Abstract
DNA methylation is an epigenetic modification essential for development. The DNA methyltransferases Dnmt3a and Dnmt3b execute de novo DNA methylation in gastrulating embryos and differentiating germline cells. It has been assumed that these enzymes generally play a role in regulating cell differentiation. To test this hypothesis, we examined the role of Dnmt3a and Dnmt3b in adult stem cells. CD34−/low, c-Kit+, Sca-1+, lineage marker− (CD34− KSL) cells, a fraction of mouse bone marrow cells highly enriched in hematopoietic stem cells (HSCs), expressed both Dnmt3a and Dnmt3b. Using retroviral Cre gene transduction, we conditionally disrupted Dnmt3a, Dnmt3b, or both Dnmt3a and Dnmt3b (Dnmt3a/Dnmt3b) in CD34− KSL cells purified from mice in which the functional domains of these genes are flanked by two loxP sites. We found that Dnmt3a and Dnmt3b function as de novo DNA methyltransferases during differentiation of hematopoietic cells. Unexpectedly, in vitro colony assays and in vivo transplantation assays showed that both myeloid and lymphoid lineage differentiation potentials were maintained in Dnmt3a-, Dnmt3b-, and Dnmt3a/Dnmt3b-deficient HSCs. However, Dnmt3a/Dnmt3b-deficient HSCs, but not Dnmt3a- or Dnmt3b-deficient HSCs, were incapable of long-term reconstitution in transplantation assays. These findings establish a critical role for DNA methylation by Dnmt3a and Dnmt3b in HSC self-renewal.
DNA methylation is a major epigenetic modification of the genome that regulates genomic function during cell differentiation (1–4). DNA methylation patterns are established during early embryogenesis and gametogenesis, and are maintained in somatic cells thereafter (5). The DNA methyltransferases Dnmt1, Dnmt3a, and Dnmt3b catalyze methylation of CpG dinucleotides in genomic DNA (6–8). Dnmt3a and Dnmt3b supposedly act as de novo methyltransferases, whereas Dnmt1 acts as a “maintenance” methyltransferase (9).
Embryogenesis is severely impaired in Dnmt1- or Dnmt3b-deficient mice (7, 10). Spermatogenesis is impaired in Dnmt3a-deficient mice (8). Embryonic stem (ES) cells without Dnmt3a and Dnmt3b are viable and maintain replication potential but progressively lose differentiation potential with repeated passage (11). These data imply that when stem cells undergo differentiation, Dnmt3a and Dnmt3b progressively establish DNA methylation status in each cell lineage, and then restrict cells' differentiation potential. We decided to examine the function of Dnmt3a and Dnmt3b in hematopoietic stem cells (HSCs) because HSCs are the best-studied stem cells and an excellent model system for studies of cell differentiation.
Because Dnmt3a-deficient mice die by 4 wk of age, and Dnmt3b-deficient mice die before birth, we conditionally disrupted Dnmt3a, Dnmt3b, or both Dnmt3a and Dnmt3b in adult mouse HSCs. We show that Dnmt3a and Dnmt3b establish DNA methylation status as a de novo DNA methyltransferase during the maturation of hematopoietic cells, and HSCs, unlike ES cells, require either Dnmt3a or Dnmt3b for self-renewal, but not for differentiation into all blood cell lineages. This work provides insights into how DNA methylation controls normal stem cell functions.
RESULTS AND DISCUSSION
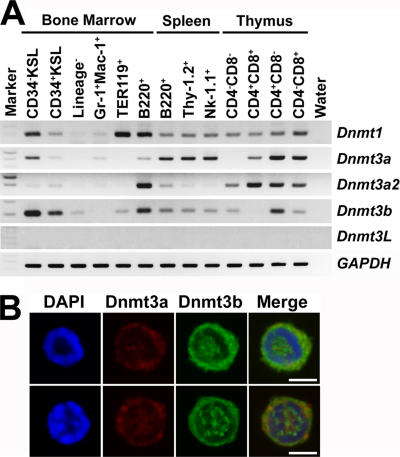
We first examined expression of Dnmt1, Dnmt3a, Dnmt3a2 (an isoform of Dnmt3a, predominantly expressed in undifferentiated ES cells; reference 12), Dnmt3b, and Dnmt3L (13) in various hematopoietic cell populations from C57BL/6 (B6) mice by RT-PCR analysis. As shown in Fig. 1 A, Dnmt1 was detected in all blood lineages. Dnmt3L, which encodes a protein lacking enzymatic activity but required for establishment of maternal methylation imprints (8, 13, 14), was undetectable in all blood lineages. Dnmt3a, Dnmt3a2, and Dnmt3b were expressed mainly in B and T lymphoid lineages. Interestingly, expression of both Dnmt3a and Dnmt3b was readily detectable in CD34−/low, c-Kit+, Sca-1+, lineage marker− (CD34− KSL) cells, a population exclusively enriched in HSCs (15). Immunocytostaining with anti-Dnmt3a or -Dnmt3b antibodies also revealed expression of these proteins in CD34− KSL cells (Fig. 1 B). Both Dnmt3a and Dnmt3b were detected in 55.9% of the cells, and either Dnmt3a or Dnmt3b was detected in, respectively, 3.4 and 40.7% of the cells. We extended this immunostaining analysis to various cells in different hematopoietic lineages. Consistent with RT-PCR data, expression of both Dnmt3a and Dnmt3b was strong in B and T lymphoid lineages, but barely detectable in neutrophil/macrophage and erythroid lineages (not depicted). These and previous data (11) predicted that these enzymes are required for functions of HSCs and lymphoid cells.
