP thymocytes were present, indicating that they passed β-selection (Fig. 2, A and C). There was a major reduction in the percentage and absolute numbers of CD4 and CD8 SP thymocyte populations (Fig. 2, A and C), suggesting that their differentiation from DP to SP thymocytes was compromised. In addition, the small numbers of BCL11B-deficient CD4 and CD8 SP thymocytes have reduced levels of TCRβ (Fig. 2 B), indicating that they are immature.
Figure 1.
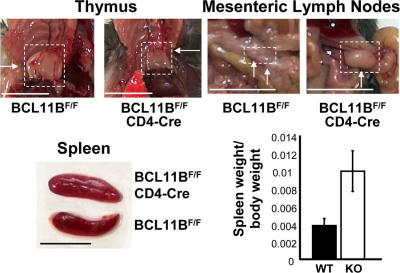
Thymus size is reduced, whereas mesenteric lymph nodes and spleen are enlarged, in BCL11BF/FCD4cre mice. Gross photographs of mutant and control thymi, lymph nodes, and spleens, indicating reduction in the thymus size and increase in the mesenteric lymph nodes and spleen of BCL11BF/FCD4cre mice. Bars, 1 cm. (bottom right) Ratios of spleen weight (g) to total body weight (g) of BCL11BF/F (WT) and BCL11BF/FCD4cre (KO) mice. P = 1.28771E-05; n = 6; 10 wk old.
Figure 2.
Removal of BCL11B in DP thymocytes results in a severe decrease in the number of CD4 and CD8 SP thymocytes. (A) Thymocytes from BCL11BF/FCD4cre (KO) and BCL11BF/F mice (WT) were stained for CD4 and CD8 and analyzed by FACS. (B) Thymocytes were stained for CD4, CD8, and TCRβ. Histograms show TCRβ on gated CD4 or CD8 SP thymocytes. Representative data from multiple experiments are shown. (C) Thymic cellularity of BCL11BF/FCD4cre and control mice. Thymocytes were stained for CD4 and CD8 coreceptors and analyzed by FACS. Total cellularity was determined by counting the live cells. Absolute numbers of cells were calculated based on the percentage of each population and represented as values ± the SD. Cell counts should be multiplied by 106. A two-tailed Student's t test was applied to determine the statistical significance. Mice in these experiments were between 5 and 6 wk of age.
The mesenteric lymph nodes and the spleens of BCL11BF/FCD4cre mice, as well as of the BCL11BF/FCD4cre/TCR transgenic mice (see bellow), were considerably enlarged (Fig. 1 and Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20070863/DC1), and their total cellularity was increased (Table I and Table II), whereas the increase in the cervical lymph nodes and total cellularity was more modest (Table I). The percentages and absolute numbers of CD4+ and CD8+ T lymphocytes were reduced in lymph nodes and spleen (Fig. 3, A and B; Table I and Table II). TCRβ levels were reduced in the small number of BCL11B-deficient peripheral T lymphocytes (Fig. 3 C), suggesting that they are immature. In addition, there was a major reduction in the percentage of naive CD62LhighCD44low CD4+ and CD8+ T lymphocyte populations, whereas the CD44high populations, comprising activated and memory-like T cells, were increased (Fig. 3 D), results which suggest a homeostatic deregulation in CD4+ and CD8+ T lymphocyte populations, which is often observed in lymphopenic mice (26). The lymph nodes and spleens of BCL11BF/FCD4cre mice presented a marked increase in percentage and absolute numbers of B220+ lymphocytes (Table I and Table II). In addition to the increased numbers of B220+ lymphocytes, the spleens of BCL11BF/FCD4cre mice presented increased numbers of macrophages, dendritic cells, NK, megakaryocytes and TCRγδ populations (Table II), suggesting alterations in homeostasis. This was not the case in a TCRα−/− background (Table S1), demonstrating that the increase in the numbers of these populations is dependent on the presence of the few BCL11B-deficient T cells present in the periphery.
Table I.
Lymph node cellularity of BCL11BF/FCD4cre mice
| Tissue | Mouse | Total | CD4+ | CD8+ | B220+ | Macrophages | Others |
|---|---|---|---|---|---|---|---|
| Mesenteric lymph nodes |
BCL11BF/F
(n = 6) |
34.8 ± 10.8 | 14.4 ± 4.1 | 7.2 ± 2.2 | 11.4 ± 5.9 | 0.4 ± 0.3 | 1.4 ± 0.9 |
| BCL11BF/FCD4cre (n = 6) |
70.5 ± 36.4 | 5.1 ± 3. | 3.6 ± 2.1 | 55.8 ± 33.7 | 2.0 ± 1 | 1.9 ± 1.2 | |
| P value | 0.00210 | 1.28E-08 | 1.97E-05 | 0.00016 | 0.00201 | 0.02717 | |
| Cervical lymph nodes |
BCL11BF/F
(n = 6) |
26.1 ± 12.8 | 9.4 ± 4.5 | 5.7 ± 2.7 | 9.2 ± 5.3 | 0.4 ± 0.1 | 1.4 ± 2 |
| BCL11BF/FCD4cre (n = 6) |
29.4 ± 17.8 | 1.6 ± 1 | 2 ± 1.5 | 23.1 ± 11.5 | 0.7 ± 0.2 | 1.9 ± 1 | |
| P value | 0.30013 | 2.017E-07 | 3.85E-05 | 0.00155 | 0.00358 | 0.54395 |
Total cellularity was determined by counting the live cells. Absolute numbers were calculated based on the percentage of each population. All mice in these experiments were between 8 and 12 weeks of age. Absolute numbers should be multiplied by 106. Two-tailed Student's t test was applied to determine the statistical significance.
Figure 3.
Peripheral T lymphocytes of BCL11BF/FCD4cre mice are reduced in number and present an activated/memory-like phenotype. Lymphocytes from lymph nodes (A) and spleen (B) of BCL11BF/FCD4cre (KO) and BCL11BF/F mice (WT) were stained for CD4 and CD8 and analyzed by FACS. (C) Lymphocytes were stained for CD4, CD8, and TCRβ. Histograms show TCRβ on gated CD4+ or CD8+ T lymphocytes. (D) Surface expression of CD44 and CD62L on gated CD4+ and CD8+ T lymphocytes. Representative data from multiple experiments are shown.
Table II.
Spleen cellularity of BCL11BF/FCD4cre mice
| Spleen
|
|
|
|
|
|
|
|
|
|
|---|---|---|---|---|---|---|---|---|---|
| Mouse | Total | CD4+ | CD8+ | B220+ | Macrophages | Dendritic cells |
NK cells | TCRγδ | TCRαβ (CD4−/CD8−) |
| BCL11BF/F
(n = 8) |
39.6 ± 14.8 | 10.1 ± 3.8 | 5.5 ± 1 | 16.7 ± 7.2 | 1.2 ± 0.5 | 1.4 ± 0.5 | 1.6 ± 0.8 | 0.54 ± 0.4 | 0.45 ± 0.37 |
| BCL11BF/FCD4cre (n = 8) |
76 ± 12.5 | 5.6 ± 2.1 | 2.1 ± 0.8 | 37.4 ± 10.7 | 5.1 ± 1.1 | 6.4 ± 1.5 | 5.8 ± 2.8 | 1.55 ± 0.5 | 2.78 ± 0.8 |
| P value | 0.001 | 0.011 | 4.24E-06 | 0.0004 | 3.73E-07 | 3.56E-06 | 0.001 | 0.012 | 0.00102 |
Total cellularity was determined by counting the live cells. Absolute numbers were calculated based on the percentage of each population. All mice in these experiments were between 8 and 12 weeks of age. Absolute numbers should be multiplied by 106. Two-tailed Student's t test was applied to determine the statistical significance.
