Abstract
Recent studies have shown that activating mutations of NOTCH1 are responsible for the majority of T cell acute lymphoblastic leukemia (T-ALL) cases. Most of these mutations truncate its C-terminal domain, a region that is important for the NOTCH1 proteasome-mediated degradation. We report that the E3 ligase FBW7 targets NOTCH1 for ubiquitination and degradation. Our studies map in detail the amino acid degron sequence required for NOTCH1–FBW7 interaction. Furthermore, we identify inactivating FBW7 mutations in a large fraction of human T-ALL lines and primary leukemias. These mutations abrogate the binding of FBW7 not only to NOTCH1 but also to the two other characterized targets, c-Myc and cyclin E. The majority of the FBW7 mutations were present during relapse, and they were associated with NOTCH1 HD mutations. Interestingly, most of the T-ALL lines harboring FBW7 mutations were resistant to γ-secretase inhibitor treatment and this resistance appeared to be related to the stabilization of the c-Myc protein. Our data suggest that FBW7 is a novel tumor suppressor in T cell leukemia, and implicate the loss of FBW7 function as a potential mechanism of drug resistance in T-ALL.
T cell acute lymphoblastic leukemia (T-ALL) is a disease induced by malignant transformation of T lymphocytes that afflicts mainly children and adolescents (1). Although treatment outcome in T-ALL has improved in recent years, patients with relapsed disease continue to have dismal prognosis, despite the use of protocols involving hematopoietic stem cell transplantation. It is thus very important to identify and study the molecular pathways that control both induction of transformation and treatment resistance in this particular type of leukemia. Recent compelling evidence demonstrated that activating mutations in the NOTCH1 gene are the trigger for cell transformation in the majority of T-ALL patients.
Notch1 is a transmembrane receptor that controls the differentiation of multiple cell types, including cells of the immune system (2). More specifically, signaling through the NOTCH1 receptor orchestrates the development of T lym phocytes from uncommitted hematopoietic stem cells (3–5). Although NOTCH1 signaling is required for T cell development (6, 7), aberrant activation of the pathway can lead to disease; >50% of T-ALL cases harbor activating mutations in the NOTCH1 gene (8). Similar mutations were found in several mouse models of T cell leukemia (9–12). Very recent evidence showed that NOTCH1 pathway activation can induce multiple downstream signaling pathways and genes/targets, including the NF-kB pathway (13–15) and the transcription factor c-Myc (16–18).
The majority of the T-ALL NOTCH1-acti vating mutations truncate the C terminus of the protein, called the PEST domain because of the high frequency of P-E-S-T amino acids. As the PEST domain is believed to be essential for proteasome-dependent degradation of NOTCH1, these findings suggest that NOTCH1 protein stability could be an important regulator of intra cellular signaling thresholds and that abrogation of the NOTCH1 degradation machinery could predispose cells for transformation. The exact mechanisms by which the PEST domain confers instability to N1-IC remain unclear. Early genetic studies in Caenorhabditis elegans identified a candidate silencer of Notch, the E3 ubiquitin ligase SEL-10 (FBW7/hAGO/hCDC4, which is referred to hereafter as FBW7) (19). FBW7 is a potent tumor suppressor (20), targeting proteins such as cyclin E, c-Myc, and c-Jun for proteasomal degradation (21–23). Several reports have demonstrated that FBW7 can also target nuclear Notch1 and Notch4 for ubiquitination and can suppress Notch signaling (24, 25); however, the detailed biochemical nature and the in vivo physiological relevance in development and cell transformation of this interaction remain unknown. Moreover, Fbw7-deficient embryos die around day 10.5 because of defects in vascular development. Two such lines were simultaneously generated by different groups, and whole embryo lysates contained elevated levels of Notch1 protein in one case, but not the other (26, 27), raising questions about the importance of Fbw7 for the degradation of Notch1 or suggesting tissue-specific interactions between the two proteins.
We demonstrate that FBW7 targets NOTCH1, and per form a detailed mapping of their interaction by identifying a degron in the NOTCH1 PEST domain, which is centered on a conserved threonine, T2512. Alteration of this degron prevents FBW7-mediated ubiquitination of nuclear NOTCH1. Furthermore, we show that loss of FBW7 occurs frequently in T-ALL, as we identify FBW7 mutations in both T-ALL cell lines and primary tumors that carry NOTCH1-activating mutations. These mutations target specific residues in the FBW7-binding pocket and render the molecule in capable of binding NOTCH1, c-Myc, and cyclin E. FBW7 mutations usually pair with NOTCH1-HD mutations, suggesting a functional cooperation between the two events in the induction of NOTCH pathway activation. Finally, we demonstrate that there is an overrepresentation of FBW7 mutations in relapse patient samples, and that all T-ALL cell lines carrying FBW7 mutations are resistant to γ-secretase inhibitor treatment, suggesting that FBW7 inactivation could serve as a mechanism of treatment resistance, potentially caused by the stabilization of the downstream NOTCH1 effector c-Myc.
RESULTS
Physical and functional interaction between FBW7 and oncogenic NOTCH1 mutants
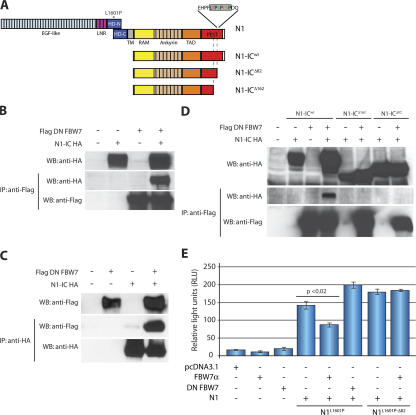
Fig. 1 A schematically displays the NOTCH1 and N1-IC proteins and maps the potential sites of the FBW7 interactions discussed in our article. To confirm that NOTCH1 and FBW7 physically interact, we performed coimmunoprecipitation studies using a dominant-negative (DN) FBW7 that contains the substrate-recognizing WD40 repeats of FBW7, but lacks the F-box domain that recruits ubiquitination machinery (Fig. 3 A). As shown in Fig. 1 (B and C), we detected both N1-IC and DN FBW7 in the same precipitated immunocomplex. Importantly, this interaction required an intact PEST domain, as two different N1-IC mutants lacking the C-terminal 162 or 82 amino acids (N1-ICΔ162 and N1-ICΔ82, respectively) failed to coimmunoprecipitate with DN FBW7 (Fig. 1 D).
Figure 1.
Physical and functional association between FBW7 and NOTCH1. (A) Schematic depiction of the NOTCH1 (N1) protein domains, and N1-IC WT (N1-ICWT) and 82 and 162 ΔPEST mutants (N1-ICΔ82 and N1-ICΔ162). Asterisk denotes L1601P mutation. The FBW7 degron is highlighted in gray. LNR, LIN-Notch repeat domain; RAM, RBJ-κ–associated module; TAD, transcriptional activation domain; PEST, P-E-S-T–rich domain; HD, heterodimerization domain. (B and C) Coimmunoprecipitation experiments using anti-Flag beads or anti-HA beads, showing interaction between DN FBW7 and N1-ICWT, but DN FBW7 (D) does not interact with N1-ICΔ82 and N1-ICΔ162. (E) Cotransfection with FBW7α repressed N1L1601P (P < 0.02), but not N1L1601P-Δ82, in reporter assays using a CSL luciferase reporter. pCDNA3.1 represents the control, “empty” expression vector. Error bars represent the SD of duplicate wells.