Figure 1.
Expression of Dnmt3a and Dnmt3b in hematopoietic cells. (A) Expression of Dnmt1, Dnmt3a, Dnmt3a2, Dnmt3b, Dnmt3L, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was examined by semiquantitative RT-PCR analysis. cDNAs were prepared from CD34− KSL cells, CD34+ KSL cells, lineage marker− cells, Gr-1+ Mac-1+ neutrophils/macrophages, TER119+ erythroblasts, and B220+ B lymphoid cells in the BM; from B220+ B lymphoid cells, Thy-1.2+ T lymphoid cells, and Nk-1.1+ NK/NKT cells in the spleen; and from CD4−CD8−, CD4+CD8+, CD4+CD8−, and CD4−CD8+ T lymphoid cells in the thymus of adult B6 mice. (B) Immunostaining detected Dnmt3a (red) and Dnmt3b (green) in the cytoplasm and nucleus (blue) of CD34− KSL cells. Bars, 4 μm.
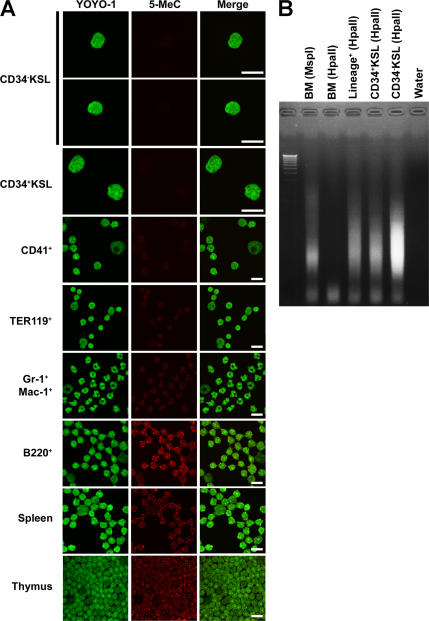
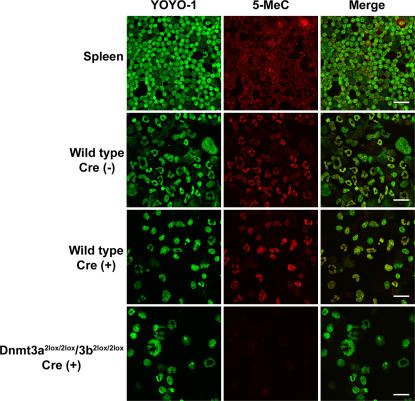
We next examined the DNA methylation status in each hematopoietic lineage using immunocytochemistry (16). The DNA methylation level in B and T lymphoid lineage or myeloid lineage cells was very high or moderate, respectively. Cells' DNA methylation levels correlated with their Dnmt3 expression levels (Fig. 2 A). The DNA methylation level in CD34− KSL cells was unexpectedly much lower than that in mature hematopoietic cells (Fig. 2 A). These data were further confirmed by a modified method of methylation-sensitive representational difference analysis using a methylation-sensitive HpaII restriction enzyme (17). After the genomic DNA of CD34− KSL cells was digested by a HpaII restriction enzyme, the digested DNA fragments were amplified by PCR and checked by electrophoresis (Fig. 2 B). These data suggest that the DNA methylation level of HSCs is low and that it gradually increases as HSC progeny mature into functional blood cells.
Figure 2.
Status of DNA methylation in hematopoietic cells. (A) Status of DNA methylation in hematopoietic cells was investigated by the detection of 5-methyl-cytosine (5-MeC). DNA methylation level was lower in undifferentiated cells and higher in mature hematopoietic cells, especially B and T lymphoid cells. Bars, 10 μm. (B) Status of DNA methylation in hematopoietic cells was confirmed by PCR using HpaII-digested genomic DNA. When PCR products are detectable, the genomic DNA is methylated low.