BCL11B–deficient DP thymocytes fail to undergo positive selection
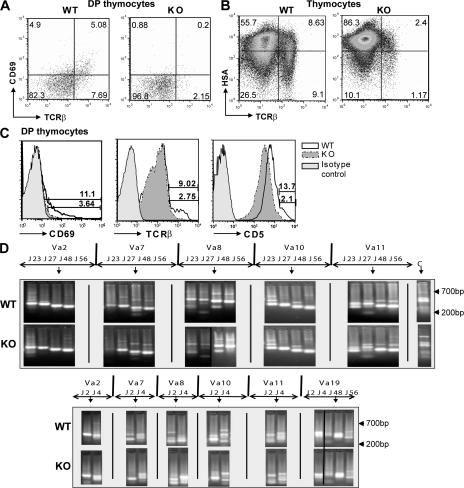
The defects in the BCL11BF/FCD4cre T lymphocytes are likely initiated by the perturbations at the DP stage at T cell development when BCL11B is removed. To obtain further insight into the mechanism underlying the altered thymic development, we first investigated whether the reduction in the numbers of SP thymocytes was caused by a defect in positive selection. During positive selection, CD69 expression is induced (4) and CD5 (5) and TCRβ (1) are up-regulated. Our results show that BCL11BF/FCD4cre mice have a marked reduction in the CD69+TCRβhigh DP thymocyte population (Fig. 4 A), indicating that they are unable to pass positive selection. We also evaluated the percentage of HSAlowTCRβhigh thymocyte population, which contains DP thymocytes that passed positive selection, as well as mature SP thymocytes, and found that it was lower in the BCL11BF/FCD4cre mice (Fig. 4 B). In addition, a detailed analysis of the surface levels of CD69, TCRβ, and CD5 showed a profound decrease in the populations expressing high levels of CD69, CD5, and TCRβ (Fig. 4 C), further demonstrating that positive selection was altered.
Figure 4.
Positive selection of BCL11BF/FCD4cre DP thymocytes is diminished without alteration in TCRα locus rearrangements. (A) Thymocytes were stained for CD4, CD8, TCRβ, and CD69 and analyzed by FACS. CD69 and TCRβ expression on gated DP thymocytes. Numbers indicate percentage of cells in each quadrant. (B) Thymocytes were stained for HSA and TCRβ and analyzed by FACS. Numbers indicate the percentage of cells in each quadrant. (C) Thymocytes were stained for CD4, CD8, TCRβ, CD69, and CD5. Histograms show TCRβ, CD69, and CD5 levels on gated DP thymocytes. (D) Genomic DNA was extracted from DP thymocytes and amplified by PCR using primers specific for the indicated Vα and Jα gene segments. Amplification of the αC region was used for normalization. Dividing lines indicate grouping of images from different parts of the same gel or different gels. This is one representative experiment out of four.
TCRα rearrangements occur normally in BCL11B-deficient DP thymocytes
Germline BCL11B-deficient mice have a block in β-selection caused by unsuccessful TCRβ rearrangements (23). Therefore, the reduction in the number of postselected DP thymocytes in BCL11BF/FCD4cre mice may be caused by the observation that these DP thymocytes do not appropriately rearrange TCRα. Analysis of TCRα rearrangements demonstrated that both early and late rearrangements proceeded similarly in BCL11B-deficient and control DP thymocytes (Fig. 4 D), indicating that the defect in positive selection is unlikely to be caused by impaired TCRα rearrangements. Thus, BCL11B does not control rearrangements at the TCRα locus, although it was previously suggested to be implicated in the control of rearrangements at the TCRβ locus (23).
Provision of transgenic TCRs favoring positive selection of CD4 or CD8 lineages failed to restore positive selection of BCL11B-deficient DP thymocytes
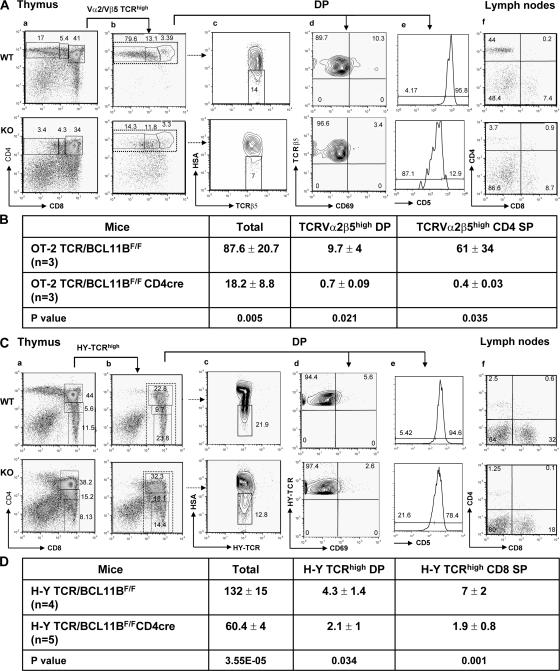
To further facilitate the examination of positive selection, we analyzed BCL11BF/FCD4cre thymocytes expressing transgenic TCRs that recognize specific antigens presented in the context of MHC class I or II. Positive selection of CD4 lineage was investigated with the OT-2 TCR transgenic mice that express a TCR composed of Vα2/Vβ5 chains, recognizing the OVA peptide 323–339 in the context of MHC class II I-Ab (27). The analysis indicated a severe decrease in the percentage of CD4 SP thymocytes (Fig. 5 A, a), as well as peripheral CD4+ T lymphocytes (Fig. 5 A, f), both in percentage and absolute numbers (Fig. 5 B). The same reduction was observed when gating on the TCRα2β5high population (Fig. 5 A, b). Moreover, both TCRβ5highCD69high DP and TCRβ5highHSAlow thymocyte populations remained low (Fig. 5 A, c and d), demonstrating that positive selection of CD4 lineage was impaired. In support of this conclusion, the number of TCRα2β5high DP thymocytes up-regulating CD5 levels was severely reduced (Fig. 5 A, e). Collectively, these results demonstrate that antigen-specific positive selection of CD4 lineage was majorly altered in BCL11BF/FCD4cre mice.
Figure 5.
Altered positive selection of CD8 and CD4 lineages in BCL11BF/FCD4cre mice expressing transgenic TCRs. (A) Positive selection analysis in OT2 mice. Thymocytes were stained for CD4, CD8, TCRVα2, TCRVβ5, and HSA (c), CD69 (d), or CD5 (e). (a) Surface expression of CD4 and CD8 on thymocytes analyzed by FACS. Boxes indicate DP, CD4+CD8low, and CD4 SP populations, and numbers represent the percentage of cells in the boxes. (b) Surface expression of CD4 and CD8 on gated Vα2/Vβ5TCRhigh thymocytes. Boxes indicate DP, CD4+CD8low, and CD4 SP populations. The dotted line rectangle contains all CD4+ Vα2/Vβ5TCRhigh thymocytes. (c) Surface expression of HSA and Vβ5TCR on gated CD4+ Vα2/Vβ5TCRhigh thymocytes. Vβ5TCRhigh/HSAlow population is indicated by a rectangle. (d) Surface expression of CD69 and Vβ5TCR on DP thymocytes. (e) Histogram showing surface expression of CD5 on Vα2/Vβ5TCRhigh DP gated populations. (f) Surface expression of CD4 and CD8 on lymphocytes. Representative data from multiple experiments are shown. (B) Thymic cellularity of OT-2 TCR/BCL11BF/FCD4cre mice. Thymocytes were stained for CD4, CD8, TCRVα2, and TCRVβ5 and analyzed by flow cytometry. Total cellularity was determined by counting the live cells. Absolute numbers of cells were calculated based on the percentage of each population and represented as values ± the SD. Cell counts should be multiplied by 106. Two-tailed Student's t test was applied to determine the statistical significance. (C) Positive selection analysis in H-Y TCR females. Thymocytes were stained for CD4, CD8, T3.70 TCR, and HSA (c), CD69 (d) or CD5 (e). (a) Surface expression of CD4 and CD8 on thymocytes. Boxes indicate DP, CD8+CD4low, and CD8 SP populations, and the numbers represent the percentage of cells in the boxes. (b) Surface expression of CD4 and CD8 on gated T3.70 TCRhigh thymocytes. Boxes indicate DP, CD8+CD4low, and CD8 SP populations. The dotted line rectangle contains all CD8+ T3.70 TCRhigh thymocytes. (c) Surface expression of HSA and T3.70 TCR on all CD8+ gated thymocytes. The T3.70 TCRhigh/HSAlow population is indicated by a rectangle. (d) Surface expression of CD69 and T3.70 TCR on DP gated populations. (e) Histogram showing surface expression of CD5 on T3.70 TCRhigh DP gated populations. (f) Surface expression of CD4 and CD8 on lymphocytes. These are representative experiments out of four. (D) Thymic cellularity of H-Y TCR/BCL11BF/FCD4cre female mice. Thymocytes were stained for HY-TCR, CD4, and CD8 coreceptors and analyzed by flow cytometry. Thymic cellularity was calculated as described in B. Cell counts should be multiplied by 106. Two-tailed Student's t test was applied to determine the statistical significance.