Figure 3.
FBW7 mutations in T-ALL preclude interaction with NOTCH1, c-Myc, and cyclin E. (A) Schematic depiction of the positions of the FBW7 binding pocket and DN FBW7. (B) Chromatograms showing mutations of FBW7 R465 and R505 found in CEM and P12-ICHIKAWA T-ALL cell lines. DND41 cells express WT FBW7. Arrows indicate mutated nucleotides. (C) Coimmunoprecipitation experiments demonstrating the inability of DN FBW7R465C, DN FBW7R479Q, and DN FBW7R505C to interact with N1-IC. (D and E) The same DN FBW7 mutants as in C do not interact with c-Myc and cyclin E. Asterisk in E indicates a nonspecific band.
We then assessed the functionality of this interaction by monitoring the ability of FBW7 (α isoform) to reduce the activation of a NOTCH1 (RBP-J/CSL)-dependent luciferase reporter. To induce NOTCH1 activation, we generated a full-length NOTCH1 construct that contains a leucine-to-proline substitution in the heterodimerization (HD) domain (N1L1601P). We and others have previously shown that this mutation found in T-ALL patients (8) can induce NOTCH1 activity (15). As evident from Fig. 1 E, FBW7α significantly (P < 0.02) blocked the activation induced by N1L1601P. Similar results were obtained using N1-IC (unpublished data). Cotransfection of DN FBW7 enhanced NOTCH1 activation (Fig. 1 E) and could antagonize the effect of the full-length FBW7α (not depicted). On the other hand, an L1601P mutant that also lacked the C-terminal 82 amino acids of the PEST domain (N1L1601P-Δ82) was unaffected by FBW7α. These observations confirm that FBW7 can target and suppress the activity of NOTCH1-HD mutants found in T-ALL, and suggest that PEST deletion is an efficient “escape” mechanism that promotes cell transformation.
A T2512-centered degron controls the interaction of NOTCH1 with FBW7
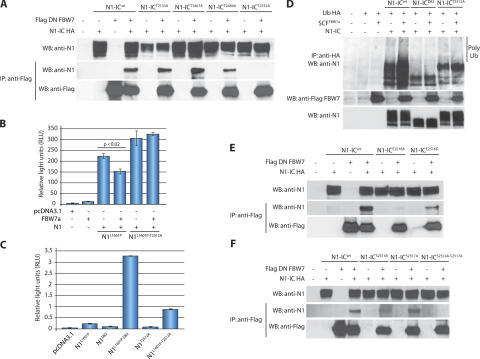
Specific F-box protein binding sites, termed degrons, have been characterized for several FBW7 substrates. In each confirmed case, the degron contains a crucial threonine residue that must be phosphorylated to allow recognition by FBW7. This in mind, we chose to analyze four threonine- containing potential degrons in N1-IC that could be targeted by FBW7. One site fell outside of the PEST domain (T2133), but was chosen because it was previously suggested as a potential GSK3-β phosphorylation site and FBW7 degron (18). The other three sites were within the PEST domain and included two potential phosphorylation sites for a proline-directed kinase (T2484 and T2512) and one nonproline-directed site (T2467). We generated four N1-IC constructs, in which one of these threonines was replaced with alanine (N1-ICT2133A, N1-ICT2467A, N1-ICT2484A, and N1-ICT2512A), and tested each one's ability to interact with DN FBW7. Of the four mutants, only N1-ICT2512A failed to coimmunoprecipitate with DN FBW7, suggesting that the N1-IC degron recognized by FBW7 depends on phosphorylation of T2512 (Fig. 2 A).
Figure 2.
Mutational analysis of the FBW7 NOTCH1 degron. (A) Coimmunoprecipitation experiment using threonine mutants of N1-IC (N1-ICT2133A, N1-ICT2467A, N1-ICT2484A, and N1-ICT2512A) and DN FBW7. Only N1-ICT2512A was unable to coimmunoprecipitate with DN FBW7. (B) Cell lysates from Bosc23 cells transfected with N1L1601P or N1L1601P-T2512A and FBW7α were used for luciferase reporter assays. N1L1601P-T2512A was not repressed by FBW7α. (C) Cell lysates from HeLa cells transfected with full-length N1 mutants were used for luciferase reporter assays. N1L1601P-Δ82 and N1L1601P-T2512A induced a 15- and 5-fold increase in transcriptional activity, respectively, compared with N1L1601P. (D) In vivo ubiquitination assay using Bosc23 cells transfected with N1-ICWT, N1-ICΔ82, or N1-ICT2512A with or without SCFFBW7α (SKP1, CUL1, ROC1, and FBW7α) and Ub-HA. SCFFBW7α enhanced polyubiquitination of N1-ICWT, but not of N1-ICΔ82 or N1-ICT2512A. (E) A negative charge is required at position 2516 (T+4). Interaction with DN FBW7 is partially preserved with N1-ICE2516D, but abolished with N1-ICE2516A. (F) Mutation of two CDK8 phosphorylation sites prevents N1-IC interaction with FBW7. A mutant lacking both S2514 and S2517 (N1-ICS2514A-S2517A) did not coimmunoprecipitate with DN FBW7. Error bars represent the SD of duplicate wells.
Next, we verified the functionality of these results using transcriptional reporter assays. In contrast to N1L1601P, FBW7α failed to suppress the activation of a NOTCH1-dependent luciferase reporter by N1L1601P that also contained a T2512A substitution (N1L1601P-T2512A; Fig. 2 B). Furthermore, DN FBW7 was unable to augment NOTCH1 activation induced by that construct in Bosc23 cells (unpublished data). Thus, mutation of a single residue (T2512) could recapitulate the effects of a ΔPEST mutation in this specific assay. Finally, we have previously hypothesized that loss of FBW7–NOTCH1 interactions could be a mechanism for enhancing NOTCH1-dependent transcriptional induction. To address this issue, we compared the activity of each mutant and found that N1L1601P-T2512A produced a five-fold increase in NOTCH1 activity in HeLa cells compared with N1L1601P (Fig. 2 C). We should note that the difference in N1L1601P-induced transcriptional activation in Fig. 2 (B and C) is caused by the utilization of two different cell lines for the luciferase reporter assays.