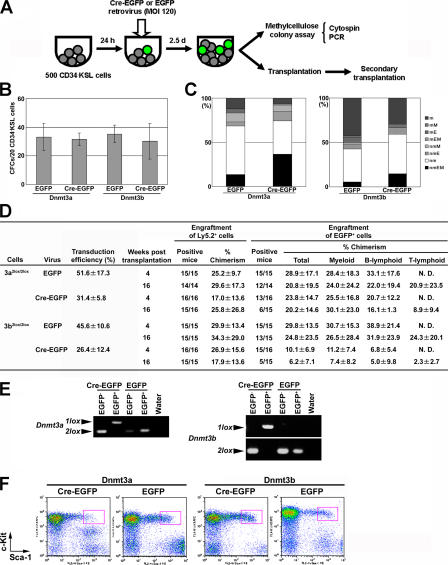
To clarify whether Dnmt3a or Dnmt3b is required for self-renewal and multilineage differentiation of HSCs, we decided to analyze mice conditional for these genes (Dnmt3a2lox/2lox, Dnmt3b2lox/2lox, or both Dnmt3a2lox/2lox and Dnmt3b2lox/2lox [Dnmt3a2lox/2lox/Dnmt3b2lox/2lox] mice) by using the retroviral Cre gene transduction system (18, 19). The conditional alleles and the Cre-mediated deletion alleles were designated as 2lox and 1lox, respectively (8, 20). As illustrated in Fig. 3 A, 500 CD34− KSL cells were isolated from the BM cells of each 2lox/2lox mouse and infected with pMSCV-Cre-IRES-enhanced GFP (Cre-EGFP) retrovirus or pMSCV-IRES-EGFP (EGFP) retrovirus at a dose of 120 multiplicity of infection (21, 22). After an additional 2.5 d of incubation with the retrovirus, the cells were divided into two aliquots to be examined by in vitro methylcellulose colony assays and in vivo competitive repopulation assays.
Figure 3.
Deletion of Dnmt3a or Dnmt3b in HSCs. (A) A study design is schematically shown. (B) Methylcellulose colony assay was performed for Dnmt3a2lox/2lox or Dnmt3b2lox/2lox CD34− KSL cells after infection with EGFP or Cre-EGFP retrovirus. 4% of the infected cells were cultured for a total of 14 d. Highly proliferative colony-forming cells, defined as cells capable of forming colonies >1 mm in diameter, were counted. Data are given as mean ± SD (n = 3). (C) EGFP+ colonies were assigned colony types on morphological examination of colony cells. (D) Competitive repopulation was performed using Dnmt3a2lox/2lox or Dnmt3b2lox/2lox CD34− KSL cells after infection with EGFP or Cre-EGFP retrovirus. Transduction efficiencies were estimated by counting EGFP+ and EGFP− colonies in each case in B. Engraftment of Ly5.2+ donor cells was assessed by the ratio of the number of mice engrafted with Ly5.2+ cells to the number of recipient mice, and by the percentage of Ly5.2+ cells in peripheral blood (mean ± SD). Contribution of EGFP+ cells to all test donor-derived cells was assessed by the ratio of the number of mice engrafted with EGFP+ cells to the number of mice engrafted with Ly5.2+ cells, the percentage of EGFP+ cells among Ly5.2+ cells, and the percentage of EGFP+ cells among each lineage marker expressing Ly5.2+ cells (mean ± SD). N.D., not detected. (E) Representative genotyping of BM cells reconstituted with Dnmt3a2lox/2lox or Dnmt3b2lox/2lox cells infected with EGFP or Cre-EGFP retrovirus is shown. EGFP− or EGFP+ cells were isolated by flow cytometry from the BM of recipient mice, and their genomic DNA was examined by PCR. (F) BM cells in reconstituted mice were analyzed by flow cytometry. Only Ly5.2+ EGFP+ cells are displayed to show whether the KSL population (red squares) is derived from transduced cells.
Numbers of colonies formed by CD34− KSL cells from Dnmt3a2lox/2lox or Dnmt3b2lox/2lox mice did not differ significantly, regardless of whether these cells were infected with Cre-EGFP or EGFP retrovirus (Fig. 3 B). To evaluate the presence of myeloid lineage cells in each colony, cells composing colonies were morphologically identified as neutrophils (n), macrophages (m), erythroblasts (E), and megakaryocytes (M). As shown in Fig. 3 C, Dnmt3a2lox/2lox and Dnmt3b2lox/2lox cells formed a variety of colony types after infection with Cre-EGFP or EGFP retrovirus. 51.6 ± 17.3% or 31.4 ± 5.8% of Dnmt3a2lox/2lox cells infected with EGFP or Cre-EGFP retrovirus exhibited EGFP fluorescence. 45.6 ± 10.6% or 26.4 ± 12.4% of Dnmt3b2lox/2lox cells infected with EGFP or Cre-EGFP retrovirus exhibited EGFP fluorescence (Fig. 3 D).
To verify deletion of the target genes in EGFP+ colonies, PCR was performed on genomic DNA extracted from colonies formed by Dnmt3a2lox/2lox or Dnmt3b2lox/2lox cells infected with Cre-EGFP retrovirus (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20060750/DC1). Each gene turned out to be deleted from all EGFP+ colonies examined (Tables S1 and S2). Among these EGFP+ and gene-deleted colonies, nmEM colonies were detected (Fig. 3 C). From these results, we concluded that the lack of Dnmt3a or Dnmt3b has little effect on in vitro myeloid colony formation by HSCs and their immediate progeny.
