Abstract
NK and T lymphocytes express both activating and inhibiting receptors for various members of the major histocompatibility complex class I superfamily (MHCISF). To evade immunologic cytotoxicity, many viruses interfere with the function of these receptors, generally by altering the displayed profile of MHCISF proteins on host cells. Using a structurally constrained hidden Markov model, we discovered an orthopoxvirus protein, itself distantly class I–like, that acts as a competitive antagonist of the NKG2D activating receptor. This orthopoxvirus MHC class I–like protein (OMCP) is conserved among cowpox and monkeypox viruses, secreted by infected cells, and bound with high affinity by NKG2D of rodents and humans (KD ∼ 30 and 0.2 nM, respectively). OMCP blocks recognition of host-encoded ligands and inhibits NKG2D-dependent killing by NK cells. This finding represents a novel mechanism for viral interference with NKG2D and sheds light on intercellular recognition events underlying innate immunity against emerging orthopoxviruses.
Viral replication alters gene expression and protein metabolism in host cells. This perturbs the profile of proteins displayed on the plasma membrane, labeling infected cells as abnormal and targeting them for destruction by cytotoxic effectors. Such perturbations fall into two categories: the display of pathogen-encoded molecules and the altered expression of host-encoded markers, including diverse members of the MHC class I superfamily (MHCISF). The former are sensed using clonotypic receptors of adaptive immunity, whereas the latter are sensed through receptors associated with innate immunity.
NKG2D is an activating receptor of NK lymphocytes, as well as of species-specific subsets of T lymphocytes (for review see reference 1). It binds host-encoded members of the MHCISF whose expression is enhanced by extracellular signals and intracellular metabolic disturbances, particularly those resulting from infection (for review see reference 1). Human and mouse CMV encode MHC class I–like proteins that block cell-surface expression of NKG2D ligands (2–7). By interfering with NKG2D-dependent responses to infected cells, these proteins permit enhanced replication of the virus in a hostile immunologic environment. Conservation of the corresponding genes by wild strains of CMV (unpublished data) demonstrates, prima facie, the importance of NKG2D-mediated immunity to CMV evolution.
In this paper, we show that zoonotic orthopoxviruses (ZPXVs) encode a secreted class I–like competitive antagonist of NKG2D, suggesting that NKG2D function also applies selective pressure to these viruses. In contrast to CMV, ZPXVs have adapted the class I domain fold to target the receptor rather than its ligands. Because receptors that direct innate cytotoxicity against epidemiologically relevant orthopoxviruses remain undefined, this finding should open new avenues of research into immunity against these emerging pathogens.
RESULTS AND DISCUSSION
Hidden Markov model (HMM) analysis reveals an orthopoxvirus MHC class I–like protein (OMCP)
NKG2D ligands adopt MHC class I–like backbone folds despite low sequence identity (for review see reference 8). The m157 protein of mouse CMV (a ligand for the Ly49H NK receptor) probably does so as well, though the Basic Local Alignment Search Tool heuristic fails to align it with any mammalian member of the MHCISF (9, 10). We hypothesized that orthopoxviruses also encoded MHCISF proteins that remained undetected because of their extreme sequence divergences. Indeed, uncharacterized class I–like open reading frames in the genomes of molluscipoxvirus, suipoxvirus, and yatapoxviruses have been predicted using local alignment methods (for review see reference 11). To search for the former, we adapted the profile HMM algorithm embodied in the Sequence Alignment and Modeling (SAM) software package from the University of California, Santa Cruz (12, 13).
To avoid bias toward classical class I sequences, we used a seed alignment consisting only of divergent class I–like platform domains; during iterative model weighting, this was left inviolate despite the eventual inclusion of classical class I sequence information. To avoid alignment errors arising from the small number of available highly divergent MHCISF members, this seed was based on secondary structure defined by x-ray diffraction.