To confirm that N1-ICT2512A cannot be ubiquitinated by FBW7, we transfected Bosc23 cells with N1-ICWT, N1-ICΔ82, or N1-ICT2512A in the presence or absence of the SCFFBW7α complex. To preserve ubiquitin conjugates, cells were treated with the proteasome inhibitor MG132 6 h before lysis in a buffer containing N-ethyl maleimide. HA-ubiquitin immuno precipitates were further probed for NOTCH1 expression (Fig. 2 D). Cotransfection of SCFFBW7α increased polyubiquitination of N1-ICWT, but not N1-ICΔ82 or N1-ICT2512A. The point mutant retained a higher level of polyubiquitination compared with N1-ICΔ82 that was not enhanced by SCFFBW7α, suggesting that an additional E3 ligase can promote ubiquitination of N1-IC. This is consistent with a recent report of an FBW7–independent destabilizing element in the PEST domain (28). These experiments identify a conserved FBW7 degron in the N1-IC PEST domain and demonstrate its ability to alter NOTCH1 signaling.
Molecular analysis of the NOTCH1 FBW7 degron
FBW7 degrons in most other substrates contain a serine residue at the T+4 position. Welcker et al. have demonstrated that a negative charge at that position—either from a phospho-serine/threonine or an acidic residue—is required for recognition by FBW7 (29, 30). To determine if this is true for recognition of NOTCH1 by FBW7, we substituted E2516 with either alanine or aspartate (N1-ICE2516A and N1-ICE2516D, respectively). As shown in Fig. 2 E, N1-ICE2516A was unable to bind to DN FBW7; however, we observed partial binding with N1-ICE2516D. Thus, although glutamate appears to be required at T+4 for optimal binding, substituting a negatively charged aspartate at that position permits at least some interaction with FBW7.
The degron we identified in N1-IC contains serine-proline motifs at T+2 and T+5, either of which could be targeted by a proline-directed kinase. Indeed, both S2514 and S2517 have been previously suggested to be CDK8 phosphorylation sites (31). To evaluate their importance for recognition by FBW7, we generated N1-IC constructs in which one or both residues were replaced with alanine (N1-ICS2514A, N1-ICS2517A, and N1-ICS2514A-S2517A). Although mutation of S2517 did not affect binding, mutation of S2514 impaired it, and binding was completely abolished in the N1-IC mutant lacking both residues (Fig. 2 F). These studies demonstrate that the FBW7 degron in NOTCH1 requires a conserved charged residue and is potentially targeted by CDK8.
PEST mutations in primary T-ALL disrupt the FBW7 degron
To further test the hypothesis that mutations of the FBW7 degron (spanning from L2511 to P2519) could be a mechanism that promotes transformation in T-ALL, we have studied the pattern of PEST mutations found in T-ALL lines and primary tumors (8). PEST mutations appear to be of great importance, as they have been found in the majority of Notch1-induced T-ALL samples of both human and mouse origin. These mutations fell into three categories relative to the FBW7 degron. The majority of the mutations introduced translational termination codons starting as early as amino acid 2342 (sample T-ALL 44), and thus deleting almost the entire PEST domain. A second category included mutations that introduced termination codons immediately upstream of the L2511 residue (sample T-ALL 63). A third category of at least nine identified mutants included mutations that specifically targeted the FBW7 degron (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20070872/DC1). For example, the mutant found in the KOPTK1 T-ALL sample replaced the WT LTPSPESP degron with a mutated LTPSRVP sequence. These findings support our hypothesis and suggest that abrogation of the FBW7-controlled NOTCH1 degradation is an important transforming event in T-ALL.
T-ALL cell lines and primary tumors carry conserved FBW7 mutations
Our previous observations suggested an “indirect” way to inactivate FBW7 by deleting the degron on NOTCH1. As PEST mutations occur in a fraction of T-ALL tumors, we wondered whether selective pressure might exist for these cancers to develop FBW7 mutations that abrogate NOTCH1 recognition. Although FBW7 mutations underlie a wide variety of solid human cancers, it is unknown whether they occur in T-ALL. To address this, we sequenced the F-box and WD40 repeat regions of FBXW7 from 13 T-ALL cell lines. We found single-nucleotide substitutions that resulted in point mutations in 4 lines (30.8%, Table S1, available at http://www.jem.org/cgi/content/full/jem.20070872/DC1). We also found that one T-ALL line (Be13) does not express any FBW7 message, likely because of a chromosomal deletion near the FBXW7 locus (32). In addition to the cell lines, we sequenced FBXW7 from 95 primary T-ALL patient samples; 15 of these samples (16%) carried an FBW7 mutation (Table I). Further analysis revealed that 12% (7/58) of the diagnostic and 22% (8/37) of the relapse samples sequenced harbored FBW7 mutations. For 7 of those relapse samples, we were able to locate and sequence their diagnostic counterparts, and we discovered that 4 (57%) of these patients acquired the mutation only at relapse (Table I), suggesting that such mutations could serve as a mechanism of drug resistance in T-ALL.
Table I.
FBW7 is mutated in a high percentage of primary human T-ALL samples
| Patient number |
Diagnostic/relapse sample |
FBXW7 sequence NM_033632.2 |
Predicted amino acid change |
|---|---|---|---|
| 602-783 | Diagnosis | C1542T | R465C |
| 603-932 | Diagnosis | C1542T | R465C |
| 1 | Diagnosis | NA | NA |
| Relapse | C1542T (homozygous) | R465C | |
| 4 | Diagnosis | WT | WT |
| Relapse | C1542T | R465C | |
| 17 | Diagnosis | C1542T | R465C |
| Relapse | C1542T | R465C | |
| 16 | Diagnosis | G1543A | R465H |
| Relapse | G1543A | R465H | |
| 602-455 | Diagnosis | G1543A | R465H |
| 601-268 | Diagnosis | G1543A | R465H |
| 27 | Diagnosis | WT | WT |
| Relapse | G1543A | R465H | |
| 4451 | Diagnosis | G1585A | R479Q |
| 602-839 | Diagnosis | G1585A | R479Q |
| 603-473 | Diagnosis | G1585A | R479Q |
| 29 | Diagnosis | C1662T | R505C |
| Relapse | C1662T | R505C | |
| 22 | Diagnosis | WT | WT |
| Relapse | C1662T | R505C | |
| 35 | Diagnosis | WT | WT |
| Relapse | G1663T | R505L |
T-ALL FBW7 mutants lose their ability to bind NOTCH1, c-Myc, and cyclin E
All of the mutations identified affected one of three conserved arginine residues (R465, R479, and R505) in WD40 repeats III and IV of FBW7 (Table I and Fig. 3, A and B). These three residues (Fig. 3 A) comprise the binding pocket of FBW7 that permits substrate recognition, and all of them contact the phosphothreonine in the consensus degron (33). Nash et al. have shown that mutations of any of these three arginine residues prevent yeast Cdc4 from associating with cyclin E (34). To determine whether the NOTCH1 degron is recognized by FBW7 in the same manner, we generated DN FBW7 mutants in which R465, R479, or R505 was replaced with cysteine (DN FBW7R465C and DN FBW7R505C) or glutamine (DN FBW7R479Q) and tested their abilities to interact with N1-IC. We chose these substitutions because they recapitulated FBW7 mutations found in our T-ALL samples. All three FBW7 mutants failed to bind (Fig. 3 C) and ubiquitinate (not depicted) N1-IC. They also failed to bind to two other characterized FBW7 substrates, c-Myc and cyclin E (Fig. 3, D and E).