We then examined repopulating activity in Dnmt3a2lox/2lox or Dnmt3b2lox/2lox CD34− KSL cells after infection with Cre-EGFP or EGFP retrovirus. To do this, 0.1 vol of infected cultured cells (equivalent to 50 culture-initiating CD34− KSL cells) from the 2lox/2lox mice (Ly5.2) was transplanted into a lethally irradiated B6-Ly5.1 mouse together with 2 × 105 competitor cells from a B6-Ly5.1/Ly5.2 F1 mouse. Recipient mice were analyzed 4 and 16 wk after transplantation. As shown in Fig. 3 D, regardless of whether CD34− KSL cells were infected with EGFP or Cre-EGFP retrovirus, EGFP+ Dnmt3a2lox/2lox or Dnmt3b2lox/2lox cells were engrafted in most recipient mice 4 wk after transplantation. Substantial numbers of mice were reconstituted in myeloid, B lymphoid, and T lymphoid lineages with Cre-EGFP–transduced Dnmt3a2lox/2lox or Dnmt3b2lox/2lox cells 16 wk after transplantation. To rule out the possibility of Cre toxicity, long-term reconstitution of hematopoiesis by Cre-overexpressing HSCs was also confirmed in Cre-EGFP–transduced wild-type cells (Table S3, available at http://www.jem.org/cgi/content/full/jem.20060750/DC1). To confirm that Dnmt3a- or Dnmt3b-deleted cells in fact contributed to long-term reconstitution, EGFP+ and EGFP− cells were isolated from the BM cells of the recipient mice. Genomic DNA was extracted from these cells and was subjected to PCR-based genotyping. The 1lox and 2lox alleles were detected in EGFP+ and EGFP− cells in Cre-EGFP–transduced Dnmt3a2lox/2lox or Dnmt3b2lox/2lox cells, respectively (Fig. 3 E). These results suggest that HSCs are able to undergo self-renewal and multilineage differentiation in the absence of Dnmt3a or Dnmt3b.
We also examined BM cells in these reconstituted recipient mice. EGFP+ KSL cells were detected among BM cells by flow cytometry (Fig. 3 F). Secondary transplantation of these BM cells again resulted in multilineage reconstitution by Dnmt3a- or Dnmt3b-deficient BM cells (Table S4, available at http://www.jem.org/cgi/content/full/jem.20060750/DC1). These data reinforce the notion that in HSCs, the lack of either Dnmt3a or Dnmt3b does not affect self-renewal and multilineage differentiation potential.
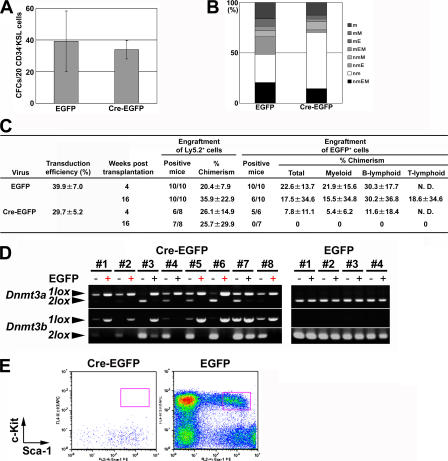
We then went on to assess the effect of the lack of both Dnmt3a and Dnmtb3b (Dnmt3a/Dnmt3b) on HSCs. In vitro methylcellulose colony assays detected no difference in numbers, sizes, and types of colonies between Dnmt3a2lox/2lox/Dnmt3b2lox/2lox CD34− KSL cells infected with Cre-EGFP retrovirus and those infected with EGFP retrovirus (Fig. 4, A and B). 39.9 ± 7.0% or 29.7 ± 5.2% of colonies made by Dnmt3a2lox/2lox/Dnmt3b2lox/2lox cells infected with EGFP or Cre-EGFP retrovirus exhibited EGFP fluorescence (Fig. 4 C). Both genes were completely deleted from two thirds of EGFP+ colonies derived from Cre recombinase-transduced cells (Table S5, available at http://www.jem.org/cgi/content/full/jem.20060750/DC1). Among these EGFP+ and gene-deleted colonies, nmEM colonies were detected (Fig. 4 B). Transplantation of Cre-EGFP–transduced Dnmt3a2lox/2lox/Dnmt3b2lox/2lox CD34− KSL cells showed that these cells were transiently able to reconstitute myeloid and B lymphoid lineages at 4 wk after transplantation but could not sustain reconstitution for 16 wk (Fig. 4 C). Most of the transient EGFP+ mature hematopoietic cells were Dnmt3a1lox/1lox/Dnmt3b1lox/1lox cells (Fig. 4 D). KSL BM cells were not reconstituted by Cre-EGFP–transduced Dnmt3a2lox/2lox/Dnmt3b2lox/2lox cells (Fig. 4 E). These data indicate that either Dnmt3a or Dnmt3b is necessary to maintain HSC self- renewal capacity after transplantation into lethally irradiated mice.