To evaluate positive selection of CD8 lineage, we used the H-Y TCR transgenic mice, which express a TCR that recognizes an antigen derived from the male-specific Smcy protein in the context of H-2Db MHC class I and support positive selection of the CD8 lineage in female mice (28). H-YTCR/BCL11BF/FCD4cre females had reduced percentages and absolute numbers of CD8 SP thymocytes (Fig. 5 C, a, and D). Similar results were obtained when gating on the T3.70high (H-YTCRhigh) population (Fig. 5, C [b] and D). The HY-TCRhigh/HSAlow and H-YTCR highCD69+ populations continued to be low, and the surface level of CD5 was significantly decreased (Fig. 5 C, c–e). Supporting the observation that positive selection was impaired, the peripheral CD8+ T lymphocyte population was diminished (Fig. 5 C, f). Collectively, these results demonstrate that, in the absence of BCL11B, positive selection of the CD8 lineage was impaired, and therefore BCL11B is essential for positive selection of both CD4 and CD8 lineages. In addition, these results suggest that the defect in positive selection is not caused by the TCR itself. A detailed analysis of the cellularity in TCR transgenic and nontransgenic BCL11BF/FCD4cre mice indicated a more severe decrease in the number of CD4 than in the number of CD8 SP thymocytes in the absence of BCL11B (Fig. 5, B and D, and Fig. 2 C), suggesting that selection of the CD4 lineage was more severely altered compared with the CD8 lineage.
Finally, provision of transgenic H-Y or OT-2 TCRs did not restore the absolute numbers of DP thymocytes (Figs. 5, B and D), suggesting that survival of BCL11B-deficient DP thymocytes (see below) remains perturbed even in conditions that favor positive selection to CD4 or CD8 lineages.
BCL11B-deficient DP thymocytes have altered ERK phosphorylation, calcium flux, and proximal events of TCR signaling
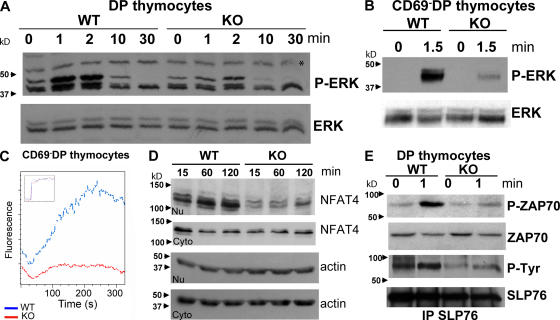
Signaling events downstream of the TCR play a critical role in positive selection. To further determine the cause of the defect in positive selection of BCL11B-deficient DP thymocytes, we examined their responses to TCR stimulation. Specifically, we assessed the Ras–MEK–ERK pathway and calcium mobilization, as these TCR responses play an essential role in positive selection (29–31). Our results demonstrated that ERK phosphorylation was altered both in total DP (Fig. 6 A), as well as in the presignaled CD69− DP thymocytes (Fig. 6 B).
Figure 6.
ERK phosphorylation, Ca flux, and proximal TCR signaling events are attenuated in BCL11BF/FCD4cre DP thymocytes. Immunoblot analysis of ERK phosphorylation kinetics in response to stimulation by TCR cross-linking in DP-enriched thymocytes (A) or sorted presignaled CD69− DP thymocytes (B). In both A and B, ERK phosphorylation was evaluated with anti–phospho-p42/p44. The asterisk in A indicates a nonspecific band. (C) Ca flux analysis in CD69− DP thymocytes. Thymocytes were loaded with Fluo4, surface stained for CD4, CD8, and CD69, and activated as described in the Supplemental materials and methods. Fluorescence was recorded for 5 min after stimulation and ionomycin was added to record the maximal calcium response. Calcium flux was analyzed in presignaled CD69− DP thymocytes using the kinetics platform of the FlowJo and represented as Fluo-4 fluorescence over time. Notably, when anti-CD4 RM4-5 antibody was used for staining and anti-CD4 GK 1.5 antibody was used for stimulation (see Materials and methods and Supplemental materials and methods), the cross-blocking reduced the potency of the anti-CD4 stimulation. (D) Immunoblot analysis of nuclear translocation of NFAT4 in response to TCR activation. (E) Immunoblot analysis of ZAP70 and SLP76 phosphorylation. Preparation of enriched DP thymocytes and their activation is described in the Supplemental materials and methods. Representative data from multiple experiments are shown. Supplemental materials and methods are available at http://www.jem.org/cgi/content/full/jem.20070863/DC1.
Similarly, calcium mobilization in response to TCR activation was attenuated in presignaled CD69− DP thymocytes (Fig. 6 C), and this was not caused by reduced levels of inositol 1,4,5-triphosphate receptor (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20070863/DC1). In DP thymocytes, calcium mobilization in response to TCR signaling results in nuclear translocation of the transcription factor NFAT4 (12). Indeed, our results showed reduced nuclear translocation of NFAT4 in the BCL11B-deficient DP thymocytes (Fig. 6 D), supporting the observation that calcium flux was altered.
To further define the defect in the TCR signaling in BCL11B-deficient DP thymocytes, we investigated proximal events of TCR signaling, including phosphorylation of ZAP70 kinase and of the adaptor molecule SLP76. Our results demonstrated that both ZAP70 and SLP76 phosphorylation was altered in the absence of BCL11B (Fig. 6 E), suggesting that early events upstream of calcium flux and ERK phosphorylation are perturbed in BCL11B-deficient DP thymocytes. Together, these results suggest that BCL11B controls proximal events of TCR signaling and/or a pathway that regulates TCR signaling.
BCL11B-deficient DP thymocytes undergo increased apoptosis even in the absence of TCR signaling
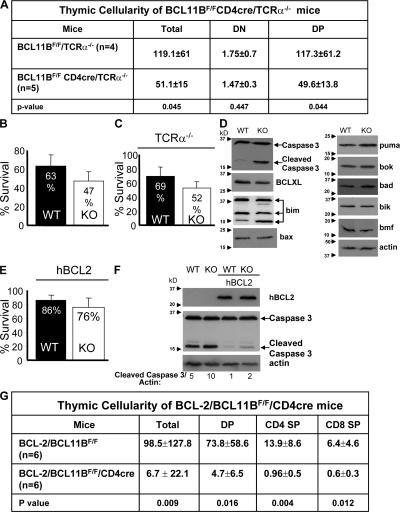
Although the defect in positive selection can explain the reduced numbers of CD4 and CD8 SP thymocytes, it does not explain the reduction in the absolute number of DP thymocytes in BCL11BF/FCD4cre mice (Fig. 2 C). This reduction suggests that BCL11B-deficient DP thymocytes die more compared with wild type. Our hypothesis was that BCL11B-deficient DP thymocytes have an intrinsic defect in survival, as opposed to dying through negative selection, because of their overall reduced capacity to signal through TCR (see above), and also because of the observation that removal of BCL11B in DN3 thymocytes and in cell lines was demonstrated to predispose them to apoptosis (23, 32). To eliminate the possibility that the reduction in DP thymocytes is caused by negative selection, we conducted the following experiments. (a) We crossed the BCL11BF/FCD4cre mice with TCRα−/− mice in which DP thymocytes fail to undergo positive or negative selection and die by neglect because of the lack of TCR signaling. The results show that total and DP thymocyte absolute numbers still remained reduced in BCL11BF/FCD4cre/TCRα−/− compared with TCRα−/− mice (Fig. 7 A), demonstrating that BCL11B-deficient DP thymocytes die more even in the absence of TCR signaling. (b) Because of the observation that the numbers of DP thymocytes were severely reduced in conditions of provision of transgenic TCRs in BCL11BF/FCD4cre mice (Fig. 5, B and D), to further eliminate the possibility that TCR signaling itself induces active cell death in TCR transgenic BCL11BF/FCD4cre mice, we conducted reconstitution experiments with OT2/BCL11BF/FCD4cre bone marrow cells in nonselective (I-Ad) versus selective (I-Ab) recipients. Our results demonstrate that even in a nonselective (I-Ad) background the absolute numbers of OT2/BCL11BF/FCD4cre DP thymocytes were still reduced compared with OT2/BCL11BF/F DP thymocytes (Table S2, available at http://www.jem.org/cgi/content/full/jem.20070863/DC1), again demonstrating that even in the absence of TCR signaling they die more. On a selective background (I-Ab), formation of CD4 SP thymocytes in mice reconstituted with OT2/BCL11BF/FCD4cre bone marrow remained impaired (Table S2), demonstrating that the defect in positive selection is cell intrinsic. Similarly, the numbers of reconstituted OT2/BCL11BF/FCD4cre DP thymocytes remained reduced compared with OT2/BCL11BF/F DP thymocytes (Table S2), despite the observation that the reconstitutions were initiated with the same numbers of bone marrow cells. One possible explanation for the enhanced cell death that we observed in BCL11BF/FCD4cre TCR transgenic mice compared with polyclonal TCR background may be related to the observation that the expression of TCR transgenes is initiated before cells become DP (33). Although in wild-type mice this may favor an earlier selection compared with a polyclonal background, it is possible that in BCL11BF/FCD4cre mice, the cells that express TCR, but fail to be selected, die. This is plausible because removal of the floxed allele in CD4-Cre model is initiated past DN3 stage and continues on during the DP stage (25).