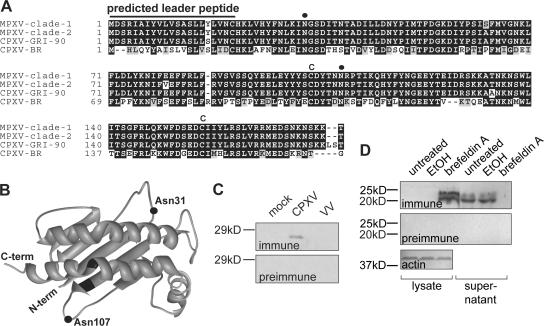
A search of National Center for Biotechnology Information (NCBI) raw viral sequence data yielded high-scoring translations encoded within the genomes of cowpox virus (CPXV; available from GenBank/EMBL/DDBJ under accession nos. AF482758 and X94355) and monkeypox virus (MPXV; available from GenBank/EMBL/DDBJ under accession no. NC_003310.1). None were found in the vaccinia or variola genomes. In every case, the corresponding open reading frames lay within genomic flanking regions, the sites of poxvirus immunomodulatory loci (for review see reference 11). These sequences correspond to a single protein (OMCP) with four variants, the first encoded by less virulent West African MPXV (clade-1) (14, 15), the second encoded by the more virulent Central African MPXV (clade-2) (14, 15), the third encoded by CPXV-GRI90, and the fourth encoded by CPXV–Brighton Red (CPXV-BR; Fig. 1 A). Clade-1 MPXV possess identical functional copies at both flanks of their genomes, whereas clade-2 viruses possess only a right-sided functional copy, with the left-sided one being a nonfunctional fragment (not depicted). Consistent with the proposal that CPXV-BR represents a distinct orthopoxvirus species (16), Brighton Red OMCP is considerably more divergent from the other sequences than they are from each other. Because of the >93% aa identity between the first three variants, they henceforth are collectively termed OMCPMPX. The Brighton Red variant henceforth is termed OMCPBR.
Figure 1.
OMCP is likely a secreted class I–like protein. (A) Multiple alignment of the four OMCP variants. Cysteines (C) and potential N-linked glycosylation sites (closed circles) are indicated. (B) Backbone model of OMCPBR derived by threading through the RAE1β structure (closed squares, cysteines). Similar results were obtained for OMCPMPX. (C) Immunoblot of culture media conditioned by mock-infected and CPXV-BR–infected B-SC-1 cells suggests secretion of OMCP. (D) Immunoblot showing that brefeldin A, but not its ethanol vehicle, prevents the appearance of OMCPBR in supernatants from infected OP9 cells (far right lane vs. second lane from right). Brefeldin A also causes the accumulation of OMCP in lysates of treated cells (third lane from left).
OMCP possesses a strongly predicted leader sequence, two cysteines (Fig. 1 A), and a common potential N-linked glycosylation site at Asn31 (CPXV-BR numbering); OMCPBR has a second potential N-linked site at Asn107 (Fig. 1 A). Backbone prediction using the FUGUE (17) threading algorithm assigns reasonable likelihood (z-score >5) to a class I–like fold for OMCP. In this model, the cysteines are close in three-dimensional space (Fig. 1 B) and are predicted to form a disulfide bond. Neither transmembrane spans nor GPI transamidation sites could be predicted in any reading frame near the ends of the coding sequences, suggesting OMCP to be a secreted protein. To test this, supernatants from both mock-infected B-SC-1 cells as well as those infected either with CPXV-BR or Western Reserve vaccinia virus (VV) were screened by immunoblot for reactivity with a polyclonal antiserum against recombinant OMCPBR (Fig. 1 C). Only supernatants from CPXV-BR–infected cells reacted with the antiserum, while neither supernatant reacted with preimmune serum. To verify that OMCP was secreted and not released from lysed cells, CPXV-BR–infected OP9 cells were treated with nothing, vehicle alone, or brefeldin A (Fig. 1 D). Treatment with brefeldin A, but not its ethanol vehicle, prevented OMCP reactivity from appearing in the supernatant. Accordingly, OMCP accumulated in brefeldin A–treated cell lysates. No supernatant reacted with preimmune serum. The basis of the two OMCP bands secreted by OP9 cells is unknown.
Because it is not associated with a cell membrane, OMCP cannot cluster its cognate receptor. Making the assumption that most ZPXV immunomodulating proteins are immunosuppressive, we hypothesized that OMCP binds an activating receptor, acting as a competitive antagonist.
OMCP binds to NKG2D with high affinity
Because NKG2D ligands adopt a class I–like fold (for review see reference 8), we tested whether NKG2D is a receptor for OMCP. To this end, Ba/F3 cells were transduced with defective dicistronic retroviruses encoding human NKG2D (huNKG2D) and enhanced GFP (EGFP), mouse NKG2D (muNKG2D) and EGFP, or EGFP alone. The NKG2D ectodomain was spliced to Ly49A transmembrane and intracellular domains to avoid the need for a signaling chain. Transductants were then treated with fluorescent tetramers and analyzed by flow cytometry. OMCP tetramers, but neither streptavidin-PE (SA-PE) alone nor irrelevant tetramers, stained these transductants (Fig. 2 A, middle and bottom rows). No tetramers stained EGFP-only transductants (Fig. 2 A, top row). Preincubation either with mAb against NKG2D or with recombinant NKG2D ligands inhibited the interactions between OMCP tetramers and NKG2D transductants; however, preincubation either with isotype control mAb or with irrelevant recombinant protein failed to inhibit these interactions (Fig. 2 B). The origin of two distinct transductant populations in certain purified cell preparations is unclear. To exclude an artifact arising from a joint binding surface presented by OMCP and SA, the inverse experiment was also performed. muNKG2D tetramers stained Ba/F3 transductants expressing chimeric OMCP-Thy1.1 on the cell surface but not cells expressing only Thy1.1 (Fig. 2 C). Thus, OMCP is a virally encoded NKG2D ligand.