Figure 4.
Deletion of both Dnmt3a and Dnmt3b in HSCs. (A) A methylcellulose colony assay was performed for Dnmt3a2lox/2lox/Dnmt3b2lox/2lox CD34− KSL cells after infection with EGFP or Cre-EGFP retrovirus. 4% of the infected cells were cultured for a total of 14 d. (B) EGFP+ colonies were assigned colony types. (C) Competitive repopulation was performed using Dnmt3a2lox/2lox/Dnmt3b2lox/2lox CD34− KSL cells after infection with EGFP or Cre-EGFP retrovirus. Transduction efficiencies were estimated by counting EGFP+ and EGFP− colonies in each case in A. N.D., not detected. (D) Genotyping of peripheral blood cells transiently reconstituted with Dnmt3a2lox/2lox/Dnmt3b2lox/2lox cells infected with EGFP or Cre-EGFP retrovirus is shown. EGFP− and EGFP+ cells were isolated by flow cytometry from the peripheral blood of recipient mice, and their genomic DNA was examined by PCR. In the recipient mice transplanted with Cre-EGFP–transduced Dnmt3a2lox/2lox/Dnmt3b2lox/2lox CD34− KSL cells, Ly5.2+/EGFP+ peripheral blood cells were detected in 8 of 10 mice. In Cre-EGFP–transduced Dnmt3a2lox/2lox/Dnmt3b2lox/2lox cells, Dnmt3a1lox/1lox/Dnmt3b1lox/1lox mature cells were detected in five (red, +) of eight recipient mice. (E) BM cells in mice transplanted with transduced cells were analyzed by flow cytometry. Only Ly5.2+ EGFP+ cells are displayed to show whether the KSL population (red squares) is derived from transduced cells.
Because both Dnmt3a and Dnmt3b were expressed in T lymphoid cells, they were suspected to have a role in T lymphoid cell development. However, it is difficult to evaluate T lymphoid lineage differentiation potential in HSCs by competitive repopulation when HSC long-term repopulating activity is impaired. To address this issue, we transplanted Cre-EGFP–transduced Dnmt3a2lox/2lox/Dnmt3b2lox/2lox CD34− KSL cells into sublethally irradiated nonobese diabetes/SCID mice without competitor cells (23). We then attempted to detect Dnmt3a/Dnmt3b-deficient T lymphoid cells in recipient mouse thymus 7 wk after transplantation. CD4+CD8− and CD4−CD8+ cells were indeed detected among EGFP+ cells (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20060750/DC1). Collectively, HSCs without Dnmt3a and Dnmt3b can differentiate into myeloid, B lymphoid, and T lymphoid lineages. Neither Dnmt3a nor Dnmt3b seems essential for lineage commitment processes.
We examined whether Dnmt3a and Dnmt3b control de novo DNA methylation activity during hematopoietic cell differentiation. Although differentiated hematopoietic cells derived from Cre-EGFP–transduced wild-type CD34− KSL cells showed a high level of DNA methylation, that of differentiated hematopoietic cells derived from Cre-EGFP–transduced Dnmt3a2lox/2lox/Dnmt3b2lox/2lox CD34− KSL cells remained low (Fig. 5). These data indicate that Dnmt3a and Dnmt3b function as de novo DNA methyltransferases genome-wide during hematopoietic differentiation. However, de novo DNA methylation does not seem necessary for lineage commitment processes, although it is still possible that these molecules play a role in functions of mature blood cells, particularly B and T lymphoid cells. Further work is required to identify the genes targeted by Dnmt3a and Dnmt3b.
Figure 5.
DNA methylation status in Dnmt3a1lox/1lox/Dnmt3b1lox/1lox hematopoietic cells. CD34− KSL cells of each type were cultured for 14 d. Cre-EGFP–transduced Dnmt3a2lox/2lox/Dnmt3b2lox/2lox cells showed a low level of DNA methylation compared with Cre-EGFP–transduced wild-type cells. Bars, 20 μm.
