Abstract
Decline of peak viremia during acute HIV-1 infection occurs before the development of vigorous adaptive immunity, and the level of decline correlates inversely with the rate of AIDS progression, implicating a potential role for the innate immune response in determining disease outcome. The combined expression of an activating natural killer (NK) cell receptor, the killer immunoglobulin-like receptor (KIR) 3DS1, and its presumed ligand, human leukocyte antigen (HLA)–B Bw4-80I, has been associated in epidemiological studies with a slow progression to AIDS. We examined the functional ability of NK cells to differentially control HIV-1 replication in vitro based on their KIR and HLA types. NK cells expressing KIR3DS1 showed strong, significant dose- and cell contact–dependent inhibition of HIV-1 replication in target cells expressing HLA-B Bw4-80I compared with NK cells that did not express KIR3DS1. Furthermore, KIR3DS1+ NK cells and NKLs were preferentially activated, and lysed HIV-1 infected target cells in an HLA-B Bw4-80I–dependent manner. These data provide the first functional evidence that variation at the KIR locus influences the effectiveness of NK cell activity in the containment of viral replication.
Despite significant advances over the past two decades with respect to our understanding of the immune response to HIV-1 infection, the precise immune correlates of protection against HIV-1 disease progression are still largely unknown. Epidemiological studies have identified several host genetic factors that are strongly associated with better HIV-1 disease outcome. A 32–base-pair deletion in CC chemokine receptor (CCR) 5 is associated with an increased resistance to infection (1–3). Furthermore, a significant protective effect from HIV-1 disease progression has been described for several human leukocyte antigen (HLA) class I alleles, whereas an association with more rapid disease progression has been described for other HLA class I alleles (4–6). These studies suggest a critical role for HIV-1–specific CD8+ T cells in the control of viral replication and have helped guide functional studies in the area of T cell immunology in HIV-1 infection. More recently, a significant association has been described between slower HIV-1 disease progression and the expression of an activating NK cell receptor, the killer immunoglobulin-like receptor (KIR) 3DS1, in conjunction with its putative ligand, HLA-B Bw4 alleles with an isoleucine at position 80 (referred to as HLA-B Bw4-80I) (7). This protective effect was independent of the expression of the respective HLA-B Bw4-80I alleles HLA-B57 or HLA-B27 (7). These epidemiological data were the first to implicate specific NK cell receptor genes in modulating HIV-1 pathogenesis, but the functional basis of the epidemiological association between these KIR/HLA compound genotypes and control of HIV-1 replication and disease progression have not yet been identified.
NK cells are the primary effector cells of the innate immune response, as they are able to lyse virally infected cells and secrete large amounts of proinflammatory cytokines/chemokines without prior antigen sensitization (8). NK cells play a critical role in the control of viral replication in several disease models (9–11), and depletions or deficiencies of NK cells lead to significantly more severe viral disease and death in both mice and humans (12, 13). In addition to the protective effect of the KIR3DS1/HLA-B Bw4-80I compound genotype in HIV-1 infection (7), the protective effects of particular KIR/HLA genotypes have been demonstrated in several other epidemiological studies in human viral infections, including hepatitis C virus (14), human papillomavirus (15), and CMV reactivation after transplantation (16), suggesting that the interaction between KIRs and their HLA class I ligands can have a profound impact on the efficacy of NK cell–mediated control of viral pathogenesis (17).
As many as 15 different KIRs have been identified (18), and this family of glycoprotein receptors expressed predominantly on NK cells can be grouped into categories based on whether they possess two or three immunoglobulin-like domains (18). They are also characterized by long or short cytoplasmic tails that transmit inhibitory or activating signals, respectively (18). KIR3DS1 has been recently shown to be expressed on the surface of NK cells (19, 20) and to associate with Dap12 (19), a characteristic of activating NK cell receptors. Although the precise ligand for KIR3DS1 has not been identified, HLA class I molecules of the HLA-B Bw4-80I family have been proposed as putative ligands, as KIR3DS1 shares >97% sequence homology in the extracellular domains with KIR3DL1, an inhibitory KIR that interacts with several HLA-B Bw4-80I molecules (18, 21–24).
We hypothesized that the observed epidemiological association of the KIR3DS1/HLA-B Bw4-80I genotype with slower HIV-1 disease progression is caused by the ability of KIR3DS1+ NK cells to strongly inhibit HIV-1 replication in HLA-B Bw4-80I+ CD4+ T cells, and we developed an in vitro viral replication inhibition assay to test this hypothesis. In this paper, we demonstrate that NK cells are able to suppress HIV-1 replication in vitro in a dose- and cell contact–dependent manner. Furthermore, we show that NK cells expressing KIR3DS1 strongly and significantly inhibited HIV-1 replication in target cells expressing HLA-B Bw4-80I relative to NK cells that do not express KIR3DS1. These data provide the first functional evidence that variation at the KIR locus influences the effectiveness of NK cell activity in the containment of HIV-1 replication.
RESULTS
NK cells can suppress HIV-1 replication in vitro
To determine the anti-HIV-1 activity of NK cells in vitro, we developed a co-culture assay to measure the level of HIV-1 replication in CD4+ T cells in the presence or absence of autologous NK cells. In this assay, purified activated CD4+ T cells were infected with HIV-1 and coincubated with purified autologous NK cells at different effector/target (E/T) ratios over a period of 14 d in the presence of IL-2, and HIV-1 replication was quantified using a p24 Gag ELISA. The initial evaluations of this in vitro system demonstrated that freshly isolated NK cells were able to inhibit HIV-1 replication in autologous HIV-1–infected CD4+ T cells. NK cells persisted throughout the 14-d culture and acquired an activated phenotype expressing high levels of CD56, KIR, NKp46, CD161, and NKG2D (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20070695/DC1).
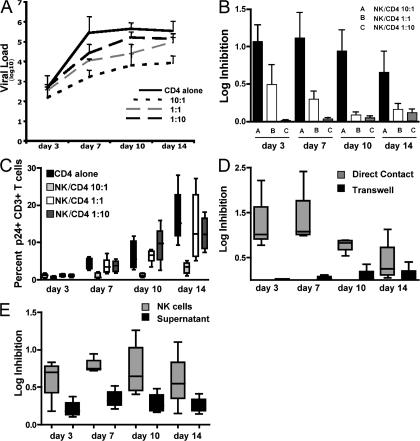
Results for the inhibition of viral replication at different E/T ratios showed high concordance when tested in three separate experiments on cells derived from the same individuals (small standard deviations), as demonstrated for a representative individual in Fig. 1 A. Furthermore, NK cell–mediated inhibition of HIV-1 replication in autologous CD4+ T cells was dependent on the number of NK cells added into CD4+ T cell cultures (Fig. 1, A and B), further suggesting that NK cells are directly responsible for the inhibition of HIV-1 replication observed in this system. The level of viral replication peaked by day 7 after infection, and at that time the strongest NK cell–mediated inhibition of HIV-1 replication was observed, expressed as the log difference in p24 Gag production between CD4+ T cells alone or in the presence of NK cells (Fig. 1, A and B). Collectively, these data demonstrate that the ability of NK cells to inhibit HIV-1 replication can be quantified in vitro and that the inhibition of viral replication depends on the NK effector cell/CD4+ T cell target ratios used.