Figure 2.
OMCP binds huNKG2D and muNKG2D. (A) Ba/F3 cells expressing EGFP only (top), huNKG2D (middle), or muNKG2D (bottom) were treated with nothing, SA-PE, or fluorescent tetramers. (B) Ba/F3 cells transduced with huNKG2D or muNKG2D were incubated with anti-NKG2D mAb, isotype-matched mAb, irrelevant soluble protein, or soluble ligand before staining with OMCPBR (rows 1 and 3) or OMCPMPX (rows 2 and 4) tetramers. (C) Ba/F3 cells transduced either with Thy1.1 (top) or OMCP-Thy1.1 (middle and bottom) were treated with SA-APC or fluorescent tetramers. Untransduced (nongreen) Ba/F3 cells were included as internal standards in A–C. Transduction vectors are dicistronic, with EGFP in the second cistron. (D, 1) SPR assay of OMCPBR (0, 2, 5, 10, 20, 50, 100, 200, 500, and 1,000 nM) binding to immobilized muNKG2D. The horizontal bar represents the OMCP injection, and the star represents the time point used for KD fitting. (D, 2) Nonlinear curve fitting of steady-state SPR data reveals high affinity for muNKG2D and a good coefficient of determination for the fit. (D, 3) One representative experiment out of four showing kinetic data (black lines) and global fit (gray lines) of OMCPBR (5, 10, 20, 50, and 100 nM) binding to muNKG2D. (D, 4) One representative experiment out of six showing kinetic data (black lines) and global fit (gray lines) of OMCPBR (0.32, 0.63, 1.25, and 2.5 nM) binding to huNKG2D. Note the very different dissociation kinetics between the data in 3 and 4.
We hypothesized that OMCP blocks the interaction between NKG2D and its target cell–expressed ligands. If so, it should bind NKG2D with high affinity to overcome the multivalency of cell-surface ligands at reasonable protein concentrations. We performed surface plasmon resonance (SPR) studies to determine the binding parameters of this interaction, assuming a 1:1 binding model for (NKG2D)2/OMCP. The KD of OMCPBR for immobilized muNKG2D was 37 ± 6 nM (n = 3) by steady-state analysis at 20°C, pH 7.5, and 150 mM NaCl (Fig. 2 D, 2). KD calculated from kinetic analysis (i.e., KD = koff/kon; Fig. 2 D, 3) was similar: 28 ± 4 nM, with koff = 1.8 ± 0.1−2 s−1 and kon = 6.8 ± 1.2 × 105 M−1s−1 (n = 4). These parameters are akin to those of the higher affinity muNKG2D ligands H60 and RAE1ɛ (18, 19). Curiously, affinity of OMCPBR for huNKG2D was strikingly higher: KD = 168 ± 33 pM (n = 6), with the difference largely caused by the lower koff (1.23 ± 0.12 × 10−4 s−1; Fig. 2 D, 4). Analogous experiments with OMCPMPX were not performed because of the inability to obtain consistent specific activity of the recombinant protein.
Next, C57BL/6 (B6) splenocytes were costained with NK markers and OMCPBR tetramers (Fig. 3 A). NK and NKT cells bound OMCPBR tetramers. Pretreatments with either anti-muNKG2D mAb (Fig. 3 A, lines 6) or the muNKG2D ligand MULT1 (lines 8) inhibited binding to both cell types; treatments with an isotype-matched mAb (lines 5) or an irrelevant protein had no effect (lines 7). Consistent with the lack of NKG2D expression on resting mouse T cells (20), OMCPBR tetramers failed to bind freshly isolated splenic T cells. Splenocytes from three other strains of mice and (within the limitations imposed by a lack of validated reagents) rats, hamsters, guinea pigs, and voles also exhibited similar staining with OMCPBR (unpublished data). This argues for the relevance of this work to rodents generally and the reservoir hosts of ZPXV in particular (21, 22).