Figure 7.
BCL11BF/FCD4cre DP thymocytes present enhanced apoptosis independent of TCR signaling, which is only partially rescued by transgenic BCL2. (A) Thymic cellularity of BCL11BF/FCD4cre/TCRα−/− and BCL11BF/F/TCRα−/− mice. Thymic cellularity was calculated as described in Fig. 2 C. Absolute cell numbers should be multiplied by 106. Two-tailed Student's t test was applied to determine the statistical significance. Mice in these experiments were between 5 and 8 wk of age. (B) In vitro survival of BCL11BF/FCD4cre DP thymocytes. Fresh or cultured thymocytes were stained for CD4 and CD8, and with AnnexinV and 7-AAD and analyzed by FACS. Survival of DP thymocytes in vitro was estimated as the frequency of live (Annexin V−/7-AAD−) cells after 16 h in culture reported to the input population, and represented as the mean ± the SD. n = 4 pairs of mice. P = 0.034. (C) In vitro survival of BCL11BF/FCD4cre/TCRα−/− DP thymocytes. Fresh or cultured thymocytes from BCL11BF/FCD4cre/TCRα−/− and BCL11BF/F/TCRα−/− mice were stained, analyzed, and represented as in B. n = 4 pairs of mice. P = 0.032. (D) Immunoblot analysis of the indicated proapoptotic and prosurvival factors. (E) In vitro survival of BCL11BF/FCD4cre/BCL2 DP thymocytes. Fresh or cultured thymocytes from BCL11BF/FCD4cre/BCL2 and BCL11BF/F/BCL2 mice were stained, analyzed, and represented as in B. n = 6 pairs of mice. P = 0.054. (F) Immunoblot analysis showing cleaved caspase 3 in BCL2 transgenic and nontransgenic BCL11BF/FCD4cre and BCL11BF/F DP thymocytes. Cleaved caspase 3 levels were quantified by densitometric analysis and normalized to actin. (G) Thymic cellularity of BCL-2/BCL11BF/FCD4cre mice. Thymocytes were stained for CD4 and CD8 and analyzed by FACS. Thymic cellularity was calculated as described in Fig. 2 C. Cell counts should be multiplied by 106. Two-tailed Student's t test was applied to determine the statistical significance.
We also evaluated the survival of BCL11B-deficient DP thymocytes in vitro and the results showed that it was lower compared with wild-type control (Fig. 7 B), indicating that BCL11B-deficient DP thymocytes are more sensitive to spontaneous apoptosis. Similarly, DP thymocytes of BCL11BF/FCD4cre/TCRα− mice also died more in vitro (Fig. 7 C), supporting the hypothesis that their enhanced apoptosis is the result of death by neglect or an intrinsic predisposition to cell death. To further support the idea that BCL11B promotes survival of T lymphocytes, we used a Jurkat cell line model that overexpresses BCL11B, generated through retroviral transduction (22). Our results demonstrate that overexpression of BCL11B conferred increased protection to apoptosis to Jurkat cells (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20070863/DC1), suggesting that BCL11B supports survival of immature and transformed T cells.
Enhanced apoptosis of BCL11B-deficient DP thymocytes is mediated through caspase 3, and only partially dependent on BCL2 family members
To investigate the mechanism by which BCL11BF/FCD4cre DP thymocytes die, we first evaluated the levels of cleaved caspase 3, which is downstream of both intrinsic and extrinsic apoptotic pathways. Cleaved caspase 3 levels were elevated in BCL11B-deficient DP thymocytes (Fig. 7 D), demonstrating the implication of this effector caspase. The reduced survival of BCL11B-deficient DP thymocytes could be caused by a decrease in the levels of BCLXL, which plays a prominent role in DP thymocyte survival (34). However, we found only a very modest decrease of BCLXL protein levels (Fig. 7 D). As survival/apoptosis is determined by the balance between prosurvival versus proapoptotic factors, we further evaluated the protein levels of several proapoptotic factors, and found only modest increases in Bim, Puma, Bok, and Bad levels (Fig. 7 D). Overall, the balance between proapoptotic/prosurvival factors was inclined toward apoptosis. To further clarify whether the imbalance in proapoptotic/prosurvival factors is the cause of enhanced apoptosis, we provided transgenic BCL2, as BCL2 and BCLXL were demonstrated to be equally potent in inhibiting thymocyte death (34–37). In vitro survival of BCL11B-deficient DP thymocytes was improved by provision of BCL2 transgene (Fig. 7 E), suggesting that BCL2 family proteins play a role in survival mediated by BCL11B. The levels of cleaved caspase 3 still remained slightly elevated (Fig. 7 F). However, the cellularity of the transgenic BCL2/BCL11B-deficient thymi, including the absolute numbers of DP thymocytes, remained severely reduced (Fig. 7 G), indicating that the survival of BCL11B-deficient DP thymocytes was not restored in vivo. These results suggest that although BCL2 family members play a role in the survival defect of BCL11B-deficient DP thymocytes, other mechanisms are also likely to be involved. Finally, provision of BCL2 did not restore the CD4 and CD8 SP thymocyte numbers or positive selection (Fig. 7 G and Fig. S5, available at http://www.jem.org/cgi/content/full/jem.20070863/DC1).
BCL11B-deficient DP thymocytes have altered expression of genes involved in TCR signaling, positive selection, and survival/apoptosis
As BCL11B is a transcription factor, we reasoned that the defects of BCL11B-deficient DP thymocytes are caused by changes in gene expression. We conducted a gene expression profiling in BCL11B-deficient DP thymocytes versus wild-type control (unpublished data) and focused our analysis on genes relevant for positive selection, signaling, and survival/apoptosis. Expression of candidate genes was confirmed by qRT-PCR (Fig. 8 A). To eliminate the possibility that the differences in the mRNA levels are a consequence of the absence of postselected DP thymocytes in BCL11BF/FCD4cre mice, for genes with relevance for TCR signaling and positive selection, we also tested the mRNA levels in TCRα−/− DP thymocytes, which predominantly consist of presignaled DP thymocytes (Fig. 8 A). mRNAs for three molecules previously linked to positive selection were found to be deregulated in BCL11B-deficient DP thymocytes, i.e., programmed cell death 1 (PD-1), p55 γ adaptor subunit of PI 3-kinase (Pik3r3), and CD5 (Fig. 8 A). PD-1, which is a member of B7/CD28 costimulatory receptors, previously demonstrated to alter positive selection when transgenically overexpressed in DP thymocytes (38). Interestingly, PD-1 mRNA and protein levels were up-regulated in BCL11B-deficient DP thymocytes (Fig. 8, A and B), thus suggesting that this molecule can be implicated in the mechanisms that alter signaling and positive selection of BCL11B-deficient DP thymocytes.