This study showed that in sharp contrast to Dnmt3a/Dnmt3b-deficient ES cells, which maintain replication potential, Dnmt3a/Dnmt3b-deficient HSCs progressively lose replication potential, but not differentiation potential. The roles of de novo DNA methylation by Dnmt3a and Dnmt3b may differ between embryonic and adult stem cells from the developmental point of view. Alternatively, epigenetic modifications may differ substantially between in vitro–manipulated cell lines and cells in vivo in a physiological environment. ES cells should have their genomic DNA globally hypermethylated to maintain their differentiation potential (9, 24). HSCs may not need such extensive genomic DNA methylation to preserve self-renewal and differentiation. Furthermore, in a recent study, it has been suggested that Dnmt3a and Dnmt3b localize to the replication complex, recognize unmethylated CpG sites, which are left untouched by Dnmt1, and restore methylation via de novo DNA methylation (9). Dnmt3a and Dnmt3b may maintain HSC-specific DNA methylation patterns via de novo methylation activity. To address this issue, we need to develop technology to analyze genome-wide DNA methylation status in a very limited number of cells. Further work is also required to address the mechanisms of how Dnmt3a and Dnmt3b act on the genome via DNA methylation to maintain self-renewal capacity in HSCs. Dnmt3a and Dnmt3b may take part in the regulation of other stem cells; indeed, realization is growing that they are expressed in a variety of tissue-specific stem cells (25). There is a possibility that Dnmt3a and Dnmt3b play a role not only in the maintenance of self-renewal capacity in adult stem cells in general, but also in clonal expansion of cancer stem cells.
MATERIALS AND METHODS
Mice.
Dnmt3a2lox/2lox, Dnmt3b2lox/2lox, and Dnmt3a2lox/2lox/Dnmt3b2lox/2lox mice (129SvJae × C57BL/6) have been described previously (8, 20). C57BL/6 mice congenic for the Ly5 locus (B6-Ly5.1) and nonobese diabetic/SCID mice were bred and maintained by Sankyo Labo Service, Tsukuba, Japan. 2lox/2lox and C57BL/6 (B6-Ly5.1/Ly5.2) mice were maintained at the Animal Research Center of the Institute of Medical Science, University of Tokyo, which is approved by the Animal Experiment Committee of the Institute.
Purification of HSCs.
CD34− KSL cells were purified from the BM of Dnmt3a2lox/2lox, Dnmt3b2lox/2lox, or Dnmt3a2lox/2lox/Dnmt3b2lox/2lox mice as described previously in detail (15, 26). Multicolor analysis and sorting were performed using a MoFlo (DakoCytomation).
Transduction of CD34− KSL cells.
Purified CD34− KSL cells were infected with pMSCV-IRES-EGFP or pMSCV-Cre-IRES-EGFP retrovirus as described previously (22). Cells were preincubated with 100 ng/ml mouse stem cell factor (SCF) and 100 ng/ml human thrombopoietin (TPO) for 24 h. Cells were infected with a titrated amount of retrovirus to avoid toxicity by overexpression of Cre recombinase (21). After culture for an additional 2.5 d, cells were subjected to methylcellulose colony assay and transplantation analysis.
In vitro colony assay.
Transduced CD34− KSL cells were cultured in a 35-mm dish containing 1 ml of 1.2% methylcellulose, 30% FCS, and 1% BSA supplemented with 10 ng/ml mouse SCF, 10 ng/ml human TPO, 10 ng/ml mouse IL-3, and 2 U/ml human erythropoietin. Cells were incubated at 37°C in a humidified atmosphere with 5% CO2 for a total of 14 d. Colonies were counted under an inverted fluorescence microscope and individually picked from methylcellulose. Half of the cells per colony were stained with May-Grünwald-Giemsa solution for morphological analysis. The remaining cells in each colony were used as sources of genomic DNA for PCR genotyping.
Competitive repopulation assay.
Competitive repopulation assay was performed by using the Ly5 congenic mouse system (26). Retrovirus-infected cultured cells from 2lox/2lox mice (Ly5.2) were transplanted into B6-Ly5.1 mice irradiated at a dose of 9.5 Gy together with 2 × 105 competitor cells from a B6-Ly5.1/Ly5.2 F1 mouse. Peripheral blood cells of the recipient mice were taken 4 and 16 wk after transplantation and analyzed by flow cytometry to determine reconstitution levels in myeloid (Mac-1+/Gr-1+), B lymphoid (B220+), and T lymphoid (CD4+/CD8+) lineages as described previously (27).
For secondary transplantation, recipient mice were killed between 16 and 20 wk after transplantation, and 2 × 106 BM cells from these recipients were transplanted into B6-Ly5.1 mice irradiated at a dose of 9.5 Gy. Peripheral blood cells of the B6-Ly5.1 recipient mice were analyzed 12 wk after secondary transplantation. EGFP− and EGFP+ cells separated from BM cell surplus to secondary-transplantation requirements were used for PCR genotyping.