Figure 3.
OMCP binds to mouse and human leukocyte subpopulations. (A) B6 splenocytes were stained with mAbs against NK markers and OMCPBR tetramers. OMCPBR staining of cells in each gate is indicated in histograms. Each histogram group shows unstained cells (lines 1), cells stained with SA-APC alone (lines 2), irrelevant tetramers (lines 3), OMCPBR tetramers (lines 4), or OMCPBR tetramers after pretreatment with 50 μg/ml of irrelevant hamster IgG (lines 5), equimolar anti-muNKG2D (lines 6), 9.5 μM of irrelevant protein (lines 7), or equimolar MULT1 (lines 8). (B) Human PBMCs were stained with NK or T cell marker mAbs and were tested for OMCP binding, as in A, for lines 1–4. Pretreatments with 50 μg/ml of mouse IgG1 (lines 5) or equimolar anti-huNKG2D mAb (lines 6) were also performed before staining with tetramers. Representative experiments are shown (n = 4). Vertical lines in histograms represent the 99th percentile of staining for irrelevant tetramer- stained cells.
To explore the potential relevance of OMCP to orthopox zoonoses, human PBMCs were tested for binding to OMCPBR tetramers. Binding to human cells was essentially the same as that to mouse cells, except that fresh CD8+ T cells bound OMCPBR (Fig. 3B), consistent with the known expression of NKG2D on these cells. Analogous to mouse splenocytes, OMCP binding to human NK and NKT cells was blocked by anti-huNKG2D mAb (Fig. 3 B, lines 6). Cumulatively, these findings indicate that OMCP binds to NKG2D with high affinity and in a manner competitive with the endogenous ligands of this receptor.
OMCP inhibits NKG2D-dependent killing as a soluble antagonist
51Cr killing assays were performed using B6 lymphokine–activated NK cells against RMA target cells. Control vector–transduced RMA cells are relatively resistant to killing by B6 NK cells because of syngeneic MHC class I expression (Fig. 4 A, open squares) (23, 24). Expression of H60 by RMA cells enhanced their susceptibility to killing by activated NK cells in an NKG2D-dependent fashion (circles). NKG2D specificity was verified by inhibition of killing upon addition of the high-affinity muNKG2D ligand MULT1 to the reaction (unpublished data) (23). Recombinant OMCPBR at concentrations from 100 nM to 1 μM (open and closed triangles, diamond, and closed squares) blocked RMA-H60 killing, whereas 1 μM of irrelevant recombinant protein did not (Fig. 4 A, X). Similar results were obtained with fresh NK cells (not depicted). This effect was not caused by down-regulation of NKG2D, as exposure of B6 NK cells to 1 μM OMCPBR had no effect on NKG2D expression (Fig. 4 B, lines 3 and 6).
Figure 4.
OMCP inhibits muNKG2D-dependent killing by activated NK cells but does not down-regulate NKG2D. (A) B6 IL2-activated NK cells were used as cytotoxic effectors against transduced RMA cells in 4-h 51Cr release assays at effector/target ratios of 1:1, 3:1, or 10:1. There was minimal killing of EGFP-only transductants (open squares). There was effective NKG2D-dependent killing of H60 transductants (circles), but 1 μM of irrelevant protein failed to inhibit NKG2D-dependent killing (X). However, OMCPBR at 1 (closed squares), 0.5 (diamond), 0.25 (closed triangle), and 0.1 μM (open triangle) did effectively inhibit NKG2D-dependent killing. Error bars were omitted to avoid crowding; coefficients of variation for all data points were <10%. (B) B6 splenocytes were cultured for 24 h with 1 μM of irrelevant protein (lines 1–3) or 1 μM OMCPBR (lines 4–6) and stained with nothing (lines 1 and 4), isotype-matched mAb (lines 2 and 5), or anti-muNKG2D (lines 3 and 6). All histograms are gated on live, CD3−, NK1.1+ cells. No differences in the scatter properties or viability of cells was noted (not depicted). (C) Immunoblotting of OMCP was performed with known amounts of recombinant protein (left blot) or CPXV-infected OP9 supernatants (right blot). The former was used to quantitate the latter using a linear fit of OMCP densitometry to quantity (plot), as detailed in Materials and methods. One experiment out of three is shown. Xs represent mean calibration data, dots represent supernatant data, the continuous line represents linear fit, and the dotted lines bound the 95% confidence interval. (D) Ba/F3 cells were treated either with mock viral (left) or CPXV-BR (right) preparations for 24 h. Cells were stained with nothing (lines 1), SA-PE (rows 2), control tetramers (rows 3), or muNKG2D tetramers alone (rows 4), as well as with 7-amino-actinomycin D. NKG2D binds infected cells, indicating up-regulation of NKG2D ligands by CPXV-BR infection. The percentage of positive cells corresponds to the percentage of productively infected cells (not depicted). Vertical lines in the histograms (B and D) represent the 99th percentile of staining for irrelevant tetramer-stained cells.