Figure 1.
NK cells are able to mediate inhibition of HIV-1 replication in a contact-dependent manner. (A) The line graph depicts the mean change in the viral load (log10 p24 Gag in supernatant) observed in a single subject from three separate experiments in wells containing CD4+ T cells alone (continuous black line), and NK cells and autologous CD4+ T cells at a ratio of 10:1 (short dashed black line), 1:1 (long dashed gray line), and 1:10 (long dashed black line). (B) The figure represents a log reduction in p24 Gag production at different NK cell/CD4+ T cell ratios (10:1, 1:1, and 1:10) for days 3, 7, 10, and 14 (n = 5). (C) The whisker box plots show the percentage of p24 Gag+ CD3+ T cells at days 3, 7, 10, and 14 in wells containing NK cells at different NK cell/CD4+ T cell ratios (10:1, 1:1, and 1:10; n = 6). (D) The figure represents the log inhibition of HIV-1 replication observed on days 3, 7, 10, and 14 in wells where NK cells and autologous HIV-1–infected CD4+ T cells were co-cultured together (gray) or separated in a transwell experiment (black) at an NK cell/CD4+ T cell ratio of 10:1 (n = 5). (E) The whisker box plots show the log inhibition of HIV-1 replication observed on days 3, 7, 10, and 14 in wells where NK cells and autologous HIV-1–infected CD4+ T cells were co-cultured together (gray) at an NK cell/CD4+ T cell ratio of 10:1, or where the supernatant was transferred from co-cultured cells to autologous HIV-1–infected CD4+ T cells (black; n = 5). Data represent the mean of at least three experiments and standard deviations.
NK cells suppress the spread of HIV-1 infection in vitro
Although the production of p24 Gag is a strong surrogate marker for the level of viral replication in vitro, it does not allow for the determination of whether the observed suppression of p24 Gag production is caused by the continuing infection of CD4+ T cell targets but reduced virion release, or by the reduction of the number of infected CD4+ T cells. We used intracellular p24 staining on infected T cells at 3–4-d intervals to quantify the number of infected cells over time and observed a significantly lower proportion of p24+ T cells in wells containing NK cells and CD4+ T cells at an E/T ratio of 10:1 compared with wells containing HIV-1–infected CD4+ T cells alone or NK cells at lower E/T ratios (Fig. 1 C and Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20070695/DC1). Given that only NK cells (CD3−CD56+/−CD16+/−) and CD4+ T cells (CD3+CD4+) cells were present in the co-cultures, we stained cells with CD3 to distinguish these two populations and assessed the level of p24 in cells expressing CD3. Overall, p24 Gag production quantified in the supernatant by ELISA was significantly correlated to the percentage of p24+ CD3+ T cells quantified by flow cytometry (r= 0.66; P = 0.002). These results demonstrate that the observed NK cell–mediated inhibition of HIV-1 replication was caused by the containment of HIV-1 infection to a small subset of CD4+ T cells.
NK cells require contact to mediate HIV-1 inhibition
NK cells are involved in the control of viral infection both via the release of antiviral cytokines and the direct lysis of target cells (25). To determine whether NK cells require contact with infected cells to mediate viral inhibition, NK cells and HIV-1–infected CD4+ T cells were separated in transwell experiments at an E/T ratio of 10:1. No significant inhibition of HIV-1 replication was observed when NK cells and autologous infected CD4+ T cells were separated by a semipermeable membrane (Fig. 1 D), demonstrating that direct cell-to-cell contact was required for the initial triggering of NK cell activity and/or for mediating their antiviral activity.
Furthermore, to determine whether secreted factors from NK cells alone were sufficient to inhibit viral replication in vitro, supernatant collected from experiments in which NK cells and HIV-1–infected CD4+ T cells were in contact during co-culture was added on days 3, 7, 10, and 14 to wells containing only autologous HIV-1–infected CD4+ T cells. Supernatant alone mediated some viral suppression over the course of the 14-d period (Fig. 1 E), albeit at significantly lower levels than in wells where the cells were in direct contact. Collectively, these data demonstrate that direct cell-to-cell contact between NK cells and autologous HIV-1–infected CD4+ T cells is required to trigger the inhibition of HIV-1 replication in this system, and that secreted factors alone are not sufficient to mediate significant inhibition.
Subjects expressing KIR3DS1 and HLA-B Bw4-80I suppress HIV-1 replication most potently compared with individuals expressing only one or none of these alleles
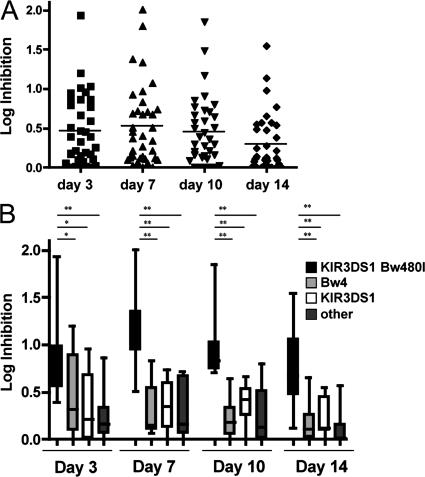
To assess the impact of the KIR/HLA compound genotype on the ability of NK cells to inhibit HIV-1 replication in vitro, NK cell–mediated inhibition of viral replication was quantified in a group of 36 HIV-1–negative study subjects expressing a diverse array of KIR/HLA genotypes. As demonstrated in Fig. 2 A, the level of NK cell–mediated inhibition of HIV-1 replication in autologous CD4+ T cells in vitro differed largely among the 36 subjects tested, ranging from 0 to 2.1 logs at day 7, when inhibition of viral replication was most pronounced.
Figure 2.
NK cells derived from subjects with KIR3DS1 and HLA-B Bw4-80I suppress viral replication more effectively in vitro. (A) The dot plot demonstrates the heterogeneity of viral suppression (given as log inhibition of p24 Gag production) observed in the total cohort of 36 study subjects at an NK cell/CD4+ T cell ratio of 10:1 (n = 36). Horizontal bars represent the mean log inhibition. (B) The whisker box plots represent the log inhibition observed in subjects with KIR3DS1 and HLA-B Bw4-80I (black; n = 8), HLA-Bw4 alone (light gray; n = 10), KIR3DS1 alone (white; n = 8), and individuals that express neither of these two alleles (dark gray; n = 10), where NK cells and CD4+ T cells were co-cultured at an E/T ratio of 10:1. Data represent the mean and SDs. **, P < 0.001; *, P < 0.05.