Analysis of the mutational status of the NOTCH1 locus in FBW7 mutants
Interestingly, subsequent analysis of the NOTCH1 locus demonstrated that none of the T-ALL lines bearing FBW7 mutations (0/5) had any NOTCH1 PEST mutations. Indeed, all of them had either NOTCH1-HD mutations (RPMI 8402, P12-ICHIKAWA, and CEM) or expressed WT NOTCH1 alleles (Be13 and Jurkat E6). Furthermore, all sequenced T-ALL lines carrying NOTCH1 PEST mutations expressed WT FBW7. These observations (Table S1) suggested that a NOTCH1 PEST mutation relieves the mutational “pressure” on the FBW7 gene. They also suggested that an FBW7 muta tion could be a mechanism of signal amplification for the NOTCH1-HD mutants that are weak transcriptional activators. This scenario was supported by our reporter analysis shown in Fig. 2 C, where introduction of a T2512 mutation abolishes FBW7 binding and induces a fivefold increase in NOTCH1 transcriptional activation. To further prove this hypothesis, we have sequenced the NOTCH1 locus in the previously described cohort of primary T-ALL patient samples. As evident from Table II, almost 43% (38/89) of the patient samples harbored NOTCH1 mutations. In agreement with our previous observations, none of the FBW7mut samples (0/15) had a PEST mutation. On the other hand, 60% (9/15) of the FBW7mut samples had NOTCH1-HD mutations. The remaining 40% of the FBW7mut samples had no NOTCH1 mutations. These results suggested that cooperation of FBW7 and NOTCH1 mutations can be found in a substantial fraction of T-ALL, and could be critical for disease progression and treatment outcome.
Table II.
Notch1 mutational analysis of the primary T-ALL patient samples
| Patient # | Notch1 | FBW7 | Patient # | Notch1 | FBW7 |
|---|---|---|---|---|---|
| 600–929 | WT | WT | 13a | WT | WT |
| 601–039 | PEST | WT | 14a | WT | WT |
| 601–068 | PEST | WT | 15a | TAD | WT |
| 601–268 | HD exon 26 | R465H | 16a | HD, exon 26 | R465H |
| 601–301 | PEST | WT | 17a | HD, exon 26 | R465C |
| 601–464 | WT | WT | 18a | WT | WT |
| 601–705 | WT | WT | 19a | WT | WT |
| 601–756 | HD exon 26, TAD | WT | 20a | TAD | WT |
| 1601–967 | WT | WT | 21a | WT | WT |
| 1601–991 | WT | WT | 22a | HD, exon 26 | R505C |
| 602–214 | WT | WT | 26a | HD, exon 26 | WT |
| 602–317 | PEST | WT | 27a | WT | R465H |
| 602–455 | WT | R465H | 28a | PEST | WT |
| 602–465 | HD exon 26 | WT | 29a | HD, exon 26 | R505C |
| 602–774 | TAD | WT | 30a | WT | WT |
| 602–787 | HD exon 26 | R465C | 31a | HD, exon 26 | WT |
| 602–818 | HD exon 26, PEST | WT | 33a | WT | WT |
| 602–839 | HD exon 27 | R479Q | 34 | PEST | WT |
| 603–007 | HD exon 26, PEST | WT | 34a | HD, exon 26, PEST | WT |
| 603–118 | WT | WT | 35 | WT | WT |
| 603–194 | HD exon 26 | WT | 35a | HD, exon 26 | R505L |
| 603–473 | WT | R479Q | 37a | WT | WT |
| 603–824 | WT | WT | 38a | WT | WT |
| 603–825 | WT | WT | 39a | WT | WT |
| 603–885 | PEST | WT | 40a | WT | WT |
| 603–888 | WT | WT | 41a | WT | WT |
| 603–895 | WT | WT | 42a | WT | WT |
| 603–932 | WT | R465C | 600–361 | WT | WT |
| 603–082 | PEST | WT | 600–619 | WT | WT |
| 604–099 | WT | WT | 600–745 | WT | WT |
| 604–198 | WT | WT | 600–971 | HD, exon 27 | WT |
| 604–436 | WT | WT | 600–484 | PEST | WT |
| 604–580 | WT | WT | 602–819 | PEST | WT |
| 604–618 | PEST | WT | 603–829 | WT | WT |
| 1a | HD, exon 26 | R465C | 600–903 | WT | WT |
| 2a | WT | WT | 2629 | WT | WT |
| 3a | PEST | WT | 8192 | WT | WT |
| 4a | WT | R465C | 8024 | HD, exon 26 | WT |
| 5a | WT | WT | 5833 | WT | WT |
| 6a | PEST | WT | 5182 | WT | WT |
| 7a | WT | WT | 747 | PEST | WT |
| 8a | WT | WT | 4451 | WT | R479Q |
| 9a | PEST | WT | 16a | HD, exon 26 | R465H |
| 10a | WT | WT | 17a | HD, exon 26 | R465C |
| 11a | WT | WT | 18a | WT | WT |
| 12a | WT | WT | 19a | WT | WT |
PEST, HD, and TAD denote the NOTCH1 domains where the mutations are located.
Restoration of FBW7 function decreases activation of the NOTCH1 pathway
If our hypothesis that FBW7 mutations in T-ALL lines contribute to NOTCH1-induced transformation is correct, then restoration of FBW7 expression should suppress NOTCH1 pathway activation in these cells. We thus used retroviral gene transfer to restore WT FBW7α expression in mutant T-ALL lines. Our initial observations, using both RT-PCR and immunofluorescence detection, was that Flag-FBW7α was efficiently expressed and had a characteristic nuclear localization (Fig. 4, A and B). To directly connect targeting of the NOTCH1 pathway by FBW7, we quantified the expression of two well-characterized direct transcriptional targets of NOTCH1, HES1 and DELTEX1 (17). As shown by real-time RT-PCR experiments (Fig. 4 C), expression of both genes was significantly (P < 0.001) decreased upon FBW7 expression. These observations demonstrate that restoration of FBW7 expression specifically targets the NOTCH1 pathway in T-ALL cells.
Figure 4.