OMCP is likely a paracrine inhibitor of NKG2D function
OMCP binds NKG2D in a manner preventing recognition of host ligands. This may be an important immune evasion strategy during ZPXV infections. Given an adequate local OMCP concentration around infected cells, inhibitory conditions analogous to those in the killing assay of Fig. 4 A might be attained. To measure the amount of OMCP produced during infection, semiquantitative immunoblotting of infected cell supernatants was performed using recombinant OMCP as a standard (Fig. 4 C). Protein was secreted at an average rate of ∼3,000 copies/cell/s during the viral incubation time. Assuming diffusion constants in interstitial fluid and basement membrane of ∼10−7 and 5 × 10−9 cm2s−1, respectively (25), this secretion rate reasonably allows inhibitory concentrations of OMCP to develop around infected cells in situ given the KD values of the NKG2D–OMCP interaction (26). We thus propose a model in which secreted OMCP acts in paracrine fashion to competitively antagonize the interaction between NKG2D on cytotoxic effectors and induced ligands on infected targets (Fig. 4 D). Because effective NKG2D cross-linking cannot then take place, activation signals promoting granule release are blunted, prolonging viral replication in infected cells. This, especially at mucosal sites and in the skin, increases the odds of transmission to another host, enhancing viral fitness. Though NKG2D ligands shed into serum may cause systemic receptor down-regulation (27), we disfavor this mode of action for OMCP because soluble OMCP fails to down-regulate NKG2D on mouse NK cells (Fig. 4 B).
Although certain virally encoded molecules are known to impair NKG2D-mediated detection of infected cells (2–7), OMCP is the first to target the receptor itself rather than its ligands. Of greater note, OMCP serves as a pointer to a receptor specifically important to the interactions between ZPXV and their hosts, guiding future studies of intercellular recognition events underlying cytotoxic immunity to these viruses.
MATERIALS AND METHODS
HMM analysis of sequences.
The SAM software package (version 3.4) was installed on a workstation running Redhat Linux 9. SAM is extensively documented at http://www.cse.ucsc.edu/research/compbio/papers/sam_doc/.
Using structural models of RAE1 (1jfm), ULBP3 (1kcg), MICA (1b3j), MICB (1je6; for review see reference 8), and MULT1 (Fremont, D., personal communication), multiple sequence alignment based on secondary structural boundaries was generated and used as a seed for the target2k script with match positions as constraints. Target2k in superfamily mode was applied to the NCBI nr protein database. This alignment was then used by the buildmodel script to produce HMMs of divergent class I–like protein sequences.
To tune parameters of the buildmodel and hmmscore procedures, a test set of 26,000 irrelevant mouse cDNA sequences spiked with the cDNAs encoding H60, m157, m154, and mill-1 was used. Parameters were chosen based on assignment of lower expect values to the four spiked sequences than to non–MHC-like sequences. The final model was built using w0.5 (global scoring for model construction) default parameters. Sequence scoring was ultimately performed using -sw2 (local) mode with a reverse-sequence null model on reading frames >60 aa from raw genomic sequences of all viruses (except CMV) available from the NCBI virus genome database as of February 2004. Sequence alignment was performed in SAM and formatted using BoxShade software (version 3.2; available at http://www.ch.embnet.org/software/BOX_doc.html).
Molecular cloning, expression, refolding, and SPR analysis of NKG2D and mature OMCP.
OMCPMPX (N3R; available from GenBank/EMBL/DDBJ under accession no. NC_003310) and OMCPBR (V018; available from GenBank/EMBL/DDBJ under accession no. AF482758) coding sequences were amplified from monkeypox Zaire 1979 DNA and CPXV-BR DNA, respectively. Amplicons excluded the predicted leaders (Fig. 1) but included C-terminal biotin ligase tags. Cloning and expression was performed as previously reported (19). OMCP and muNKG2D refolding, purification, biotinylation, characterization, and SPR analysis took place as previously described (19), except that the refolding time was extended to >72 h. A huNKG2D protein from …ESYCG to the C terminus, fused to an N-terminal BirA tag, was similarly produced, except that refolding was performed in 2.5 M proline/0.1 M Tris-Cl, pH 7.2/0.01% Na-azide/5 mM of reduced glutathione/0.5 mM of oxidized glutathione, as previously described (28). Regeneration of the huNKG2D/OMCP SPR chips was performed using 30-s injections of 170 μM Triton X-100/0.5 M LiCl/20 mM Na-citrate, pH 4.8.