To test the initial hypothesis that NK cells derived from individuals with the KIR3DS1/HLA-B Bw4-80I compound genotype mediate strong antiviral activity, the study subjects were grouped according to their genotype (KIR3DS1/Bw4-80I, n = 8; KIR3DS1 alone, n = 8; Bw4 alone, n = 10; and neither gene, n = 10). As early as day 3, NK cells derived from subjects possessing both KIR3DS1 and HLA-B Bw4-80I alleles exhibited a superior level of viral inhibition compared with NK cells derived from subjects who had one of the two or neither of the alleles (Fig. 2 B); these differences reached statistical significance at days 7, 10, and 14 (P < 0.01 for comparisons of each group versus KIR3DS1+/Bw4-80I+). In contrast, the ability of HIV-1 to replicate in CD4+ T cells in the absence of NK cells did not differ across the four genotypic groups (P > 0.3 for comparisons of p24 Gag levels between each group versus KIR3DS1+/Bw4-80I+; Fig. S2 C), demonstrating that the inhibition of viral replication was caused by differences in the antiviral activity of the added NK cells and not by differences in the ability of the virus to replicate in the different CD4+ T cell populations studied. Data presented in Fig. 2 were derived from experiments using a CCR5-tropic HIV-1 strain derived from a clinical sample; however, similar suppressive activity was observed with CCR5-tropic laboratory-adapted strains and CXCR4-tropic strains (unpublished data). These data demonstrate that NK cells derived from subjects with the compound genotype KIR3DS1/HLA-B Bw4-80I exhibit strong anti–HIV-1 activity in vitro and for the first time provide functional data in support of a plausible mechanistic basis for the described epidemiological association between this genotype and slower HIV-1 disease progression.
KIR3DS1+ NK cells are mainly responsible for the suppression of HIV-1 replication
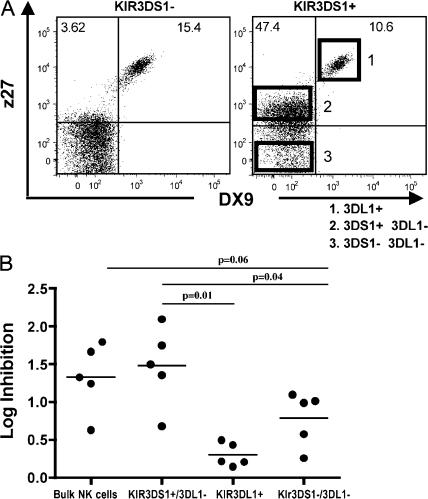
KIR molecules are expressed in a stochastic, variegated manner on NK cells within a given individual, giving rise to NK cell clones with distinct KIR expression profiles in that individual (26). This results in the presence of both KIR3DS1pos and KIR3DS1neg NK cell populations in an individual who has the allele encoding for KIR3DS1. To determine whether the KIR3DS1pos NK cell populations from subjects expressing KIR3DS1 and HLA-B Bw4-80I were responsible for the observed in vitro suppression of HIV-1 replication, we used two monoclonal antibodies—one directed against KIR3DL1 (DX9) and one directed against both KIR3DL1 and KIR3DS1 (z27) (19)—to sort the individual NK cell populations ex vivo. Although the DX9 antibody only labels KIR3DL1pos NK cells, z27 labels both KIR3DS1pos NK cells with a low mean fluorescence intensity (MFI) and KIR3DL1pos NK cells with a high MFI (19, 27, 28). The combination of these antibodies with distinct specificities allowed for the dissection of three separate NK cell populations, as previously described (19, 20, 29): (a) NK cells that express neither KIR3DS1 nor KIR3DL1 (z27negDX9neg; Fig. 3), (b) NK cells expressing KIR3DS1 alone (z27loDX9neg; Fig. 3), and (c) NK cells expressing KIR3DL1 alone or in conjunction with KIR3DS1 (z27hiDX9pos, as both of these populations are included in the medium-high MFI population [19]; Fig. 3 A).
Figure 3.
NK cells expressing KIR3DS1 inhibit HIV-1 replication more effectively and respond stronger to Bw4-80I+ HIV-1–infected target cells. (A) The flow plots demonstrate the segregation of KIR3DL1+ NK cells (1), KIR3DS1+/KIR3DL1− NK cells (2), and NK cells expressing neither of these receptors (3), using the combination of the z27 and DX9 antibodies for a subject that does not encode KIR3DS1 (left) and one that does encode KIR3DS1 (right). Numbers represent the proportion of events present within that quadrant. (B). The dot plot represents the log inhibition of HIV-1 replication (given as log inhibition of p24 Gag production) in co-cultures of autologous HIV-1–infected Bw4-80I+ CD4+ T cells with bulk NK cells, sorted populations of KIR3DS1+/3DL1− NK cells, KIR3DL1+ NK cells, and NK cells expressing neither of these markers at an NK cell/CD4+ T cell ratio of 10:1. All experiments were performed in triplicate in five different KIR3DS1+/KIR3DL1+ subjects. Horizontal bars represent the mean log inhibition.
The respective NK cell populations were individually sorted from PBMCs derived from five HLA-B Bw4-80I+ individuals expressing both KIR3DS1 and KIR3DL1 using an extreme gating strategy to guarantee the sorting of highly purified subpopulations of NK cells (Fig. 3 A), and they were used separately as effector cells in a viral replication inhibition assay with autologous HIV-1–infected CD4+ T cells at an E/T ratio of 10:1. Notably, inhibition of HIV-1 replication mediated by the KIR3DS1posKIR3DL1neg NK cell populations was significantly greater than that mediated by the KIR3DL1pos subset of NK cells (1.48 vs. 0.3 logs; P = 0.01; Fig. 3 B). The KIR3DS1pos subset also mediated greater viral inhibition than the KIR3DS1negKIR3DL1neg NK cells (1.48 vs. 0.79 logs; P = 0.04). Furthermore, the NK cell population that did not contain KIR3DS1+ NK cells (the KIR3DL1−/3DS1− population) inhibited HIV-1 replication to a lesser extent than bulk NK cells (0.8 vs. 1.33 logs; P = 0.06; Fig. 3 B). The residual antiviral activity of the KIR3DS1negKIR3DL1neg NK cells suggests that other NK cell receptors in addition to KIR3DS1 are also able to restrict HIV-1 replication to some extent, specifically in the absence of KIR3DL1. Overall, these data demonstrate that KIR3DS1pos NK cells are largely responsible for the NK cell–mediated suppression of in vitro HIV-1 replication observed in individuals expressing the KIR3DS1/HLA-B Bw4-80I genotype.