Restoration of FBW7 expression in CEM cells (FBW7MUT) suppresses NOTCH1 target genes. (A and B) Efficient restoration of FBW7WT in CEM T-ALL cells after transduction with a Flag-FBW7α–expressing retrovirus (MigR1-FBW7α). Cells were infected with empty MigR1 vector as a control. (A) RT-PCR amplified Flag-FBW7α only in cells infected with MigR1-FBW7α. (B) Immunofluorescence staining of FACS-sorted CEM cells infected with MigR1-FBW7α using FITC-conjugated anti-Flag antibody shows nuclear localization of the construct. (C) Real-time RT-PCR was used to quantify the expression of the NOTCH1 target genes, DELTEX1, and HES1 in CEM cells infected as in A Infection with MigR1-FBW7α suppressed the two genes (P < 0.0001 for both). Error bars represent the SD of triplicate wells.
c-Myc stabilization in FBW7MUT as a mechanism of resistance to γ-secretase inhibition
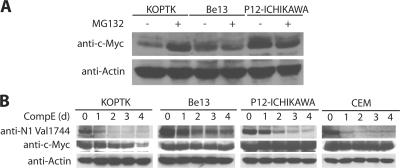
In line with the hypothesis that FBW7 mutations could be a novel mechanism of treatment resistance, we have seen that all T-ALL lines (5/5) bearing FBW7 mutations were resistant to treatment with γ-secretase inhibitors (GSIs; unpublished data) (8). The most attractive mechanism of resistance was the stabilization of c-Myc, as c-Myc was previously shown to be a direct target of NOTCH1 activation (16–18). Thus, we decided to study the regulation of c-Myc in T-ALL lines that bear FBW7 mutations. Initially, to demonstrate that c-Myc is regulated by FBW7 in T-ALL, we have shown that the mutated FBW7 proteins fail to interact with c-Myc (Fig. 3, D and E). Moreover, treatment of WT FBW7 (FBW7WT) T-ALL lines with the proteasome inhibitor MG132 for 6 h resulted in the accumulation of c-Myc caused by the inhibition of SCFFBW7-mediated proteolysis. On the other hand, similar treatment of mutant FBW7 (FBW7MUT) T-ALL lines showed no c-Myc protein accumulation (Fig. 5 A). To directly address the mechanism of γ-secretase inhibitor resistance in the FBW7MUT T-ALL lines, we treated KOPTK (FBW7WT), Be13 (FBW7MUT), P12-ICHICAWA (FBW7MUT), and CEM (FBW7MUT) cells with γ-secretase inhibitor (Compound E) for 1–4 d. As shown in Fig. 5 B, N1-IC protein expression was drastically and rapidly down-regulated in all four lines because the inhibition of γ-secretase–mediated cleavage. c-Myc protein declined after 1 d of Compound E treatment in KOPTK (FBW7WT) cells (note the ∼2-fold increase in protein loading in days 1–4 compared with day 0). In contrast, c-Myc protein levels remained stable in the 3 FBW7MUT lines during the 4 d of treatment. Together, these studies suggest that blocking NOTCH1 signaling alone cannot down-regulate c-Myc protein expression in the context of a coexisting FBW7 mutation. As a result, FBW7-inactivating mutations could provide an “escape” mechanism for GSI-treated T-ALLs caused by the stabilization of c-Myc.
Figure 5.
FBW7 mutations as a mechanism of resistance to γ-secretase inhibition. (A) T-ALL cell lines were treated with MG132 (20 μM for 6 h) and analyzed for expression of c-Myc and actin. Treatment with MG132 resulted in accumulation of c-Myc in KOPTK cells, which express WT FBW7, but not in Be13 and P12-ICHIKAWA cells, which express no and mutant FBW7, respectively. (B) Expression of N1-IC and c-Myc after treatment with Compound E (CompE) for 1–4 d. c-Myc expression declines only in KOPTK1 cells, which are sensitive to this treatment, but not in Be13 and P12-ICHIKAWA cells, which are resistant. N1-IC expression decreased in all of the cell lines, reflecting inhibition of γ-secretase–mediated cleavage of NOTCH1. Actin expression was used as a loading control for both A and B.
DISCUSSION
The discovery that hyperactivation of the NOTCH1 pathway contributes to the majority of T-ALL cases revolutionized both scientific and clinical perspectives in this field, and has spurred a search for NOTCH1-silencing therapeutic agents. At the core of this search is the need for a more complete understanding of NOTCH1 protein stability regulation, as the majority of the mutations affect proteasome-mediated degradation. We demonstrate that FBW7 targets NOTCH1 for ubiquitination and map in detail the amino acid degron sequence required for this interaction. Furthermore, we identify inactivating FBW7 mutations in ∼40% of human T-ALL lines and 16% of primary leukemias. These mutations abolish the ability of FBW7 to interact with its targets, including NOTCH1, c-Myc, and cyclin E.
Initially, using a series of amino acid point mutants, we have mapped the N1-IC–FBW7 interaction to a degron centered around T2512. We further analyzed the degron, demonstrating that a T+4 acidic glutamate is necessary for interaction with FBW7. The reason for this requirement is not clear. Possibly, a negative charge at position 2516 either facilitates the phosphorylation of T2512 or directly promotes interaction with FBW7. Current evidence suggests that phosphorylation of N1-IC by CDK8 plays a key role in recognition by FBW7. Work by Fryer et al. showed that MAML1 recruits CDK8 to the N1-IC transcriptional activation complex, and this kinase phosphorylates N1-IC to promote its degradation (31). Two of the three CDK8 phosphorylation sites reported in that study are within the T2512 degron we identified. Although mutation of either S2514 or S2517 by itself allowed at least some interaction with FBW7 (albeit compromised in the case of S2514), a N1-IC with both residues mutated lost all ability to bind FBW7. Together with the work of Fryer et al., our observations suggest that CDK8 must phosphorylate N1-IC (minimally at S2514) for it to be efficiently recognized by FBW7, either by priming the site for a different kinase that phosphorylates T2512, or by directly facilitating physical interaction with FBW7. We do not know the identity of the kinase that phosphorylates T2512; CDK8 might be responsible for this, too, but that possibility has not been directly tested. Although the N1-IC degron identified here is both similar to and different from degrons in other FBW7 substrates, it appears to occupy the canonical FBW7-binding pocket. Crystal structures of Cdc4 bound to a cyclin E peptide show that the phospho-T in the degron makes physical contact with three arginine residues, R465, R479, and R505, in WD40 repeats III and IV (numbering based on FBW7α isoform) (33). We have found that the FBW7 point mutations identified in T-ALL cell lines and primary leukemia affect these arginine residues, which is consistent with findings in solid human tumors (35, 36). It has been suggested that Fbw7 is a haploinsufficient tumor suppressor in p53+/− mice (37), which supports the idea that decreased gene dosage stabilizes FBW7 targets in vivo. However, it is also possible that these mutations act as DNs because their inabilities to interact with the NOTCH1 and c-Myc degrons might create a pool of nonfunctional SCFFBW7 complexes.
Although previously detected in certain solid tumors, FBW7-inactivating mutations do not appear to be a general oncogenic trigger in leukemia. Indeed, a recent sequencing of a large number of acute myeloid leukemia (AML) patient samples failed to detect any such mutations (38). Thus, it was a surprise to detect FBW7-inactivating mutations in both T-ALL lines and primary T-ALL samples. These findings suggested that FBW7 mutations could be frequent in T-ALL because of the ability of FBW7 to recognize, bind to, and degrade important oncogenes, including N1-IC and its transcriptional target c-Myc. Interestingly, it seems that FBW7 deficiency could either activate the NOTCH pathway in the absence of NOTCH1 mutations or amplify its signaling strength by cooperating with already existing NOTCH1-HD mutations. If loss of FBW7 is, indeed, a NOTCH1 signal amplifier that facilitates NOTCH1-induced transformation, one would predict that selective pressure to acquire an FBW7 mutation preferentially affects lymphocytes in which the NOTCH1 PEST domain remains intact, as no increase in NOTCH1 signaling capacity could be gained by eliminating FBW7 function in the context of an existing PEST deletion. In agreement with this hypothesis, none of the 13 T-ALL cell lines or 96 primary samples showed any coexisting FBW7 and NOTCH1 PEST mutations. On the other hand, 60% of FBW7mut samples had mutations in the NOTCH1-HD domain. These results suggest that, in T-ALL, the selective pressure to increase NOTCH1 stability and signaling strength is a primary force that drives oncogenesis. However, we do not want to imply that FBW7-mediated transformation depends solely on NOTCH1 activation. Both c-Myc and cyclin E could also be the mediators of this process. However, c-Myc is also a direct target of NOTCH signaling, so it is possible that FBW7 could impinge on both signaling pathways. The specific contribution and the importance of each FBW7 target in the induction of T-ALL remains to be analyzed using in vivo genetic approaches.