Figure 8.
Expression of genes implicated in positive selection, TCR signaling, and apoptosis is altered in BCL11B-deficient DP thymocytes. (A) Ratios of relative mRNA levels of genes involved in positive selection and survival/apoptosis in BCL11B-deficient DP thymocytes versus wild type. mRNA levels were quantified by qRT-PCR, as described in the Materials and methods (22), in BCL11BF/FCD4cre (KO) and BCL11BF/F (WT), and in BCL11BF/FCD4cre/TCRα−/− (KO) and BCL11BF/F/TCRα−/− (WT) mice. Representative data is presented from multiple experiments. PD-1 (B) and CD5 (C) surface levels in thymocyte populations. Total thymocytes from BCL11BF/FCD4cre (KO) and BCL11BF/F (WT) mice were stained for CD4, CD8, CD69, and PD-1 (B) or CD5 (C) and analyzed by flow cytometry. Histograms showing PD-1 (B) and CD5 (C) levels on gated CD69− DP, CD69+ DP, CD4, and CD8 SP thymocytes. Data are representative of four independent experiments.
Another candidate gene associated with TCR signaling was the γ adaptor subunit (Pik3r3), which is an enzyme previously shown to be implicated in positive selection downstream of TCR engagement and upstream of calcium mobilization (39).
In addition, CD5 mRNA levels were reduced in BCL11B-deficient DP thymocytes (Fig. 8 A), and this was not just a consequence of the failure to pass positive selection, as the surface and intracellular levels of CD5 were also reduced in BCL11B-deficient presignaled CD69− DP thymocytes (Fig. 8 C and not depicted). As expected, CD5 remained reduced in postselected CD69+ DP thymocytes, as well as in CD4 and CD8 SP thymocytes (Fig. 8 C). These results collectively demonstrate that BCL11B-deficient DP thymocytes have multiple defects in expression of genes associated with positive selection, consistent with the functional defects of BCL11B-deficient DP thymocytes.
In regard to survival/apoptosis, mRNA levels of BCLXL and BCL2 were modestly down-regulated, suggesting that they may be candidate target genes, whereas the mRNA levels for the proapoptotic factors deregulated at protein level (Fig. 7 D) were found unchanged (not depicted), suggesting that these genes are not candidate targets. In addition, the gene encoding pleckstrin homology-like domain, family A, member 1 (Phlda1), also known as T cell death associated gene 51 (Tdag51), which was previously found to induce cell death in different cellular systems (40), was highly expressed in BCL11B-deficient thymocytes (Fig. 8 A), suggesting that it may play a role in enhanced apoptosis.
In addition to genes associated with positive selection and survival, two genes encoding membrane-associated proteins, CD160 and Ly6c, which are normally not expressed in DP thymocytes, were up-regulated BCL11B-deficient DP thymocytes (Fig. 8 A). CD160 (BY55) is an Ig-like receptor expressed in NK, NKT, and cytotoxic T cells, and has broad specificity binding for classical and nonclassical MHC class I molecules (41–43). Ly6c is a GPI-anchored cell surface protein known to mediate inhibitory signals on effector CD4+ T cells (44–46). These results suggest that potentially inappropriate signals may be initiated in BCL11B-deficient DP thymocytes, which may interfere with TCR signaling and the main program at this stage of T cell development.
DISCUSSION
In this study, we investigated how selective removal of BCL11B at the DP stage of T cell development impacts positive selection and survival of DP thymocytes. Positive selection of both CD4 and CD8 lineages was compromised in BCL11BF/FCD4cre mice. However, this was not a consequence of a failure to rearrange TCRα, as rearrangements proceeded normally in the absence of BCL11B. The induction of CD69 expression and up-regulation of CD5 and TCRβ, which are hallmarks of proficient progression through positive selection (1), were altered in BCL11B-deficient DP thymocytes. The reduction in the formation and implicit selection of the CD8 lineage was less severe than that of the CD4 lineage. Interestingly, these results are similar to the phenotypes described in mice with defects in genes encoding TCR signaling components, such as those of the Ras–MEK–ERK pathway, including MEK1 (29), Ras (47), and ERK1 and ERK2 (31). Nevertheless, provision of a transgenic TCR that favors positive selection of the CD8 lineage did not restore the selection and formation of CD8 lineage, strongly suggesting that they are altered in the absence of BCL11B. Positive selection and formation of the CD4 lineage was severely compromised, and provision of a transgenic TCR that favors selection of the CD4 lineage did not restore it either. Therefore, these studies suggest that the defect in positive selection is not caused by the TCR itself, but likely by molecules implicated in TCR signaling and/or a pathway that affects TCR signaling. This idea is also supported by previous observations, which demonstrated that provision of TCRαβ in the germline BCL11B-deficient mice only partially rescued the developmental arrest at the β-selection (48), altogether suggesting that BCL11B may directly or indirectly control signaling components downstream of the TCR and/or a pathway that impacts TCR signaling. In support of this hypothesis, our results demonstrate that signaling through the two major pathways, which are activated through TCR during positive selection, Ras–MEK–ERK pathway, and Ca2+ flux (1, 29–31, 47, 49), is altered in BCL11B-deficient DP thymocytes. Moreover, phosphorylation of ZAP70 and SLP76 was also altered in BCL11B-deficient DP thymocytes, suggesting a defect in proximal TCR signaling and/or in a pathway that proximally impacts TCR signaling. There are several transcription factors with roles in positive selection known to act distally in TCR signaling, downstream of ERK or calcium signaling activation. Such examples include SAP1, which is a direct target of ERK phosphorylation (9), its target EGR1 (10), as well as EGR1's target Id3 (11), and NFAT4, which is a target for calcium signaling (12). Different from these transcription factors, BCL11B seems to have a very unique position, controlling proximal events of TCR signaling, and/or a pathway that impacts TCR signaling. How does BCL11B specifically direct transcriptional events required for successful positive selection of DP-positive thymocytes? We identified several “candidate” genes previously implicated in positive selection and signaling with altered mRNA levels in BCL11B-deficient DP thymocytes. One such candidate up-regulated in BCL11B-deficient DP thymocytes, both at mRNA and protein level, is PD-1, which is a member B7/CD28 costimulatory receptors (50–52). PD-1 was shown to diminish TCR signaling and ERK phosphorylation, and to alter positive selection (38) by diminishing Zap70 activity by recruitment of the inhibitory SHP-1 and -2 phosphatases through immunoreceptor-tyrosine inhibition motif (38, 53). Indeed, not only calcium flux and ERK phosphorylation were defective in BCL11B-deficient DP thymocytes but also phosphorylation of ZAP70, which is an indication of its altered function. Thus, the high levels of PD-1 in BCL11B-deficient DP thymocytes may account for altered TCR signaling, and implicitly defective positive selection. Another candidate gene found to be down-regulated in BCL11B-deficient DP thymocytes is Pik3r3, which is an understudied adaptor subunit of PI3-kinase. Previous findings implicated PI3-kinase in positive selection through the catalytic subunit p110α (39). However, it was reported that despite a major block in B lymphocyte development, no disruption of positive selection was observed in the absence of the α (Pik3r1) and β (Pik3r2) adaptor subunits (54, 55). Interestingly, in thymocytes of these mice, the expression of the third adaptor subunit, γ or Pik3r3, was found to be up-regulated. Therefore, it is tempting to speculate that the adaptor subunit γ (Pik3r3), identified by us as a candidate gene down-regulated in BCL11B-deficient DP thymocytes, may be the adaptor subunit of PI 3-kinase responsible for positive selection of DP thymocytes. Thus, the lower Pik3r3 levels in BCL11B-deficient DP thymocytes may participate in the altered calcium flux. In addition to the two molecules implicated in TCR signaling, CD5 was found to be reduced both at mRNA and protein levels, indicating that positive selection is compromised at multiple levels and in a complex manner. To this end, it is also possible that the defect in positive selection in BCL11B-deficient DP thymocytes is not only the result of direct alterations in TCR signaling. Rather, other surface molecules may provide signals that perturb TCR signaling, compromising positive selection. In this regard, we found increased mRNA levels for two membrane-associated proteins, CD160 and Ly6c, which are normally not expressed in DP thymocytes, but which are able to initiate signaling in NK, CD8+, and CD4+ T lymphocytes (41, 45, 46). Such signals initiated in DP thymocytes may interfere with the normal TCR signaling and the program of DP thymocytes.