KIR3DS1+ NK cells preferentially lyse HIV-1–infected HLA-B Bw4-80I–expressing target cells
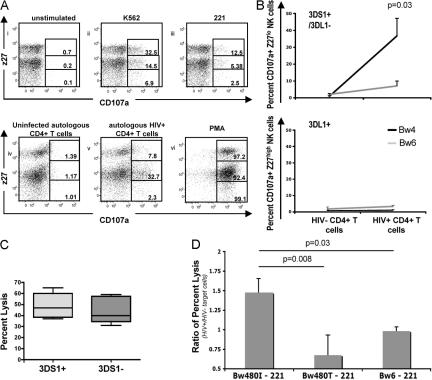
To further evaluate whether NK cells expressing KIR3DS1 in the absence of KIR3DL1 were preferentially activated by HIV-1–infected Bw4+ CD4+ T cells, we monitored KIR3DS1pos/3DL1neg and KIR3DL1pos NK cell degranulation in bulk NK cells derived from heterozygous KIR3DL1+/3DS1+ individuals (30) after 7 d of co-culture with autologous Bw4+ or Bw6+ HIV-1–infected CD4+ T cells. CD107a expression was increased fivefold in KIR3DS1pos/3DL1neg NK cells after exposure to HIV-1–infected Bw4+ CD4+ T cells but increased less than twofold after coincubation with HIV-1–infected Bw6+ CD4+ T cells (P = 0.03; Fig. 4, A and B). In contrast, very little increase in CD107a expression was observed on KIR3DL1pos NK cells after coincubation with autologous HIV-1–infected CD4+ T cells irrespective of the HLA-B Bw genotype of the individual (Fig. 4 B, bottom). KIR3DL1+ NK cells responded more potently to the MHC-devoid target cells K562 or 221 than to KIR3DS1+ NK cells, suggesting that the difference observed after stimulation with HIV-1–infected autologous target cells was specific to the KIR3DS1+ NK cell subpopulation (Fig. 4 A). To eliminate the possibility that bulk NK cell populations derived from subjects that express KIR3DS1 were just intrinsically more potent at recognizing target cells, we further measured the lytic activity of bulk NK cell populations against HLA-devoid chromium-labeled 221 cells. There was no difference in the level of killing of 221 cells among subjects that possessed or did not possess the KIR3DS1 gene (Fig. 4 C). Collectively, these data demonstrate that KIR3DS1+ NK cells can recognize HIV-1–infected HLA-Bw4+ CD4+ T cells.
Figure 4.
KIR3DS1+ NK cells are activated and lyse HIV-1–infected cells in the presence of HLA-B Bw4-80I alleles. (A) Using the combination staining of DX9 and z27, gates were set along the z27 axis to gauge the level of degranulation on z27hi (DX9+z27+ or KIR3DL1+), z27dim (DX9−z27+ or KIR3DS1+), or z27neg (DX9−z27− or double negative) NK cells after stimulation with (a) medium alone, (b) K562, (c) 221 cells, (d) uninfected autologous CD4+ T cells, (e) autologous HIV+ CD4+ T cells, and (f) PMA/ionomycin. NK cells and target cells were plated at a ratio of 10:1 for all experiments. (B) The line graphs show the percentage of CD107a+ NK cells derived from KIR3DS1+/HLA-B Bw4+ (black lines) and KIR3DS1+/HLA-B Bw6+ (gray lines) individuals after the culture of NK cell populations in the presence of autologous HIV-1–uninfected or –infected CD4+ T cells (at a ratio of 10:1). Gating strategies were used to calculate the percentage of CD107a+ NK cells within the KIR3DS1+ NK cell population (z27+DX9−; top) or within the KIR3DL1+ NK cell population (z27+DX9+; bottom). Only KIR3DS1+/3DL1− NK cells co-cultured with Bw4+ HIV-1–infected CD4+ T cells significantly (P = 0.03) up-regulated CD107a expression. (C) The whisker box plot represents the percentage of untransfected 221 cells that were lysed in a 6-h chromium assay using bulk NK cells derived from KIR3DS1+ subjects (light gray; n = 6) and KIR3DS1− individuals (dark gray; n = 6). NK cells were co-cultured with Cr51-labeled targets at a ratio of 20:1. (D) The bar graph displays the ratio of the percent lysis of HIV-1–infected versus –noninfected, HLA-transfected 221 cells (with 52% of cells staining for p24 by intracellular flow at the maximal time of infection) by a KIR3DS1-transfected NKL (NKL/221 target cell, 20:1; n = 3). Data represent the mean of at least three experiments and standard deviations.
To further evaluate the ability of KIR3DS1+ cells to recognize HIV-1–infected target cells in the context of HLA-B Bw4-80I, we infected 221 cells that were either untransfected or transfected with individual HLA class I alleles with a pseudotyped R5 virus (JRCSF). HIV-1–infected and –uninfected 221 cells were chromium labeled and placed in co-culture with the NK tumor cell line NKL expressing either KIR3DS1, KIR3DL1, or neither of the receptors for 6 h (19, 21). The level of virus-associated target cell lysis by the effector NK cells was calculated as a ratio of the percent lysis of HIV-1–infected versus –uninfected HLA-transfected 221 cells. Target cell lysis, irrespective of the expressed HLA class I allotype, did not significantly differ between infected and uninfected 221 cells when exposed to KIR3DL1+ or KIR3DL1neg/KIR3DS1neg NKLs (mean ratios of lysis of infected/uninfected cells were 1.01, 0.82, and 0.98 for KIR3DL1+ NKLs, and 0.93, 0.88, and 1.01 for KIR3DL1neg/KIR3DS1neg NKLs with respect to the lysis of HLA-B Bw4-80I–, Bw4-80T–, and Bw6-transfected 221 cells, respectively). Furthermore, there was no increase in the level of killing by KIR3DS1+ NKLs of HIV-1–infected 221 cells transfected with either Bw4-80T or Bw6 compared with uninfected 221 cells (Fig. 4 D). In contrast, KIR3DS1+ NKLs lysed HIV-1–infected 221 cells expressing HLA-B Bw4-80I significantly more efficiently than uninfected Bw4-80I+ 221 cells (Fig. 4 D and Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20070695). Collectively, these data using both primary NK cells and KIR-transfected NKLs demonstrate that KIR3DS1+ NK cells can recognize and lyse HIV-1–infected target cells in the context of HLA-B Bw4-80I.
DISCUSSION
NK cells play a crucial role in the control of several human viral infections, but their functional role in HIV-1 pathogenesis is not known. In this paper, we demonstrate that NK cells expressing KIR3DS1, which has recently been shown to be expressed on the surface of NK cells (20, 29) and to confer activating signals to NK cells (19), are strongly activated by autologous HIV-1–infected Bw4-80I+ CD4+ T cells and can significantly inhibit HIV-1 replication in these cells in vitro compared with NK cells expressing the inhibitory receptor KIR3DL1, or neither of these receptors. These data provide the first functional correlation and a plausible mechanistic basis for the epidemiological studies that described a protective effect of the KIR3DS1/HLA-B Bw4-80I compound genotype on HIV-1 disease progression (7), implicating NK cells directly in the control of HIV-1 pathogenesis.