Mice and cell culture.
C57BL/6 mice were obtained from the National Cancer Institute. RMA, Ba/F3, and activated NK cells were used as previously described (23, 29). B-SC-1 monkey kidney cells and OP9 marrow fibroblastic cells were obtained from American Type Culture Collection. Mouse work was approved by the Animal Studies Committee.
Retroviral constructs and generation of transduced cell lines.
HuNKG2D-expressing cells were produced by amplifying the huNKG2D ectodomain sequence from FNQEV… to …MQRTV, the muNKG2D ectodomain sequence from …KETFQ to …MKRAV, and the Ly49A transmembrane and intracellular domains from MSEQE… to …SVLAI, with primers such that the NKG2D N-termini overlapped with the Ly49A C terminus, and vice versa. These PCR products were PCR-spliced to produce chimeric NKG2D-Ly49A. For OMCP-expressing cells, OMCP was fused to the mature N terminus of Thy1.1. Full-length OMCPBR and OMCPMPX were amplified with primers encoding a C-terminal linker of Ser(Gly)4Ser. Thy1.1 was amplified from sequence QKVT… to …DFISL. Primers amplifying the OMCP C termini overlapped with the N-terminal Thy1.1 primer, and vice versa, allowing these amplicons to be spliced together. Full-length Thy1.1 was amplified from sequence MNPAI… to …DFISL. Amplicons were used to produce retrovirally transduced cell lines sorted to homogeneity for expression of the transgenes, as previously described (23).
Antibodies and antiserum.
The following antibodies were purchased from eBioscience: anti-CD3 PE-Cy5, anti-CD4 FITC, mouse IgG1, anti-muNKG2D–allophycocyanin (APC; clone CX5), and rat IgG1-APC. Anti-CD3 FITC, anti-CD8 PE, anti-CD3 CyChrome, anti-CD56 PE, anti-NK1.1 PE, and anti-huNKG2D (clone 1D11), and hamster IgG were purchased from BD Biosciences. Anti-muNKG2D (clone 3C7) has been previously described (29). Rabbit OMCPBR antiserum was obtained from Washington Biotechnology after immunization with refolded OMCPBR.
Viruses.
CPXV-BR and VV were obtained from American Type Culture Collection and were propagated in B-SC-1 cells using standard techniques.
Western blot analyses.
0.5 × 106 B-SC-1 cells/sample were infected with CPXV-BR and/or VV at 5 PFU/cell in 1 ml of medium. The supernatant was collected at 24 h, centrifuged at 13,000 g for 10 min, mixed with loading buffer, resolved by SDS-PAGE, and transferred to nitrocellulose using standard techniques. Membranes were incubated with 1:2,000 dilutions of OMCPBR antiserum, preimmune serum, or anti–mouse actin (Chemicon), followed by 0.5 μg/ml of horseradish peroxidase–labeled goat anti–rabbit Ig antibody and visualized with enhanced chemiluminescence (GE Healthcare).
Measurement of OMCP secretion was performed in sixwell plates by infection of 300,000 OP9 cells with CPXV-BR (multiplicity of infection = 5) for 20 h (>90% infected though intact cells at this point) in 0.85 ml of medium. Immunoblots of culture supernatants and recombinant OMCP standards were measured directly by fluorescence densitometry on a scanner (Typhoon Trio) using ImageQuant software (both from GE Healthcare) and uniform image rectangle background subtraction. Data of triplicate standards were fit to a linear model of antibody binding to blotted OMCP using Prism software (version 4; GraphPad) to obtain a standard line. Three experiments with independent standards and infections, each in triplicate, were plotted onto these lines to obtain the mean ± SD mass of OMCP secreted. Synthesis rate (copies per second) was obtained from secreted mass as follows: rate = mass × (NA)/[(20 h × 3,600 s/h) × (m.w.OMCP)].
Flow cytometry.