Moreover, we found that FBW7 mutations were more frequent in relapse samples and that more than half of the patients with FBW7 mutations at relapse did not carry the mutation at diagnosis, suggesting that FBW7 mutation might be an event that confers resistance to treatment. Interestingly, all T-ALL lines carrying FBW7 mutations were resistant to treatment with GSI. FBW7 is a very attractive treatment resistance candidate because of its ubiquitin ligase function and its ability to target c-Myc protein stability. c-Myc is a direct tran scriptional target of NOTCH1, and overexpression of c-Myc can rescue most T-ALL lines treated with GSI (16–18, 39). As c-Myc is a posttranslational target of FBW7 (40), but—at the same time—a transcriptional target of NOTCH activity (16–18), the transforming ability of FBW7 inactivation could be twofold; by losing FBW7 function, a cell should not only stabilize c-Myc but also produce more of it because of increased NOTCH1 signaling. In agreement with this hypothesis, we have shown that T-ALL FBW7 mutants lose their ability to recognize c-Myc. Also, our experiments demonstrate that c-Myc protein remains stable upon GSI treatment of lines carrying FBW7 inhibitors. Although these experiments strongly argue for a role for FBW7, further experimentation, preferably in vivo, is required to prove the role of FBW7 deficiency in GSI resistance. Moreover, it is possible that there are multiple resistance mechanisms, as we have found that a T-ALL line (PF382) was FBW7WT, yet resistant to GSI treatment.
We report the identification of FBW7 mutations in human T-ALL and their direct connection to the NOTCH1–c-Myc oncogenic pathway caused by the ability of FBW7 to recognize, bind, and ubiquitinate these target proteins. These mutations target conserved arginines that form the binding pocket of FBW7 and abolish its binding to its targets. We believe that the identification of FBW7 mutations is of unique importance for the design of future molecular therapies for T-ALL. Indeed, our data suggest that these treatments should be multifaceted, covering both NOTCH1 activation and potential escape mechanisms, such as FBW7 mutations, and their effects on the stability of downstream target proteins.
MATERIAL AND METHODS
FBW7 and NOTCH1 mutational analysis.
Genomic DNA extracted from cryopreserved lymphoblast samples provided by collaborating institutions in the United States (St. Jude Children's Research Hospital, Memphis, TN) and Canada (Hospital for Sick Children, Toronto, Canada). Analysis of FBW7 mutations in DNA samples from paired diagnostic and relapse T-ALLs was performed in samples from patients enrolled in Associazione Italiana Ematologia Oncologia Pediatrica–Berlin-Frankfurt-Munster Study Group protocols (41).
Recombinant DNA constructs and retrovirus production.
Expression vectors carrying Flag-tagged NOTCH1, NOTCH1L1601P-Δ82, and NOTCH1L1601P-T2512A mutants were created by multistep insertion of PCR-amplified human NOTCH1 sequences into pcDNA3.1 (Invitrogen). The ΔPEST mutants (Δ82 and Δ162) were generated by PCR and cloned into the pCS2-N1-IC HA. The Flag-tagged FBW7α and DN FBW7 expression plasmids were a gift from B. Clurman (Fred Hutchinson Cancer Research Center, Seattle, WA); the cyclin E, Ub-HA, and SCF (CUL1, ROC1, and SKP1) mammalian expression vectors were gifts from M. Pagano (New York University, New York, NY). The Flag-tagged DN FBW7 and N1-IC HA point mutants were generated by site-directed mutagenesis following the manufacturer's protocol (Stratagene). Integrity of all constructs was verified by sequencing and protein expression was confirmed by Western blotting.
Retroviral supernatants were generated as previously described (42), concentrated by 2.5 h centrifugation at 16,500 rpm, and resuspended in ∼200 μl of serum-free medium.
Western blot and coimmunoprecipitation.
Bosc23 cells were plated in 6-cm plates and transfected with expression plasmids using FuGENE 6 Transfection Reagent, according to the manufacturer's protocols (Roche). Cells were lysed using TENT buffer (50 mM Tris pH 8, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 1 mM PMSF, 1 mM NaF, 1 mM Na3VO4, and 0.2% protease inhibitor cocktail [Sigma-Aldrich]). Total cell extracts were incubated with HA or Flag beads and rocked for 1 h at 4°C. Bead-bound proteins were collected by centrifugation, and after being washed three times they were loaded in 8% SDS-based gels and analyzed by Western blotting using Flag M2 (Sigma-Aldrich), HA probe (Santa Cruz Biotechnology), and Notch1 (C20; Santa Cruz Biotechnology) antibodies. The T-ALL lines cultured at 37°C under 5% CO2 in RPMI 1640 supplemented with 10% FBS, penicillin, and streptomycin were lysed using TENT buffer after incubation with 1 μM Compound E or 20 μM MG132, as specified in the text. Total cell lysates were loaded and run in 12% SDS gels and analyzed by Western blot using antibody against Notch1 Val1744 (Cell Signaling Technology), c-Myc (N-262; Santa Cruz Biotechnology), cyclin E (HE12; Santa Cruz Biotechnology), and actin (CHEMICON International, Inc.).
RT-PCR, sequencing, and real-time RT-PCR analysis.