The expression of specific KIRs in conjunction with their HLA ligands has been shown to have an impact on disease outcome in several epidemiological studies in different human viral infections, including protection against HIV (7, 31), hepatitis C virus (14), CMV reactivation (16), and, more recently, protection against seroconversion in a group of female sex workers that were repeatedly exposed to HIV but remained seronegative (32). However, it is still unclear from these epidemiological studies what mechanism accounts for NK cell–mediated control of viral replication. In this paper, we demonstrate that NK cells are able to suppress HIV-1 viral replication in vitro. Furthermore, we show that KIR3DS1+ NK cells exert their antiviral activity preferentially in the presence of HLA-B Bw4-80I on HIV-1–infected target cells. Collectively, these data provide the first functional evidence that NK cells can contribute directly to the control of HIV-1 replication in a receptor–ligand–specific manner.
KIRs are stochastically expressed on NK cells. Thus, in a given KIR3DS1+/KIR3DL1+ individual, a subset of NK cell clones expresses KIR3DS1 alone, a small subset may express KIR3DS1 in combination with KIR3DL1 (20), and others do not express KIR3DS1 at all. To address whether KIR3DS1+ NK cells preferentially mediated the inhibitory effect on HIV-1 replication observed for subjects that encode this receptor, we used a gating strategy based on the combination of two KIR3DS1/KIR3DL1-specific antibodies (DX9 and z27)(19, 20, 27–29) to sort populations of NK cells that expressedKIR3DS1 and not KIR3DL1 (DX9− and z27lo), KIR3DL1 (DX9+ and z27hi), or neither of these two KIRs (DX9− and z27−). Thus, using these individual NK cell populations derived from the same individual, we demonstrate that KIR3DS1+/KIR3DL1− NK cells were significantly more potent in mediating inhibition of HIV-1 replication relative to the other NK cell subpopulations. Furthermore, the NK cell population that did not include KIR3DS1+ NK cells (KIR3DL1−/3DS1−) inhibited HIV-1 replication to a lesser extent than bulk NK cells. The observation that KIR3DS1−/KIR3DL1− NK cells were also able to mediate some viral inhibition (albeit at lower levels than KIR3DS1+ NK cells) was in line with previous studies demonstrating that additional NK cell receptors, including natural cytotoxicity receptors, may also be involved in the recognition of HIV-1–infected target cells (33, 34).
In contrast, little inhibition of HIV-1 replication was mediated by NK cell populations expressing the inhibitory receptor KIR3DL1 (DX9+ and z27hi) despite the fact that these DX9/z27 double-positive NK cells may also include a small fraction of NK cells that express both KIR3DL1 and KIR3DS1 (20). Recent data demonstrate that particular KIR3DL1 haplotypes are in fact associated with slower HIV-1 disease progression (31), suggesting that specific KIR3DL1 subtypes may also be critically involved in the recognition of HIV-1–infected target cells, perhaps by monitoring for changes in the downmodulation of MHC class I alleles by HIV Nef. Further studies are required to determine the underlying mechanism of the protective effect of high expressing KIR3DL1 with HLA-B Bw4 80I alleles in HIV-1 disease progression.
In line with the better in vitro inhibition of viral replication, KIR3DS1+ NK cells were preferentially activated by HIV-1–infected Bw4-80I+ target cells, because NK cells expressing KIR3DS1 (DX9negz27lo) specifically degranulated in the presence of HIV-1–infected autologous CD4+ T cells. Furthermore, only KIR3DS1-transfected NKLs and not KIR3DL1 or untransfected NKLs preferentially lysed HIV-1–infected Bw4-80I+ cells, but not uninfected Bw4-80I+ cells. How might KIR3DS1+ NK cells recognize HIV-1–infected CD4+ T cells? The activating NK cell receptor KIR3DS1 and the inhibitory receptor KIR3DL1 segregate as alleles of the same gene. KIR3DL1 binds to HLA-B molecules with the Bw4 motif and preferentially to the Bw4-80I subset of alleles (21, 22). Allele-specific, as well as peptide-dependent, binding between HLA class I molecules from the Bw4-80I family and KIR3DL1 have been recently described in the context of several different HLA class I epitopes derived from HIV-1 (22, 35). KIR3DS1 and KIR3DL1 share ∼97% amino-acid similarity in their extracellular domains (7) and may, therefore, also share a similar set of ligands, as described for other families of activating and inhibitory KIRs (18, 35–37). The observation that KIR3DS1+ NK cell populations, as well as KIR3DS1-transfected NKLs, responded to HLA-B Bw4-80I+ HIV-infected target cells raises the possibility that specific modifications of the HLA-B Bw4 80I–peptide complex occur during HIV-1 infection, leading to KIR3DS1 recognition/activation. Several reports have demonstrated that amino-acid changes at positions 7 and 8 within an epitope can have a profound effect on inhibitory KIR recognition and, thus, NK cell activation (38–40). Along the same line, it is possible that activating KIR signaling may also be influenced by specific peptides within the HLA-binding groove. Thus, it is plausible that viral or stress peptides produced during HIV-1 infection may be recognized by KIR3DS1 in the context of HLA-B Bw4-80I molecules, either directly or in conjunction with other receptors (41).
Although individuals with the KIR3DS1/HLA-B Bw4 80I haplotype suppressed HIV-1 replication more effectively in vitro than subjects that expressed only one or neither of these two alleles, there was considerable heterogeneity in NK cell–mediated viral inhibition among these individuals. One potential explanation for this heterogeneity is that the quality of the KIR3DS1+ NK cell–mediated inhibition of HIV-1 might be significantly modulated by the presence of additional receptors expressed on these NK cells (33, 34). Our data show that KIR3DS1+/3DL1neg express and transcribe additional inhibitory KIR and NKG2A (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20070695). The expression of these additional self-recognizing NK cell receptors, as well as natural cytotoxicity receptors, is necessary for the efficient education/licensing of these NK cells (42, 43), as well as their cross talk with DCs (44), both of which ultimately modulate their antiviral activity. Larger studies are required to investigate the influence of these additional NK cell receptors on the quality of KIR3DS1-mediated protection from HIV-1 disease.
To date, immunological studies in HIV-1 infection have largely focused on virus-specific T cell and antibody responses. Our functional data suggest that, in addition to these adaptive immune responses, NK cells can contribute to the antiviral immunity against HIV-1, as suggested by recent epidemiological studies (7, 31). NK cells have been shown to play a critical role in the initial containment of viral replication in several viral disease models (25, 45). In acute HIV-1 infection, NK cells are significantly expanded before the development of vigorous adaptive immunity (46), indicating a potential role for the innate immune response in the initial control of viral replication. Studies aimed at investigating the role of CD8+ T cells in the control of SIV/HIV infection demonstrated a lack of control over viral replication in rhesus macaques depleted of CD8+ cells, and the results from these studies were interpreted as demonstrating that CD8+ T cells are critical for control over SIV replication (47, 48). However, the antibody used for these studies, CM-T807 (47, 48), depleted all cells expressing CD8 on their surface, as acknowledged by the authors. In monkeys, >80% of NK cells express CD8, resulting in their depletion in this experimental system. Thus, it is possible that not only the depletion of CD8+ T cells alone but also of NK cells might have been responsible for or contributed to the loss of control of HIV-1 replication observed in these studies.