B6 splenocytes were RBC cleared and passed through a 70-μm mesh. Human PBMCs were obtained from healthy donors using Histopaque 1077 (Sigma-Aldrich). Cells were incubated with mAbs for 20 min on ice with gentle agitation after 10 min. For blocking experiments, cells were incubated with proteins (50 μg/ml mAbs, 9.5 μM MULT1, or 125 μM ULBP4) for 15 min before the addition of tetramers. Both monobiotinyl-BSA and West Nile virus glycoprotein DIII (30) were used for irrelevant control tetramers with identical results. Live cell gates were based on forward and side scatter plots and 7-amino-actinomycin D fluorescence. Data were collected on a FACSCalibur (Becton Dickinson) and analyzed with FloJo software (TreeStar, Inc.).
Chromium release assays.
B6 NK cells stimulated with IL-2 for 7 d were incubated for 10 min with media supplemented with nothing, MULT1, OMCPBR, or irrelevant recombinant protein (BSA or West Nile virus glycoprotein DIII with identical results) at the concentrations indicated in the figures. These were used in triplicate 51Cr killing assays, as previously described (23).
Acknowledgments
We thank Daved Fremont and Grant Nybakken for help with the seed secondary structural alignment, Michael Diamond for reading the paper and for the DIII expression plasmid, Mark Buller for monkeypox genomic DNA, Minji Byun for CPXV-BR DNA, Toshio Kitamura for the pMXs retroviral vector system, Darryl Higuchi for administrative support, and Chris Yee for excellent technical help.
J.A. Campbell is paid by National Heart, Lung, and Blood Institute training grant T32-HL07317 and a Kauffman Bioentrepreneurship award, D.S. Trossman was paid by the federal work-study program, W.M. Yokoyama is an investigator of the Howard Hughes Medical Institute; and L.N. Carayannopoulos is paid by National Institute of Allergy and Infectious Diseases grant K08 AI57361. This project was funded by National Institutes of Health grant U54 AI057160 to the Midwest Center of Excellence for Biodefense and Emerging Infectious Disease Research.
The authors have no conflicting financial interests.
References
- 1.Ogasawara, K., and L.L. Lanier. 2005. NKG2D in NK and T cell-mediated immunity. J. Clin. Immunol. 25:534–540. [DOI] [PubMed] [Google Scholar]
- 2.Krmpotic, A., D.H. Busch, I. Bubic, F. Gebhardt, H. Hengel, M. Hasan, A.A. Scalzo, U.H. Koszinowski, and S. Jonjic. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat. Immunol. 3:529–535. [DOI] [PubMed] [Google Scholar]
- 3.Welte, S.A., C. Sinzger, S.Z. Lutz, H. Singh-Jasuja, K.L. Sampaio, U. Eknigk, H.G. Rammensee, and A. Steinle. 2003. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur. J. Immunol. 33:194–203. [DOI] [PubMed] [Google Scholar]
- 4.Dunn, C., N.J. Chalupny, C.L. Sutherland, S. Dosch, P.V. Sivakumar, D.C. Johnson, and D. Cosman. 2003. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J. Exp. Med. 197:1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lodoen, M.B., G. Abenes, S. Umamoto, J.P. Houchins, F. Liu, and L.L. Lanier. 2004. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J. Exp. Med. 200:1075–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krmpotic, A., M. Hasan, A. Loewendorf, T. Saulig, A. Halenius, T. Lenac, B. Polic, I. Bubic, A. Kriegeskorte, E. Pernjak-Pugel, et al. 2005. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J. Exp. Med. 201:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenac, T., M. Budt, J. Arapovic, M. Hasan, A. Zimmermann, H. Simic, A. Krmpotic, M. Messerle, Z. Ruzsics, U.H. Koszinowski, et al. 2006. The herpesviral Fc receptor fcr-1 down-regulates the NKG2D ligands MULT-1 and H60. J. Exp. Med. 203:1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strong, R.K., and B.J. McFarland. 2004. NKG2D and Related Immunoreceptors. Adv. Protein Chem. 68:281–312. [DOI] [PubMed] [Google Scholar]
- 9.Arase, H., E.S. Mocarski, A.E. Campbell, A.B. Hill, and L.L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326. [DOI] [PubMed] [Google Scholar]
- 10.Smith, H.R., J.W. Heusel, I.K. Mehta, S. Kim, B.G. Dorner, O.V. Naidenko, K. Iizuka, H. Furukawa, D.L. Beckman, J.T. Pingel, et al. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. USA. 99:8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seet, B.T., J.B. Johnston, C.R. Brunetti, J.W. Barrett, H. Everett, C. Cameron, J. Sypula, S.H. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377–423. [DOI] [PubMed] [Google Scholar]