Total cellular RNA treated with DNaseI was extracted as described in the QIAGEN handbook (QIAGEN). cDNA was prepared according to the First Strand cDNA Synthesis kit protocol (Invitrogen). RT-PCR was performed in 25-μl reactions containing 1.5 μl cDNA, 1 μl Taq Red DNA Polymerase (Sigma-Aldrich), and 200 nM forward and reverse primers. For detection of Flag-FBW7α, the following primers were used: Flag forward 5′-TACAAGGATGACGATGACAAGC-3′ and FBW7α reverse 5′-CACTCTCCTGGTCCATCTCC-3′. For sequencing of FBW7 in human T-ALL lines, ∼1.2 kb of cDNA that encodes the FBW7 F-box and WD40 repeats domains was amplified using the FastStart High Fidelity PCR kit (Roche) according to manufacturer's instructions, using the following primers: FB2 forward 5′-AACAACTTTTGGGGACCTC-3′ and WD1 reverse 5′-ACACAACTCCCCCACTCCC-3′. PCR products were run on 1% Agarose gels, and the appropriate amplicon was purified and sent for direct bidirectional sequencing with the same primers. Sequencing of FBW7 from primary human samples was done by amplifying exons 9 and 10 from genomic DNA with the following primers: exon 9 forward 5′-TGATGGGATCATTTTATACGGATG-3′, exon 9 reverse 5′-GACAAAACGCTATGGCTTTCC-3′, exon 10 forward 5′-CCC AACTTCCCATTCCCTTA-3′, and exon 10 reverse 5′-TTTCTTCATGCCAATTTTAACG-3′. The amplicon was sequenced with the following primers: exon 9seq forward 5′-TTTAAATCACTTTTCCTTTCTACCC-3′ and exon 10seq forward 5′-TGACTAAATCTACCATGTTTTCTCA. Quantitative RT-PCR was performed in a 50-μl reaction containing 2 μl of cDNA, 12.5 μl of the SYBR Green SuperMix (Bio-Rad Laboratories), 200 nM forward and reverse primers. Real-time RT-PCR amplification was performed on the iCycler iQ detection system, and the data were collected and analyzed using iCycler iQ version 3.1 software (Bio-Rad Laboratories). The primers used were as follows: DELTEX1 forward 5′-TGGTCACAGCATCAGGCTAC-3′ and DELTEX1 reverse 5′-TGGTCTGGGTATCAGGGAAG-3′; HES1 forward 5′-TGAGCCAGCTGAAAACACTG-3′ and HES1 reverse 5′-CATTGATCTGGGTCATGCAG-3′; and ACTIN forward 5′-TCGTGCGTGACATTAAGGAG-3′ and ACTIN reverse 5′-AGC ACTGTGTTGGCGTACAG-3′.
Immunofluorescence analysis.
The CEM T-ALL line cells infected with FBW7α were washed three times with PBS and fixed in 4% paraformaldehyde in PBS for 15 min. The cell membrane was permeabilized by treatment with 0.5% Triton X-100 for 12 min. The cells were washed with PBS, and nonspecific antibody binding was blocked with 5% bovine serum albumin (Sigma-Aldrich). The cells were treated with anti–Flag-FITC antibody (Sigma-Aldrich) diluted 1:100 for 1 h. After washing with PBS, the cells were stained with 4,6-diamidino-2-phenylindole, and treated with a prolonged antifading medium (Invitrogen). Images were acquired on a confocal station (Leica) and analyzed using ImageJ (National Institutes of Health) software.
Luciferase assays.
Bosc23 or HeLa cells cultured at 37°C under 5% CO2 in IMDM supplemented with 10% FBS, penicillin, and streptomycin were seeded on 24-well plates at 8 × 104 cells per well and transiently transfected with expression constructs using FuGENE 6 Transfection Reagent (Roche), according to the manufacturer's protocols. Equivalent DNA concentrations were maintained by adding the appropriate amounts of pCDNA3.1 vector. Whole-cell lysates were prepared 48 h after transfection, and luciferase activity was determined using a Dual-Luciferase Reporter Assay System (Promega).
Online supplemental material.
Table S1 shows that FBW7 mutations are only present in human T-ALL cell lines with HD mutations or WT NOTCH1. Fig. S1 shows a comparative analysis of NOTCH1 PEST domain mutations found in human T-ALL lines and patients. The online version of this article is available at http://www.jem.org/cgi/content/full/jem.20070872/DC1.
Supplemental Material
Acknowledgments
We would like to thank Michele Pagano, Piers Nash, William Carroll, Elizabeth Raetz, and Jane Skok for discussions and technical guidance. We also thank Bruce Clurman and Tasuku Honjo for sharing materials, and Mark Minden and A. Thomas Look for valuable clinical samples and communicating unpublished results.
This work was supported by the Fondazione Città Della Speranza (G. Basso), the WOLF Foundation (A. Ferrando), and National Institutes of Health (NIH) grant CA120196 (A. Ferrando). I. Aifantis is supported by the Cancer Research Institute, the Sidney Kimmel Foundation for Cancer Research, the G&P Foundation for Cancer Research, the Penelope London Fund, the Friedman Fund for Childhood Leukemia, and NIH grant R01CA105129. S. Buonamici is supported by the New York University Molecular Oncology and Immunology Training grant. A. Ferrando is a Leukemia and Lymphoma Society Scholar.
The authors declare that they have no competing interests
Abbreviations used: AML, acute myeloid leukemia; DN, dominant-negative; GSI, γ-secretase inhibitors; HD, heterodimerization; T-ALL, T cell acute lymphoblastic leukemia.
B.J. Thompson and S. Buonamici contributed equally to this paper.
References
- 1.Grabher, C., H. von Boehmer, and A.T. Look. 2006. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 6:1–13. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas, S., M.D. Rand, and R.J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. [DOI] [PubMed] [Google Scholar]
- 3.Radtke, F., F. Schweisguth, and W. Pear. 2005. The Notch ‘gospel’. EMBO Rep. 6:1120–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maillard, I., T. Fang, and W.S. Pear. 2005. Regulation of lymphoid development, differentiation, and function by the notch pathway. Annu. Rev. Immunol. 23:945–974. [DOI] [PubMed] [Google Scholar]
- 5.Rothenberg, E.V., and T. Taghon. 2005. Molecular genetics of T cell development. Annu. Rev. Immunol. 23:601–649. [DOI] [PubMed] [Google Scholar]
- 6.Radtke, F., A. Wilson, G. Stark, M. Bauer, J. van Meerwijk, H.R. MacDonald, and M. Aguet. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. [DOI] [PubMed] [Google Scholar]
- 7.Ciofani, M., and J.C. Zuniga-Pflucker. 2005. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 6:881–888. [DOI] [PubMed] [Google Scholar]
- 8.Weng, A.P., A.A. Ferrando, W. Lee, J.P. Morris IV, L.B. Silverman, C. Sanchez-Irizarry, S.C. Blacklow, A.T. Look, and J.C. Aster. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271. [DOI] [PubMed] [Google Scholar]