In conclusion, a better understanding of the factors that contribute to the control of HIV-1 replication and the speed of HIV-1 disease progression is needed to guide the design of effective immunotherapeutic interventions or vaccines. In this paper, we show compelling evidence that NK cells can limit HIV-1 replication in vitro and provide an initial functional correlate for the protective effect of the KIR3DS1/HLA-B Bw4-80I compound genotype from HIV-1 disease progression described in epidemiological studies (7). These data suggest a more important role of effector cells of the innate immune system in the control of HIV-1 replication than traditionally thought, which will help to guide the development of antiviral strategies directly targeting innate immunity.
MATERIALS AND METHODS
Subjects.
36 HIV-1–negative subjects were recruited for this study. The study was approved by the local Institutional Review Boards of the participating institutions, and all individuals gave informed consent for participation in this study.
HLA and KIR typing.
Genomic DNA was extracted from whole PBMCs and was typed for both KIR and HLA class I alleles. In brief, HLA typing was performed after amplification of genomic DNA using locus-specific primers that flanked exons 2 and 3. The PCR products were blotted on nylon membranes and hybridized with sequence-specific oligonucleotide (thianthrene 5-oxide [SSO]) probes. We assigned alleles by the reaction patterns of the SSO probes. Ambiguous SSO probe typing results were resolved by sequencing analysis, as previously described (5). For KIR typing, genomic DNA was typed for the presence or absence of KIR3DL1 and KIR3DS1 by PCR with sequence-specific priming. PCR amplification was performed with two pairs of specific primers for each locus. Internal control primers for a fragment of 796 bp of the third intron of DRB1 were also included in each PCR. We amplified 20−50 ng DNA in a volume of 20 liters containing 200 M dNTP, 100−500 nM of specific primer, 100 nM of internal control primer, 2 mM MgCl2, 67mM Tris, 16.6 mM (NH4)2SO4, and 0.5 U of Taq polymerase. Cycling was performed in a thermal cycler (GeneAmp PCR System 9700; Applied Biosystems) as follows: 1 min at 96°C; 5 cycles of 96°C for 25 s, 65°C for 45 s, and 72°C for 30 s; 21 cycles of 96°C for 25 s, 60°C for 45 s, and 72°C for 30 s; 5 cycles of 96°C for 25 s, 55°C for 1 min, and 72°C for 2 min; and a final extension step of 10 min at 72°C. PCR products were separated in a 1.5% agarose gels containing ethidium bromide and visualized under ultraviolet light, as previously described (7).
Cell purification.
PBMCs were stimulated for 4 d with a bispecific CD3/CD8 antibody in RPMI 1640 (Sigma-Aldrich) containing 10% FBS (Atlanta Biologicals), 2 mM l-glutamine (Cellgro), and 50 IU/ml penicillin (Cellgro), supplemented with 100 U/ml IL-2. At day 4, the purity of the expanded CD4+ T cell population was verified by flow cytometry (mean of 92% CD4+ T cells, with no contaminating CD8+ T cells). Autologous NK cells were purified using the NK cell enrichment RosetteSep (StemCell Technologies Inc.) with a mean purity of 95.3%, again with <0.01% CD8+ T cells.
Inhibition of HIV-1 replication assay.
CD4+ T cells that were generated after 4 d in culture with the bispecific antibody to CD3/CD8 were infected with a clinical or lab strain R5 (JRCSF) and/or X4 (IIIB) virus at a multiplicity of infection of 0.01 for 4 h at 37°C. Cells were washed two times. Equal numbers of CD4+ T cells were plated at NK cell/CD4+ T cell ratios of 10:1, 1:1, and 1:10, or alone for 14 d in the presence of 50 U/ml IL-2. Supernatant was collected every 3–4 d for quantification of p24 Gag production by ELISA (p24 ELISA; Perkin Elmer), and in a subset of experiments, 100 μl of cells was collected for intracellular p24 Gag staining (KC57-RD1; Beckman Coulter) using CD3-allophycocyanin (BD Biosciences) to identify the proportion of infected CD4+CD3+ T cells in co-cultures containing CD3+CD4+ T cells and CD3−CD56+ NK cells (Fig. S2 A). This strategy was chosen for all experiments shown, as previous studies have shown that CD4 expression on HIV-1–infected CD4+ T cells is reduced. To determine whether contact was required for NK cell–mediated inhibition of HIV-1 replication, autologous NK cells were physically separated from infected CD4+ T cells in a transwell experiment, and medium was collected every 3–4 d for p24 Gag quantification by ELISA, as well as for the supernatant add-back experiments.
For the supernatant add-back experiments, 1 ml of supernatant was transferred from wells containing NK cells co-cultured with autologous HIV-1–infected CD4+ T cells (at a 10:1 NK cell/CD4+ T cell ratio) to wells containing the same HIV-1–infected CD4+ T cells in the absence of NK cells on days 3, 7, 10, and 14. Supernatant was collected on days 3, 7, 10, and 14 for both the wells containing CD4+ T cells and those containing NK cells using the first day of co-culture as a baseline, as well as wells containing CD4+ T cells alone using the first day of addition of supernatant as a baseline. Levels of p24 Gag in the supernatant were quantified by ELISA.
Assessment of NK cell activation by flow cytometry.
To examine which subsets of NK cells were preferentially activated in the presence of HIV-1–infected autologous CD4+ T cells, we monitored the level of CD107a up-regulation on KIR3DS1+ or KIR3DL1+ NK cells. Using the combination of DX9 and z27 staining, gates were set along the z27 axis to separate z27hi (KIR3DL1+), z27lo (KIR3DS1+), or z27neg (KIR3DL1−/KIR3DS1−) NK cells. The level of degranulation was assessed as the proportion of CD107a+ NK cells belonging to each individual subset. Purified NK cells were co-cultured in the presence of autolgous HIV-1–uninfected or –infected CD4+ T cells for 7 d. Monensin was added to co-cultures on day 7 at 0.3 μg/ml in the presence of 20 μl CD107a–PE-Cy5 for 6 h. Cells were washed and stained with CD3-allophycocyanin, CD56-FITC (BD Biosciences), and z27-PE (Beckman Coulter) for 30 min, washed, and fixed in 1% paraformaldehyde until flow cytometric analysis was performed (FACSCalibur; BD Biosciences).
Cell lysis assay.