- 12.Hughey, R., and A. Krogh. 1996. Hidden Markov models for sequence analysis: extension and analysis of the basic method. Comput. Appl. Biosci. 12:95–107. [DOI] [PubMed] [Google Scholar]
- 13.Karplus, K., C. Barrett, and R. Hughey. 1998. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 14:846–856. [DOI] [PubMed] [Google Scholar]
- 14.Likos, A.M., S.A. Sammons, V.A. Olson, A.M. Frace, Y. Li, M. Olsen-Rasmussen, W. Davidson, R. Galloway, M.L. Khristova, M.G. Reynolds, et al. 2005. A tale of two clades: monkeypox viruses. J. Gen. Virol. 86:2661–2672. [DOI] [PubMed] [Google Scholar]
- 15.Chen, N., G. Li, M.K. Liszewski, J.P. Atkinson, P.B. Jahrling, Z. Feng, J. Schriewer, C. Buck, C. Wang, E.J. Lefkowitz, et al. 2005. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology. 340:46–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gubser, C., S. Hue, P. Kellam, and G.L. Smith. 2004. Poxvirus genomes: a phylogenetic analysis. J. Gen. Virol. 85:105–117. [DOI] [PubMed] [Google Scholar]
- 17.Shi, J., T.L. Blundell, and K. Mizuguchi. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310:243–257. [DOI] [PubMed] [Google Scholar]
- 18.O'Callaghan, C.A., A. Cerwenka, B.E. Willcox, L.L. Lanier, and P.J. Bjorkman. 2001. Molecular competition for NKG2D: H60 and RAE1 compete unequally for NKG2D with dominance of H60. Immunity. 15:201–211. [DOI] [PubMed] [Google Scholar]
- 19.Carayannopoulos, L.N., O.V. Naidenko, J. Kinder, E.L. Ho, D.H. Fremont, and W.M. Yokoyama. 2002. Ligands for murine NKG2D display heterogeneous binding behavior. Eur. J. Immunol. 32:597–605. [DOI] [PubMed] [Google Scholar]
- 20.Diefenbach, A., A.M. Jamieson, S.D. Liu, N. Shastri, and D.H. Raulet. 2000. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol. 1:119–126. [DOI] [PubMed] [Google Scholar]
- 21.Khodakevich, L., M. Szczeniowski, D. Manbu-ma-Disu, Z. Jezek, S. Marennikova, J. Nakano, and D. Messinger. 1987. The role of squirrels in sustaining monkeypox virus transmission. Trop. Geogr. Med. 39:115–122. [PubMed] [Google Scholar]
- 22.Chantrey, J., H. Meyer, D. Baxby, M. Begon, K.J. Bown, S.M. Hazel, T. Jones, W.I. Montgomery, and M. Bennett. 1999. Cowpox: reservoir hosts and geographic range. Epidemiol. Infect. 122:455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carayannopoulos, L.N., O.V. Naidenko, D.H. Fremont, and W.M. Yokoyama. 2002. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J. Immunol. 169:4079–4083. [DOI] [PubMed] [Google Scholar]
- 24.Karre, K., H.G. Ljunggren, G. Piontek, and R. Kiessling. 1986. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defense strategy. Nature. 319:675–678. [DOI] [PubMed] [Google Scholar]
- 25.Filion, R.J., and A.S. Popel. 2005. Intracoronary administration of FGF-2: a computational model of myocardial deposition and retention. Am. J. Physiol. Heart Circ. Physiol. 288:H263–H279. [DOI] [PubMed] [Google Scholar]
- 26.Francis, K., and B.O. Palsson. 1997. Effective intercellular communication distances are determined by the relative time constants for cyto/chemokine secretion and diffusion. Proc. Natl. Acad. Sci. USA. 94:12258–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groh, V., J. Wu, C. Yee, and T. Spies. 2002. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 419:734–738. [DOI] [PubMed] [Google Scholar]
- 28.Samuel, D., T.K. Kumar, G. Ganesh, G. Jayaraman, P.W. Yang, M.M. Chang, V.D. Trivedi, S.L. Wang, K.C. Hwang, D.K. Chang, and C. Yu. 2000. Proline inhibits aggregation during protein refolding. Protein Sci. 9:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho, E.L., L.N. Carayannopoulos, J. Poursine-Laurent, J. Kinder, B. Plougastel, H.R. Smith, and W.M. Yokoyama. 2002. Costimulation of multiple NK cell activation receptors by NKG2D. J. Immunol. 169:3667–3675. [DOI] [PubMed] [Google Scholar]
- 30.Nybakken, G.E., T. Oliphant, S. Johnson, S. Burke, M.S. Diamond, and D.H. Fremont. 2005. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 437:764–769. [DOI] [PMC free article] [PubMed] [Google Scholar]