BCL11B-deficient DP thymocytes also presented enhanced apoptosis. Interestingly, other previously reported deficiencies in positive selection, associated with defects in CD3δ (56), Lck (57, 58, 59), MEK1 (29), Ras (47), Raf-1 (60), and ERK1 and ERK2 (31), were not accompanied by a major impairment in survival, which suggests that the reduced survival of BCL11B-deficient DP thymocytes may be independent of the defect in positive selection. This hypothesis is supported by the observation that BCL11B-deficient DP thymocytes continued to exhibit enhanced death even in the absence of TCR signaling, as demonstrated by the experiments with TCRα−/− mice. Interestingly, BCL11B-deficient DP thymocytes still survive long enough to rearrange TCRα. This phenotype is different from the RORγ deficiency, in which altered survival of DP thymocytes impairs TCRα rearrangements (61, 62). Of interest, BCL11B-deficient DN3 thymocytes were also more susceptible to apoptosis (23). Thus, it is possible that BCL11B may control thymocyte survival both at the DP and at earlier stages of T cell development. We observed enhanced levels of cleaved caspase 3 and an imbalanced proapoptotic/prosurvival factors in BCL11B-deficient DP thymocytes, and extensively evaluated the mRNA levels of multiple proapoptotic and prosurvival factors, including BCLXL, BCL2, Bad, Bok, Puma, and Bim. Except for the modest down-regulation of BCLXL and BCL2, we failed to see any differences in the mRNA levels of the aforementioned proapoptotic factors, suggesting that these genes are not candidate targets. The observation that transgenic BCL2 failed to rescue cellularity in vivo strongly suggests the implication of other pathways in the enhanced apoptosis of BCL11B-deficient DP thymocytes. In this regard, the mRNA for Phlda1/Tdag51 was found to be up-regulated in BCL11B-deficient DP thymocytes. Interestingly, Phlda1/Tdag51 was found to act both on death receptor-dependent and -independent pathways (40, 63), raising the possibility that this factor may be implicated in the enhanced apoptosis of BCL11B-deficient DP thymocytes.
Overall, the results presented here demonstrate a complex and unique role of the BCL11B transcription factor in the control of both positive selection and survival of DP thymocytes.
MATERIALS AND METHODS
Mice.
Mice carrying the floxed BCL11B alleles (BCL11BF/F) were generated in 129S3 background and will be described elsewhere (unpublished data). BCL11BF/F mice were backcrossed on C57BL/6 background for 5 generations using the “Speed Congenics” Technology, provided by The Jackson Laboratory, which allowed us to reach a 98% C57BL/6 background. BCL11BF/F mice were crossed with C57BL/6 CD4Cre transgenic mice (line 4196; Taconic) (25) to generate conditional BCL11BF/FCD4cre mice. To obtain H-Y TCR/- and OT-2 TCR/BCL11BF/FCD4cre transgenic mice, were crossed with H-Y TCR (28) (Taconic) or OT-2 TCR mice (27) (The Jackson Laboratory). To obtain BCL2/BCL11BF/FCD4cre transgenic mice, we crossed with BCL2 transgenic mice (34) (A. Singer, National Cancer Institute, Bethesda, MD). TCRα−/− mice were purchased from The Jackson Laboratory. Experiments were conducted with 5–6 wk old mice, unless specified. Mice were kept under specific pathogen–free conditions in the animal care facility of Albany Medical College, and all animal procedures were approved by the Institutional Animal Care and Use Committee.
Cell culture, antibodies, and flow cytometry.
Primary cells were maintained in RPMI 1640. DP cells were enriched using anti-CD8 magnetic beads (Miltenyi Biotec), resulting in ∼95% DP thymocytes. The following antibodies were used for surface staining: anti-CD4 (clones RMA4-5 and GK1.5), -CD8 (clone 53–6.7), -TCRβ (clone H57-597), -TCRα2 (clone B20.1), -HYTCR (clone T3.70), -HSA (clone M1/69), -CD69 (clone H1.2F3), -CD5 (clone 53–7.3), -CD3 (clone 145-2C11), -TCRγδ (clone GL3), -B220 (clone RA3-6B2), -F4/80 (clone BM8), -CD11c (clone N418), -CD49b (clone DX5), -CD41 (clone MW Reg30; eBioscience), and -TCRβ5 antibodies (clone MR9-4; BD Biosciences). Intranuclear staining of BCL11B with the previously described anti-BCL11B antibodies (22) was conducted after staining for surface CD4 and CD8, followed by fixation and permeabilization with 1% Triton X-100. Determination of apoptosis in fresh or 16-h–cultured thymocytes was conducted by staining with AnnexinV (BD Biosciences) and 7-AAD (eBioscience) after surface staining for CD4 and CD8. Flow cytometry analysis was conducted on a FACSCanto instrument (BD Biosciences). Sorting of thymocytes was performed on a FACSAria instrument. Flow cytometry data was analyzed using FlowJo software (Tree Star, Inc).
qRT-PCR.
For evaluation of gene expression, DP thymocytes were either sorted or purified by selection on anti-CD8 magnetic beads. qRT-PCR was conducted as previously described (22). The primers for q-PCR were chosen so as to extend products under 200 bp with no formation of primer dimers. The relative abundance of each message was normalized to actin and calculated as: 2−(Ct gene−Ct actin), where Ct represents the threshold cycle for each transcript. Sequence of the primers is available upon request.
Statistical analysis.
Differences between various BCL11BF/FCD4cre and control mice were analyzed by the two-tailed Student t test and expressed as the mean ± the SD. P ≤ 0.05 was considered significant for all analyses.
Online supplemental material.
Fig. S1 shows selective removal of BCL11B starting with DP thymocytes. Fig. S2 shows that BCL11BF/FCD4cre/OT2 and BCL11BF/FCD4cre/HYTCR mice present enlarged spleens and mesenteric lymph nodes. Fig. S3 shows that inositol 1,4,5-triphosphate receptor expression is not affected in BCL11B-deficient DP thymocytes. Fig. S4 shows that BCL11B-overexpressing cells are more resistant to dexamethasone-induced cell death. Fig. S5 shows that provision of BCL2 does not restore positive selection of BCL11BF/FCD4cre DP thymocytes and CD4 and CD8 SP thymocyte populations. Table S1 shows splenic Cellularity of BCL11BF/FCD4Cre/TCRα−/− mice. Table S2 shows the absolute number of thymocytes in mice reconstituted with bone marrow from TCR OT2 transgenics. A Supplemental materials and methods is available, describing in detail the experimental procedures for TCRα rearrangements, Western blot analysis, and intracellular calcium flux determination. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20070863/DC1.
Supplemental Material
Acknowledgments
We greatly acknowledge Drs. Leszek Ignatowicz (Medical College of Georgia), Jonathan Harton, Jim Drake, Michelle Lennartz, Kevin Pumiglia, and Karen Duus, and all members of Avram laboratory (Albany Medical College) for valuable suggestions. We thank Dr. Mark Preissler (AMC) for advice with calcium flux, and Dr. Al Singer (NCI) for the BCL2 transgenic mice. We thank Dr. Douglas Cohn and Ms. Betsy Bashaw for care of the mice, and Ms. Debbie Moran for secretarial assistance. We acknowledge the contribution of the Albany Medical College flow cytometry facility, and thank Drs. Sridar Chittur and Scott Tenenbaum (Center for Functional Genomics, State University of New York at Albany) for the help with the microarray analysis.
This work was supported by K01 AR-02194 (National Institute of Arthritis and Musculoskeletal and Skin Diseases) and RSG-04-01-MGO (American Cancer Society) grants (to D. Avram) and training grant National Institutes of Health T32-HL-07194 (to D.I. Albu).
The authors have no conflicting financial interests.
Abbreviations used: DP, double-positive; qRT-PCR, quantitative RT-PCR; SP, single-positive.