The lytic activity of NKLs against 221 target cells was assessed by a standard chromium release assay, as previously described (49). Thus, 2 × 106 221 cells were labeled with 50 μCi Na2(51CrO4) (1 Ci = 37 GBq; New England Nuclear) for 1 h at 37°C, 5% CO2. Target cells were washed three times and plated with effector NKLs at an E/T ratio of 20:1. Supernatant was harvested after a 6-h incubation at 37°C, 5% CO2. The percent lysis was calculated as follows: ([sample count − spontaneous release]/[maximal release − spontaneous release]) × 100.
Statistical analysis.
All experiments represent the mean of at least three experiments and standard deviations. To test for differences in the mean between several populations, a one-way analysis of variance with a Tukey's correction was used for all comparisons that were below P < 0.05. P < 0.05 was considered significant.
Online supplemental material.
Fig. S1 shows data on NK cell numbers and activation markers over the 14-d co-culture period. Fig. S2 depicts the gating strategy used for the identification of p24+ T cells (A), the loss of p24+ T cells in wells containing autologous NK cells (B), as well as the overall p24 levels in infected T cells among HLA-KIR compound genotypes (C). Fig. S3 demonstrates the raw data for the NKL lysis assay presented in Fig. 4 D. Fig. S4 shows the protein (A) and mRNA (B) expression of additional inhibitory NK cell receptors in bulk NK cells and sorted subpopulations. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20070695/DC1).
Supplemental Material
Acknowledgments
We thank Dan McVicar, Eriko Yamada, and Douglas Schneider for their helpful discussions and technical support.
This work was supported by National Institutes of Health (NIH) grant R01-A1067031. This project has been funded in part with federal funds from the National Cancer Institute (NCI), NIH, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the United States Government. This research was supported in part by the Intramural Research Support Program of the Center for Cancer Research, NCI, NIH.
The authors have no conflicting financial interests.
Abbreviations used: CCR, CC chemokine receptor; E/T, effector/target; HLA, human leukocyte antigen; KIR, killer immunoglobulin-like receptor; MFI, mean fluorescence intensity; SSO, thianthrene 5-oxide.
References
- 1.Liu, R., W.A. Paxton, S. Choe, D. Ceradini, S.R. Martin, R. Horuk, M.E. MacDonald, H. Stuhlmann, R.A. Koup, and N.R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 86:367–377. [DOI] [PubMed] [Google Scholar]
- 2.Martin, M.P., M. Dean, M.W. Smith, C. Winkler, B. Gerrard, N.L. Michael, B. Lee, R.W. Doms, J. Margolick, S. Buchbinder, et al. 1998. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 282:1907–1911. [DOI] [PubMed] [Google Scholar]
- 3.Smith, M.W., M. Dean, M. Carrington, C. Winkler, G.A. Huttley, D.A. Lomb, J.J. Goedert, T.R. O'Brien, L.P. Jacobson, R. Kaslow, et al. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 277:959–965. [DOI] [PubMed] [Google Scholar]
- 4.Carrington, M., and S.J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535–551. [DOI] [PubMed] [Google Scholar]
- 5.Gao, X., G.W. Nelson, P. Karacki, M.P. Martin, J. Phair, R. Kaslow, J.J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, et al. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668–1675. [DOI] [PubMed] [Google Scholar]
- 6.Kaslow, R.A., M. Carrington, R. Apple, L. Park, A. Munoz, A.J. Saah, J.J. Goedert, C. Winkler, S.J. O'Brien, C. Rinaldo, et al. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405–411. [DOI] [PubMed] [Google Scholar]
- 7.Martin, M.P., X. Gao, J.H. Lee, G.W. Nelson, R. Detels, J.J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, J. Trowsdale, et al. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429–434. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, M.A., T.A. Fehniger, and M.A. Caligiuri. 2001. The biology of human natural killer-cell subsets. Trends Immunol. 22:633–640. [DOI] [PubMed] [Google Scholar]
- 9.Bukowski, J.F., and R.M. Welsh. 1986. The role of natural killer cells and interferon in resistance to acute infection of mice with herpes simplex virus type 1. J. Immunol. 136:3481–3485. [PubMed] [Google Scholar]
- 10.Reading, P.C., P.G. Whitney, D.P. Barr, M.J. Smyth, and A.G. Brooks. 2006. NK cells contribute to the early clearance of HSV-1 from the lung but cannot control replication in the central nervous system following intranasal infection. Eur. J. Immunol. 36:897–905. [DOI] [PubMed] [Google Scholar]
- 11.Daniels, K.A., G. Devora, W.C. Lai, C.L. O'Donnell, M. Bennett, and R.M. Welsh. 2001. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med. 194:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biron, C.A., G. Sonnenfeld, and R.M. Welsh. 1984. Interferon induces natural killer cell blastogenesis in vivo. J. Leukoc. Biol. 35:31–37. [DOI] [PubMed] [Google Scholar]
- 13.Biron, C.A., K.S. Byron, and J.L. Sullivan. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 320:1731–1735. [DOI] [PubMed] [Google Scholar]
- 14.Khakoo, S.I., C.L. Thio, M.P. Martin, C.R. Brooks, X. Gao, J. Astemborski, J. Cheng, J.J. Goedert, D. Vlahov, M. Hilgartner, et al. 2004. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 305:872–874. [DOI] [PubMed] [Google Scholar]
- 15.Carrington, M., S. Wang, M.P. Martin, X. Gao, M. Schiffman, J. Cheng, R. Herrero, A.C. Rodriguez, R. Kurman, R. Mortel, et al. 2005. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J. Exp. Med. 201:1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, C., M. Busson, V. Rocha, M.L. Appert, V. Lepage, N. Dulphy, P. Haas, G. Socie, A. Toubert, D. Charron, and P. Loiseau. 2006. Activating KIR genes are associated with CMV reactivation and survival after non-T-cell depleted HLA-identical sibling bone marrow transplantation for malignant disorders. Bone Marrow Transplant. 38:437–444. [DOI] [PubMed] [Google Scholar]
- 17.Bashirova, A.A., M.P. Martin, D.W. McVicar, and M. Carrington. 2006. The Killer Immunoglobulin-Like Receptor Gene Cluster: Tuning the Genome for Defense (*). Annu. Rev. Genomics Hum. Genet. 7:277–300. [DOI] [PubMed] [Google Scholar]
- 18.Carrington, M., and P. Norman. 2003. The KIR Gene Cluster. NCBI, Bethesda, MD. 48 pp.