Semiquantitative RT-PCR.
cDNAs were normalized with GAPDH copy numbers calculated based on quantitative PCR data using TaqMan rodent GAPDH control reagent (PerkinElmer; reference 28). PCR for Dnmts was performed using a 34-cycle program of 20 s at 94°C, 20 s at 65°C, and 30 s at 72°C per cycle. For Dnmt1 amplification, F1 primer (5′-CGGTCATTCCAGATGATTCCTC-3′) and R1 primer (5′-TGCTGTGGATGT-AGGAAAGCTG-3′) were used. For Dnmt3a amplification, F4 primer (5′-TCCCGGGGCCGACTGCGA-3′) and R1 primer (5′-TCCCCCACACCAGCTCTC-3′) were used. For Dnmt3a2 amplification, F5 primer (5′-AGGGGCTGCACCTGGCCTT-3′) and R1 primer were used. For Dnmt3b amplification, F2052 primer (5′-GAACATGCGCCTGCAAGA-3′) and R2252 primer (5′-GCACAGACTTCGGAGGCAAT-3′) were used. For GAPDH amplification, F primer (5′-CTTCACCACCATGGAGAAGGC-3′) and R primer (5′-GGCATGGACTGTGGTCATGAG-3′) were used. PCR for Dnmt3L was performed using a 34-cycle program of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C per cycle. For Dnmt3L amplification, F primer (5′-CGGCACCAGCTGAAGGCCTT-CCATG-3′) and R primer (5′-AGGCAGCGCATACTGCAGGATCCGG-3′) were used (29).
Immunocytostaining for Dnmt3a, Dnmt3b, and 5-MeC.
Cells were directly sorted by flow cytometry into a droplet of medium on a poly-l-lysine–coated glass slide. After fixation with 2% paraformaldehyde, cells were incubated for 12 h at 4°C with anti-Dnmt3a antibody (clone 64B1446; IMGENEX) at a dilution of 1:400 and anti-Dnmt3b antibody (Abcam) at a dilution of 1:200. After washes, cells were reacted for 30 min at room temperature with Alexa Fluor 488–labeled goat anti–rabbit IgG or Alexa Fluor 647–labeled goat anti–mouse IgG secondary antibody (Invitrogen) at a dilution of 1:500. Immunocytochemistry with anti–5-MeC antibody (supernatant from hybridoma cultures) was performed as described previously (16, 30). In this protocol, each cell was cytospun onto slide glasses. An anti–5-MeC antibody was detected by an Alexa Fluor 647–labeled goat anti–mouse IgG secondary antibody (Invitrogen) at a dilution of 1:500. A Leica TCS SP2 AOBS confocal microscope (Leica Microscopy System) was used to visualize fluorescent signals.
Detection of DNA methylation status from PCR products.
DNA of each hematopoietic cell (103 cells per sample) was digested with HpaII or MspI overnight. After their digestion products were purified, they were ligated to RHpa adaptor (17). 0.1 vol of the ligation products was amplified with RHpa24 primer (5′-AGCACTCTCCAGCCTCTCACCGAC-3′) using a 25-cycle program of 1 min at 95°C and 3 min at 72°C per cycle. DNA methylation status of each cell was checked by electrophoresing their PCR products in 1% agarose gel.
Online supplemental material.
Fig. S1 shows detection of deleted alleles in blood colonies. Fig. S2 shows detection of Dnmt3a1lox/1lox/Dnmt3b1lox/1lox T lymphoid cells. Table S1 shows detection of deleted alleles in blood colonies formed by Dnmt3a2lox/2lox cells infected with Cre-EGFP retrovirus. Table S2 shows detection of deleted alleles in blood colonies formed by Dnmt3b2lox/2lox cells infected with Cre-EGFP retrovirus. Table S3 shows efficiency of competitive repopulation in Cre-EGFP–transduced wild-type CD34− KSL cells. Table S4 shows secondary transplantation of Cre-EGFP–transduced Dnmt3a2lox/2lox or Dnmt3b2lox/2lox BM cells. Table S5 shows detection of deleted alleles in blood colonies formed by Dnmt3a2lox/2lox/Dnmt3b2lox/2lox cells infected with Cre-EGFP retrovirus. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20060750/DC1.
Supplemental Material
Acknowledgments
We thank Y. Yamazaki for flow cytometer operation and A.S. Knisely for critical reading of the manuscript.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology.
The authors have no conflicting financial interests.