N. A. Jenkins and N.G. Copeland's present address is Institute of Molecular and Cell Biology, Proteos, Singapore 138673.
References
- 1.Aliahmad, P., and J. Kaye. 2006. Commitment issues: linking positive selection signals and lineage diversification in the thymus. Immunol. Rev. 209:253–273. [DOI] [PubMed] [Google Scholar]
- 2.von Boehmer, H., and P. Kisielow. 2006. Negative selection of the T-cell repertoire: where and when does it occur? Immunol. Rev. 209:284–289. [DOI] [PubMed] [Google Scholar]
- 3.Bosselut, R., T.I. Guinter, S.O. Sharrow, and A. Singer. 2003. Unraveling a revealing paradox: Why major histocompatibility complex I–signaled thymocytes “paradoxically” appear as CD4+8lo transitional cells during positive selection of CD8+ T cells. J. Exp. Med. 197:1709–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swat, W., M. Dessing, H. von Boehmer, and P. Kisielow. 1993. CD69 expression during selection and maturation of CD4+8+ thymocytes. Eur. J. Immunol. 23:739–746. [DOI] [PubMed] [Google Scholar]
- 5.Tarakhovsky, A., S.B. Kanner, J. Hombach, J.A. Ledbetter, W. Muller, N. Killeen, and K. Rajewsky. 1995. A role for CD5 in TCR-mediated signal transduction and thymocyte selection. Science. 269:535–537. [DOI] [PubMed] [Google Scholar]
- 6.Azzam, H.S., A. Grinberg, K. Lui, H. Shen, E.W. Shores, and P.E. Love. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188:2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linette, G.P., M.J. Grusby, S.M. Hedrick, T.H. Hansen, L.H. Glimcher, and S.J. Korsmeyer. 1994. Bcl-2 is upregulated at the CD4+ CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity. 1:197–205. [DOI] [PubMed] [Google Scholar]
- 8.Sudo, T., S. Nishikawa, N. Ohno, N. Akiyama, M. Tamakoshi, H. Yoshida, and S. Nishikawa. 1993. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc. Natl. Acad. Sci. USA. 90:9125–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello, P.S., R.H. Nicolas, Y. Watanabe, I. Rosewell, and R. Treisman. 2004. Ternary complex factor SAP-1 is required for Erk-mediated thymocyte positive selection. Nat. Immunol. 5:289–298. [DOI] [PubMed] [Google Scholar]
- 10.Bettini, M., H. Xi, J. Milbrandt, and G.J. Kersh. 2002. Thymocyte development in early growth response gene 1-deficient mice. J. Immunol. 169:1713–1720. [DOI] [PubMed] [Google Scholar]
- 11.Rivera, R.R., C.P. Johns, J. Quan, R.S. Johnson, and C. Murre. 2000. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 12:17–26. [DOI] [PubMed] [Google Scholar]
- 12.Oukka, M., I.C. Ho, F.C. de la Brousse, T. Hoey, M.J. Grusby, and L.H. Glimcher. 1998. The transcription factor NFAT4 is involved in the generation and survival of T cells. Immunity. 9:295–304. [DOI] [PubMed] [Google Scholar]
- 13.Egawa, T., R.E. Tillman, Y. Naoe, I. Taniuchi, and D.R. Littman. 2007. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204:1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun, G., X. Liu, P. Mercado, S.R. Jenkinson, M. Kypriotou, L. Feigenbaum, P. Galera, and R. Bosselut. 2005. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6:373–381. [DOI] [PubMed] [Google Scholar]
- 15.He, X., X. He, V.P. Dave, Y. Zhang, X. Hua, E. Nicolas, W. Xu, B.A. Roe, and D.J. Kappes. 2005. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 433:826–833. [DOI] [PubMed] [Google Scholar]
- 16.Taniuchi, I., M. Osato, T. Egawa, M.J. Sunshine, S.C. Bae, T. Komori, Y. Ito, and D.R. Littman. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633. [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson, B., J.Y. Chen, P. Han, K.M. Rufner, O.D. Goularte, and J. Kaye. 2002. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat. Immunol. 3:272–280. [DOI] [PubMed] [Google Scholar]
- 18.Aliahmad, P., E. O'Flaherty, P. Han, O.D. Goularte, B. Wilkinson, M. Satake, J.D. Molkentin, and J. Kaye. 2004. TOX provides a link between calcineurin activation and CD8 lineage commitment. J. Exp. Med. 199:1089–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avram, D., A. Fields, K. Pretty On Top, D.J. Nevrivy, J.E. Ishmael, and M. Leid. 2000. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J. Biol. Chem. 275:10315–10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avram, D., A. Fields, T. Senawong, A. Topark-Ngarm, and M. Leid. 2002. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem. J. 368:555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cismasiu, V.B., K. Adamo, J. Gecewicz, J. Duque, Q. Lin, and D. Avram. 2005. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 24:6753–6764. [DOI] [PubMed] [Google Scholar]
- 22.Cismasiu, V.B., S. Ghanta, J. Duque, D.I. Albu, H.M. Chen, R. Kasturi, and D. Avram. 2006. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 108:2695–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakabayashi, Y., H. Watanabe, J. Inoue, N. Takeda, J. Sakata, Y. Mishima, J. Hitomi, T. Yamamoto, M. Utsuyama, O. Niwa, et al. 2003. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat. Immunol. 4:533–539. [DOI] [PubMed] [Google Scholar]
- 24.Wakabayashi, Y., J. Inoue, Y. Takahashi, A. Matsuki, H. Kosugi-Okano, T. Shinbo, Y. Mishima, O. Niwa, and R. Kominami. 2003. Homozygous deletions and point mutations of the Rit1/Bcl11b gene in gamma-ray induced mouse thymic lymphomas. Biochem. Biophys. Res. Commun. 301:598–603. [DOI] [PubMed] [Google Scholar]
- 25.Lee, P.P., D.R. Fitzpatrick, C. Beard, H.K. Jessup, S. Lehar, K.W. Makar, M. Perez-Melgosa, M.T. Sweetser, M.S. Schlissel, S. Nguyen, et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. [DOI] [PubMed] [Google Scholar]
- 26.Singh, N.J., and R.H. Schwartz. 2006. The lymphopenic mouse in immunology: from patron to pariah. Immunity. 25:851–855. [DOI] [PubMed] [Google Scholar]
- 27.Barnden, M.J., J. Allison, W.R. Heath, and F.R. Carbone. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34–40. [DOI] [PubMed] [Google Scholar]
- 28.Kisielow, P., H. Bluthmann, U.D. Staerz, M. Steinmetz, and H. von Boehmer. 1988. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 333:742–746. [DOI] [PubMed] [Google Scholar]
- 29.Alberola-Ila, J., K.A. Forbush, R. Seger, E.G. Krebs, and R.M. Perlmutter. 1995. Selective requirement for MAP kinase activation in thymocyte differentiation. Nature. 373:620–623. [DOI] [PubMed] [Google Scholar]
- 30.Neilson, J.R., M.M. Winslow, E.M. Hur, and G.R. Crabtree. 2004. Calcineurin B1 is essential for positive but not negative selection during thymocyte development. Immunity. 20:255–266. [DOI] [PubMed] [Google Scholar]
- 31.Fischer, A.M., C.D. Katayama, G. Pages, J. Pouyssegur, and S.M. Hedrick. 2005. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 23:431–443. [DOI] [PubMed] [Google Scholar]
- 32.Grabarczyk, P., G.K. Przybylski, M. Depke, U. Volker, J. Bahr, K. Assmus, B.M. Broker, R. Walther, and C.A. Schmidt. 2006. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene. 26:3797–3810. [DOI] [PubMed] [Google Scholar]