- 9.Reschly, E.J., C. Spaulding, T. Vilimas, W.V. Graham, R.L. Brumbaugh, I. Aifantis, W.S. Pear, and B.L. Kee. 2006. Notch1 promotes survival of E2A-deficient T cell lymphomas through Pre-T cell receptor dependent and independent mechanisms. Blood. 107:4115–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumortier, A., R. Jeannet, P. Kirstetter, E. Kleinmann, M. Sellars, N.R. dos Santos, C. Thibault, J. Barths, J. Ghysdael, J.A. Punt, et al. 2006. Notch activation is an early and critical event during T-cell leukemogenesis in Ikaros-deficient mice. Mol. Cell. Biol. 26:209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin, Y.W., R.A. Nichols, J.J. Letterio, and P.D. Aplan. 2006. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 107:2540–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Neil, J., J. Calvo, K. McKenna, V. Krishnamoorthy, J.C. Aster, C.H. Bassing, F.W. Alt, M. Kelliher, and A.T. Look. 2006. Activating Notch1 mutations in mouse models of T-ALL. Blood. 107:781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin, H.M., L.M. Minter, O.H. Cho, S. Gottipati, A.H. Fauq, T.E. Golde, G.E. Sonenshein, and B.A. Osborne. 2006. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 25:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilimas, T., J. Mascarenhas, T. Palomero, M. Mandal, S. Buonamici, F. Meng, B. Thompson, C. Spaulding, S. Macaroun, M.L. Alegre, et al. 2006. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat. Med. 13:70–77 [DOI] [PubMed] [Google Scholar]
- 15.Vilimas, T., J. Mascarenhas, T. Palomero, M. Mandal, S. Buonamici, F. Meng, B. Thompson, C. Spaulding, S. Macaroun, M.L. Alegre, et al. 2007. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat. Med. 13:70–77. [DOI] [PubMed] [Google Scholar]
- 16.Weng, A.P., J.M. Millholland, Y. Yashiro-Ohtani, M.L. Arcangeli, A. Lau, C. Wai, C. Del Bianco, C.G. Rodriguez, H. Sai, J. Tobias, et al. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20:2096–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palomero, T., W.K. Lim, D.T. Odom, M.L. Sulis, P.J. Real, A. Margolin, K.C. Barnes, J. O'Neil, D. Neuberg, A.P. Weng, et al. 2006. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA. 103:18261–18266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klinakis, A., M. Szabolcs, K. Politi, H. Kiaris, S. Artavanis-Tsakonas, and A. Efstratiadis. 2006. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl. Acad. Sci. USA. 103:9262–9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard, E.J., G. Wu, J. Kitajewski, and I. Greenwald. 1997. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11:3182–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama, K.I., and K. Nakayama. 2005. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin. Cell Dev. Biol. 16:323–333. [DOI] [PubMed] [Google Scholar]
- 21.Welcker, M., A. Orian, J. Jin, J.E. Grim, J.W. Harper, R.N. Eisenman, and B.E. Clurman. 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA. 101:9085–9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei, W., J. Jin, S. Schlisio, J.W. Harper, and W.G. Kaelin Jr. 2005. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 8:25–33. [DOI] [PubMed] [Google Scholar]
- 23.Koepp, D.M., L.K. Schaefer, X. Ye, K. Keyomarsi, C. Chu, J.W. Harper, and S.J. Elledge. 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 294:173–177. [DOI] [PubMed] [Google Scholar]
- 24.Gupta-Rossi, N., O. Le Bail, H. Gonen, C. Brou, F. Logeat, E. Six, A. Ciechanover, and A. Israel. 2001. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 276:34371–34378. [DOI] [PubMed] [Google Scholar]
- 25.Oberg, C., J. Li, A. Pauley, E. Wolf, M. Gurney, and U. Lendahl. 2001. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276:35847–35853. [DOI] [PubMed] [Google Scholar]
- 26.Tetzlaff, M.T., W. Yu, M. Li, P. Zhang, M. Finegold, K. Mahon, J.W. Harper, R.J. Schwartz, and S.J. Elledge. 2004. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proc. Natl. Acad. Sci. USA. 101:3338–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsunematsu, R., K. Nakayama, Y. Oike, M. Nishiyama, N. Ishida, S. Hatakeyama, Y. Bessho, R. Kageyama, T. Suda, and K.I. Nakayama. 2004. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 279:9417–9423. [DOI] [PubMed] [Google Scholar]
- 28.Chiang, M.Y., M.L. Xu, G. Histen, O. Shestova, M. Roy, Y. Nam, S.C. Blacklow, D.B. Sacks, W.S. Pear, and J.C. Aster. 2006. Identification of a conserved negative regulatory sequence that influences the leukemogenic activity of NOTCH1. Mol. Cell. Biol. 26:6261–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welcker, M., J. Singer, K.R. Loeb, J. Grim, A. Bloecher, M. Gurien-West, B.E. Clurman, and J.M. Roberts. 2003. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell. 12:381–392. [DOI] [PubMed] [Google Scholar]
- 30.Welcker, M., and B.E. Clurman. 2005. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. J. Biol. Chem. 280:7654–7658. [DOI] [PubMed] [Google Scholar]
- 31.Fryer, C.J., J.B. White, and K.A. Jones. 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell. 16:509–520. [DOI] [PubMed] [Google Scholar]
- 32.Galili, U., A. Peleg, Y. Milner, and N. Galili. 1984. Be13, a human T-leukemia cell line highly sensitive to dexamethasone-induced cytolysis. Cancer Res. 44:4594–4601. [PubMed] [Google Scholar]
- 33.Orlicky, S., X. Tang, A. Willems, M. Tyers, and F. Sicheri. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 112:243–256. [DOI] [PubMed] [Google Scholar]
- 34.Nash, P., X. Tang, S. Orlicky, Q. Chen, F.B. Gertler, M.D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 414:514–521. [DOI] [PubMed] [Google Scholar]
- 35.Kemp, Z., A. Rowan, W. Chambers, N. Wortham, S. Halford, O. Sieber, N. Mortensen, A. von Herbay, T. Gunther, M. Ilyas, and I. Tomlinson. 2005. CDC4 mutations occur in a subset of colorectal cancers but are not predicted to cause loss of function and are not associated with chromosomal instability. Cancer Res. 65:11361–11366. [DOI] [PubMed] [Google Scholar]
- 36.Strohmaier, H., C.H. Spruck, P. Kaiser, K.A. Won, O. Sangfelt, and S.I. Reed. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 413:316–322. [DOI] [PubMed] [Google Scholar]
- 37.Mao, J.H., J. Perez-Losada, D. Wu, R. Delrosario, R. Tsunematsu, K.I. Nakayama, K. Brown, S. Bryson, and A. Balmain. 2004. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 432:775–779. [DOI] [PubMed] [Google Scholar]
- 38.Nowak, D., M. Mossner, C.D. Baldus, O. Hopfer, E. Thiel, and W.K. Hofmann. 2006. Mutation analysis of hCDC4 in AML cells identifies a new intronic polymorphism. Int J Med Sci. 3:148–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma, V.M., J.A. Calvo, K.M. Draheim, L.A. Cunningham, N. Hermance, L. Beverly, V. Krishnamoorthy, M. Bhasin, A.J. Capobianco, and M.A. Kelliher. 2006. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol. Cell. Biol. 26:8022–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minella, A.C., M. Welcker, and B.E. Clurman. 2005. Ras activity regulates cyclin E degradation by the Fbw7 pathway. Proc. Natl. Acad. Sci. USA. 102:9649–9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Germano, G., L. del Giudice, L. Lo Nigro, K. Polato, E. Giarin, M. Paganin, and G. Basso. 2005. Comparative sequence analysis of incomplete DJH and TCR gene rearrangements in children with relapses of T-ALL. Leukemia. 19:1687–1689. [DOI] [PubMed] [Google Scholar]
- 42.Ory, D.S., B.A. Neugeboren, and R.C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA. 93:11400–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.