- 19.Carr, W.H., D.B. Rosen, H. Arase, D.F. Nixon, J. Michaelsson, and L.L. Lanier. 2007. Cutting edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J. Immunol. 178:647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trundley, A., H. Frebel, D. Jones, C. Chang, and J. Trowsdale. 2007. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur. J. Immunol. 37:780–787. [DOI] [PubMed] [Google Scholar]
- 21.Carr, W.H., M.J. Pando, and P. Parham. 2005. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 175:5222–5229. [DOI] [PubMed] [Google Scholar]
- 22.Thananchai, H., G. Gillespie, M.P. Martin, A. Bashirova, N. Yawata, M. Yawata, P. Easterbrook, D.W. McVicar, K. Maenaka, P. Parham, et al. 2007. Cutting edge: allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J. Immunol. 178:33–37. [DOI] [PubMed] [Google Scholar]
- 23.Gumperz, J.E., L.D. Barber, N.M. Valiante, L. Percival, J.H. Phillips, L.L. Lanier, and P. Parham. 1997. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J. Immunol. 158:5237–5241. [PubMed] [Google Scholar]
- 24.Cella, M., A. Longo, G.B. Ferrara, J.L. Strominger, and M. Colonna. 1994. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 180:1235–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welsh, R.M., C.A. Biron, J.F. Bukowski, K.W. McIntyre, and H. Yang. 1984. Role of natural killer cells in virus infections of mice. Surv. Synth. Pathol. Res. 3:409–431. [DOI] [PubMed] [Google Scholar]
- 26.Moretta, A., C. Bottino, M.C. Mingari, R. Biassoni, and L. Moretta. 2002. What is a natural killer cell? Nat. Immunol. 3:6–8. [DOI] [PubMed] [Google Scholar]
- 27.Vitale, M., S. Sivori, D. Pende, R. Augugliaro, C. Di Donato, A. Amoroso, M. Malnati, C. Bottino, L. Moretta, and A. Moretta. 1996. Physical and functional independency of p70 and p58 natural killer (NK) cell receptors for HLA class I: their role in the definition of different groups of alloreactive NK cell clones. Proc. Natl. Acad. Sci. USA. 93:1453–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zambello, R., M. Falco, M. Della Chiesa, L. Trentin, D. Carollo, R. Castriconi, G. Cannas, S. Carlomagno, A. Cabrelle, T. Lamy, et al. 2003. Expression and function of KIR and natural cytotoxicity receptors in NK-type lymphoproliferative diseases of granular lymphocytes. Blood. 102:1797–1805. [DOI] [PubMed] [Google Scholar]
- 29.O'Connor, G.M., K.J. Guinan, R.T. Cunningham, D. Middleton, P. Parham, and C.M. Gardiner. 2007. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J. Immunol. 178:235–241. [DOI] [PubMed] [Google Scholar]
- 30.Alter, G., J.M. Malenfant, and M. Altfeld. 2004. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods. 294:15–22. [DOI] [PubMed] [Google Scholar]
- 31.Martin, M.P., Y. Qi, X. Gao, E. Yamada, J.N. Martin, F. Pereyra, S. Colombo, E.E. Brown, W.L. Shupert, J. Phair, et al. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 39:733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jennes, W., S. Verheyden, C. Demanet, C.A. Adje-Toure, B. Vuylsteke, J.N. Nkengasong, and L. Kestens. 2006. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J. Immunol. 177:6588–6592. [DOI] [PubMed] [Google Scholar]
- 33.Vieillard, V., J.L. Strominger, and P. Debre. 2005. NK cytotoxicity against CD4+ T cells during HIV-1 infection: a gp41 peptide induces the expression of an NKp44 ligand. Proc. Natl. Acad. Sci. USA. 102:10981–10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward, J., M. Bonaparte, J. Sacks, J. Guterman, M. Fogli, D. Mavilio, and E. Barker. 2007. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood. 110:1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stewart-Jones, G.B., K. di Gleria, S. Kollnberger, A.J. McMichael, E.Y. Jones, and P. Bowness. 2005. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur. J. Immunol. 35:341–351. [DOI] [PubMed] [Google Scholar]
- 36.Biassoni, R., C. Cantoni, M. Falco, S. Verdiani, C. Bottino, M. Vitale, R. Conte, A. Poggi, A. Moretta, and L. Moretta. 1996. The human leukocyte antigen (HLA)–C–specific “activatory” or “inhibitory” natural killer cell receptors display highly homologous extracellular domains but differ in their transmembrane and intracytoplasmic portions. J. Exp. Med. 183:645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart, C.A., F. Laugier-Anfossi, F. Vely, X. Saulquin, J. Riedmuller, A. Tisserant, L. Gauthier, F. Romagne, G. Ferracci, F.A. Arosa, et al. 2005. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. USA. 102:13224–13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malnati, M.S., M. Peruzzi, K.C. Parker, W.E. Biddison, E. Ciccone, A. Moretta, and E.O. Long. 1995. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 267:1016–1018. [DOI] [PubMed] [Google Scholar]
- 39.Peruzzi, M., K.C. Parker, E.O. Long, and M.S. Malnati. 1996. Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J. Immunol. 157:3350–3356. [PubMed] [Google Scholar]
- 40.Peruzzi, M., N. Wagtmann, and E.O. Long. 1996. A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J. Exp. Med. 184:1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altfeld, M., and P. Goulder. 2007. “Unleashed” natural killers hinder HIV. Nat. Genet. 39:708–710. [DOI] [PubMed] [Google Scholar]
- 42.Kim, S., J. Poursine-Laurent, S.M. Truscott, L. Lybarger, Y.J. Song, L. Yang, A.R. French, J.B. Sunwoo, S. Lemieux, T.H. Hansen, and W.M. Yokoyama. 2005. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 436:709–713. [DOI] [PubMed] [Google Scholar]
- 43.Anfossi, N., P. Andre, S. Guia, C.S. Falk, S. Roetynck, C.A. Stewart, V. Breso, C. Frassati, D. Reviron, D. Middleton, et al. 2006. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 25:331–342. [DOI] [PubMed] [Google Scholar]
- 44.Mavilio, D., G. Lombardo, A. Kinter, M. Fogli, A. La Sala, S. Ortolano, A. Farschi, D. Follman, R. Gregg, C. Kovacs, et al. 2006. Characterization of the defective interaction between a subset of natural killer cells and dendritic cells in HIV-1 infection. J. Exp. Med. 203:2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lodoen, M.B., and L.L. Lanier. 2006. Natural killer cells as an initial defense against pathogens. Curr. Opin. Immunol. 18:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alter, G., N. Teigen, R. Ahern, H. Streeck, A. Meier, E.S. Rosenberg, and M. Altfeld. 2007. Evolution of innate and adaptive effector cell functions during acute HIV-1 infection. J. Infect. Dis. 195:1452–1460. [DOI] [PubMed] [Google Scholar]
- 47.Schmitz, J.E., M.J. Kuroda, S. Santra, V.G. Sasseville, M.A. Simon, M.A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B.J. Scallon, et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 283:857–860. [DOI] [PubMed] [Google Scholar]
- 48.Jin, X., D.E. Bauer, S.E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C.E. Irwin, J.T. Safrit, J. Mittler, L. Weinberger, et al. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus–infected macaques. J. Exp. Med. 189:991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, O.O., and B.D. Walker. 1997. CD8+ cells in human immunodeficiency virus type I pathogenesis: cytolytic and noncytolytic inhibition of viral replication. Adv. Immunol. 66:273–311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.