- 33.Baldwin, T.A., M.M. Sandau, S.C. Jameson, and K.A. Hogquist. 2005. The timing of TCRα expression critically influences T cell development and selection. J. Exp. Med. 202:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao, D.T., G.P. Linette, L.H. Boise, L.S. White, C.B. Thompson, and S.J. Korsmeyer. 1995. Bcl-xL and Bcl-2 repress a common pathway of cell death. J. Exp. Med. 182:821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costello, P.S., S.C. Cleverley, R. Galandrini, S.W. Henning, and D.A. Cantrell. 2000. The GTPase rho controls a p53-dependent survival checkpoint during thymopoiesis. J. Exp. Med. 192:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ioannidis, V., F. Beermann, H. Clevers, and W. Held. 2001. The beta-catenin–TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat. Immunol. 2:691–697. [DOI] [PubMed] [Google Scholar]
- 37.Biju, M.P., A.K. Neumann, S.J. Bensinger, R.S. Johnson, L.A. Turka, and V.H. Haase. 2004. Vhlh gene deletion induces Hif-1-mediated cell death in thymocytes. Mol. Cell. Biol. 24:9038–9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keir, M.E., Y.E. Latchman, G.J. Freeman, and A.H. Sharpe. 2005. Programmed death-1 (PD-1):PD-ligand 1 interactions inhibit TCR-mediated positive selection of thymocytes. J. Immunol. 175:7372–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbee, S.D., and J. Alberola-Ila. 2006. Phosphatidylinositol 3-kinase improves the efficiency of positive selection. Int. Immunol. 18:921–930. [DOI] [PubMed] [Google Scholar]
- 40.Hayashida, N., S. Inouye, M. Fujimoto, Y. Tanaka, H. Izu, E. Takaki, H. Ichikawa, J. Rho, and A. Nakai. 2006. A novel HSF1-mediated death pathway that is suppressed by heat shock proteins. EMBO J. 25:4773–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maeda, M., C. Carpenito, R.C. Russell, J. Dasanjh, L.L. Veinotte, H. Ohta, T. Yamamura, R. Tan, and F. Takei. 2005. Murine CD160, Ig-like receptor on NK cells and NKT cells, recognizes classical and nonclassical MHC class I and regulates NK cell activation. J. Immunol. 175:4426–4432. [DOI] [PubMed] [Google Scholar]
- 42.Rey, J., J. Giustiniani, F. Mallet, V. Schiavon, L. Boumsell, A. Bensussan, D. Olive, and R.T. Costello. 2006. The co-expression of 2B4 (CD244) and CD160 delineates a subpopulation of human CD8+ T cells with a potent CD160-mediated cytolytic effector function. Eur. J. Immunol. 36:2359–2366. [DOI] [PubMed] [Google Scholar]
- 43.Tsujimura, K., Y. Obata, Y. Matsudaira, K. Nishida, Y. Akatsuka, Y. Ito, A. Demachi-Okamura, K. Kuzushima, and T. Takahashi. 2006. Characterization of murine CD160+ CD8+ T lymphocytes. Immunol. Lett. 106:48–56. [DOI] [PubMed] [Google Scholar]
- 44.Hanninen, A., I. Jaakkola, M. Salmi, O. Simell, and S. Jalkanen. 1997. Ly-6C regulates endothelial adhesion and homing of CD8(+) T cells by activating integrin-dependent adhesion pathways. Proc. Natl. Acad. Sci. USA. 94:6898–6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaakkola, I., M. Merinen, S. Jalkanen, and A. Hanninen. 2003. Ly6C induces clustering of LFA-1 (CD11a/CD18) and is involved in subtype-specific adhesion of CD8 T cells. J. Immunol. 170:1283–1290. [DOI] [PubMed] [Google Scholar]
- 46.Yamanouchi, S., K. Kuwahara, A. Sakata, T. Ezaki, S. Matsuoka, J. Miyazaki, S. Hirose, T. Tamura, H. Nariuchi, and N. Sakaguchi. 1998. A T cell activation antigen, Ly6C, induced on CD4+ Th1 cells mediates an inhibitory signal for secretion of IL-2 and proliferation in peripheral immune responses. Eur. J. Immunol. 28:696–707. [DOI] [PubMed] [Google Scholar]
- 47.Swan, K.A., J. Alberola-Ila, J.A. Gross, M.W. Appleby, K.A. Forbush, J.F. Thomas, and R.M. Perlmutter. 1995. Involvement of p21ras distinguishes positive and negative selection in thymocytes. EMBO J. 14:276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue, J., T. Kanefuji, K. Okazuka, H. Watanabe, Y. Mishima, and R. Kominami. 2006. Expression of TCR alpha beta partly rescues developmental arrest and apoptosis of alpha beta T cells in Bcl11b−/− mice. J. Immunol. 176:5871–5879. [DOI] [PubMed] [Google Scholar]
- 49.Dower, N.A., S.L. Stang, D.A. Bottorff, J.O. Ebinu, P. Dickie, H.L. Ostergaard, and J.C. Stone. 2000. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat. Immunol. 1:317–321. [DOI] [PubMed] [Google Scholar]
- 50.Ishida, Y., Y. Agata, K. Shibahara, and T. Honjo. 1992. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 11:3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharpe, A.H., E.J. Wherry, R. Ahmed, and G.J. Freeman. 2007. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat. Immunol. 8:239–245. [DOI] [PubMed] [Google Scholar]
- 52.Sharpe, A.H., and A.K. Abbas. 2006. T-cell costimulation–biology, therapeutic potential, and challenges. N. Engl. J. Med. 355:973–975. [DOI] [PubMed] [Google Scholar]
- 53.Sheppard, K.A., L.J. Fitz, J.M. Lee, C. Benander, J.A. George, J. Wooters, Y. Qiu, J.M. Jussif, L.L. Carter, C.R. Wood, and D. Chaudhary. 2004. PD-1 inhibits T-cell receptor induced phosphorylation of the ZAP70/CD3zeta signalosome and downstream signaling to PKCtheta. FEBS Lett. 574:37–41. [DOI] [PubMed] [Google Scholar]
- 54.Fruman, D.A., S.B. Snapper, C.M. Yballe, L. Davidson, J.Y. Yu, F.W. Alt, and L.C. Cantley. 1999. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science. 283:393–397. [DOI] [PubMed] [Google Scholar]
- 55.Deane, J.A., M.G. Kharas, J.S. Oak, L.N. Stiles, J. Luo, T.I. Moore, H. Ji, C. Rommel, L.C. Cantley, T.E. Lane, and D.A. Fruman. 2007. T cell function is partially maintained in the absence of class IA phosphoinositide 3-kinase signaling. Blood. 109:2894–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delgado, P., E. Fernandez, V. Dave, D. Kappes, and B. Alarcon. 2000. CD3delta couples T-cell receptor signalling to ERK activation and thymocyte positive selection. Nature. 406:426–430. [DOI] [PubMed] [Google Scholar]
- 57.Molina, T.J., K. Kishihara, D.P. Siderovski, W. van Ewijk, A. Narendran, E. Timms, A. Wakeham, C.J. Paige, K.U. Hartmann, A. Veillette, et al. 1992. Profound block in thymocyte development in mice lacking p56lck. Nature. 357:161–164. [DOI] [PubMed] [Google Scholar]
- 58.Hernandez-Hoyos, G., S.J. Sohn, E.V. Rothenberg, and J. Alberola-Ila. 2000. Lck activity controls CD4/CD8 T cell lineage commitment. Immunity. 12:313–322. [DOI] [PubMed] [Google Scholar]
- 59.Legname, G., B. Seddon, M. Lovatt, P. Tomlinson, N. Sarner, M. Tolaini, K. Williams, T. Norton, D. Kioussis, and R. Zamoyska. 2000. Inducible expression of a p56Lck transgene reveals a central role for Lck in the differentiation of CD4 SP thymocytes. Immunity. 12:537–546. [DOI] [PubMed] [Google Scholar]
- 60.O'Shea, C.C., T. Crompton, I.R. Rosewell, A.C. Hayday, and M.J. Owen. 1996. Raf regulates positive selection. Eur. J. Immunol. 26:2350–2355. [DOI] [PubMed] [Google Scholar]
- 61.Sun, Z., D. Unutmaz, Y.R. Zou, M.J. Sunshine, A. Pierani, S. Brenner-Morton, R.E. Mebius, and D.R. Littman. 2000. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 288:2369–2373. [DOI] [PubMed] [Google Scholar]
- 62.Guo, J., A. Hawwari, H. Li, Z. Sun, S.K. Mahanta, D.R. Littman, M.S. Krangel, and Y.W. He. 2002. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat. Immunol. 3:469–476. [DOI] [PubMed] [Google Scholar]
- 63.Park, C.G., S.Y. Lee, G. Kandala, S.Y. Lee, and Y. Choi. 1996. A novel gene product that couples TCR signaling to Fas(CD95) expression in activation-induced cell death. Immunity. 4:583–591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.