References
- 1.Jaenisch, R., and A. Bird. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33:245–254. [DOI] [PubMed] [Google Scholar]
- 2.Wolffe, A.P., and M.A. Matzke. 1999. Epigenetics: regulation through repression. Science. 286:481–486. [DOI] [PubMed] [Google Scholar]
- 3.Bird, A.P., and A.P. Wolffe. 1999. Methylation-induced repression–belts, braces, and chromatin. Cell. 99:451–454. [DOI] [PubMed] [Google Scholar]
- 4.Reik, W., W. Dean, and J. Walter. 2001. Epigenetic reprogramming in mammalian development. Science. 293:1089–1093. [DOI] [PubMed] [Google Scholar]
- 5.Chaillet, J.R., T.F. Vogt, D.R. Beier, and P. Leder. 1991. Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell. 66:77–83. [DOI] [PubMed] [Google Scholar]
- 6.Li, E., C. Beard, and R. Jaenisch. 1993. Role for DNA methylation in genomic imprinting. Nature. 366:362–365. [DOI] [PubMed] [Google Scholar]
- 7.Okano, M., D.W. Bell, D.A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99:247–257. [DOI] [PubMed] [Google Scholar]
- 8.Kaneda, M., M. Okano, K. Hata, T. Sado, N. Tsujimoto, E. Li, and H. Sasaki. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 429:900–903. [DOI] [PubMed] [Google Scholar]
- 9.Chen, T., Y. Ueda, J.E. Dodge, Z. Wang, and E. Li. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23:5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, E., T.H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 69:915–926. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, M., A. Krassowska, N. Gilbert, T. Chevassut, L. Forrester, J. Ansell, and B. Ramsahoye. 2004. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol. Cell. Biol. 24:8862–8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, T., Y. Ueda, S. Xie, and E. Li. 2002. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 277:38746–38754. [DOI] [PubMed] [Google Scholar]
- 13.Bourc'his, D., G.L. Xu, C.S. Lin, B. Bollman, and T.H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science. 294:2536–2539. [DOI] [PubMed] [Google Scholar]
- 14.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 129:1983–1993. [DOI] [PubMed] [Google Scholar]
- 15.Osawa, M., K. Hanada, H. Hamada, and H. Nakauchi. 1996. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 273:242–245. [DOI] [PubMed] [Google Scholar]
- 16.Santos, F., B. Hendrich, W. Reik, and W. Dean. 2002. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241:172–182. [DOI] [PubMed] [Google Scholar]
- 17.Ushijima, T., K. Morimura, Y. Hosoya, H. Okonogi, M. Tatematsu, T. Sugimura, and M. Nagao. 1997. Establishment of methylation-sensitive-representational difference analysis and isolation of hypo- and hypermethylated genomic fragments in mouse liver tumors. Proc. Natl. Acad. Sci. USA. 94:2284–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres, R.M., and R. Kühn. 1997. Laboratory Protocols for Conditional Gene Targeting. Oxford University Press, New York. 180 pp.
- 19.Opferman, J.T., H. Iwasaki, C.C. Ong, H. Suh, S. Mizuno, K. Akashi, and S.J. Korsmeyer. 2005. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 307:1101–1104. [DOI] [PubMed] [Google Scholar]
- 20.Dodge, J.E., M. Okano, F. Dick, N. Tsujimoto, T. Chen, S. Wang, Y. Ueda, N. Dyson, and E. Li. 2005. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J. Biol. Chem. 280:17986–17991. [DOI] [PubMed] [Google Scholar]
- 21.Loonstra, A., M. Vooijs, H.B. Beverloo, B.A. Allak, E. van Drunen, R. Kanaar, A. Berns, and J. Jonkers. 2001. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. USA. 98:9209–9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwama, A., H. Oguro, M. Negishi, Y. Kato, Y. Morita, H. Tsukui, H. Ema, T. Kamijo, Y. Katoh-Fukui, H. Koseki, et al. 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 21:843–851. [DOI] [PubMed] [Google Scholar]
- 23.Wang, J.C.Y., C. Dorrell, C.Y. Ito, T. Inamitsu, G. Guenechea, O.I. Gan, and J.E. Dick. 2001. Normal and leukemic human stem cells assayed in immune-deficient mice. In Hematopoiesis: A Developmental Approach. L.I. Zon, editor. Oxford University Press, New York. 99–118 pp.
- 24.Lei, H., S.P. Oh, M. Okano, R. Juttermann, K.A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 122:3195–3205. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe, D., I. Suetake, S. Tajima, and K. Hanaoka. 2004. Expression of Dnmt3b in mouse hematopoietic progenitor cells and spermatogonia at specific stages. Gene Expr. Patterns. 5:43–49. [DOI] [PubMed] [Google Scholar]
- 26.Ema, H., K. Sudo, J. Seita, A. Matsubara, Y. Morita, M. Osawa, K. Takatsu, S. Takaki, and H. Nakauchi. 2005. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev. Cell. 8:907–914. [DOI] [PubMed] [Google Scholar]
- 27.Sudo, K., H. Ema, Y. Morita, and H. Nakauchi. 2000. Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 192:1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osawa, M., T. Yamaguchi, Y. Nakamura, S. Kaneko, M. Onodera, K. Sawada, A. Jegalian, H. Wu, H. Nakauchi, and A. Iwama. 2002. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood. 100:2769–2777. [DOI] [PubMed] [Google Scholar]
- 29.Sakai, Y., I. Suetake, F. Shinozaki, S. Yamashina, and S. Tajima. 2004. Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr. Patterns. 5:231–237. [DOI] [PubMed] [Google Scholar]
- 30.Reynaud, C., C. Bruno, P. Boullanger, J. Grange, S. Barbesti, and A. Niveleau. 1992. Monitoring of urinary excretion of modified nucleosides in cancer patients using a set of six monoclonal antibodies. Cancer Lett. 61:255–262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.