Abstract
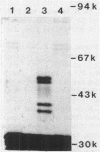
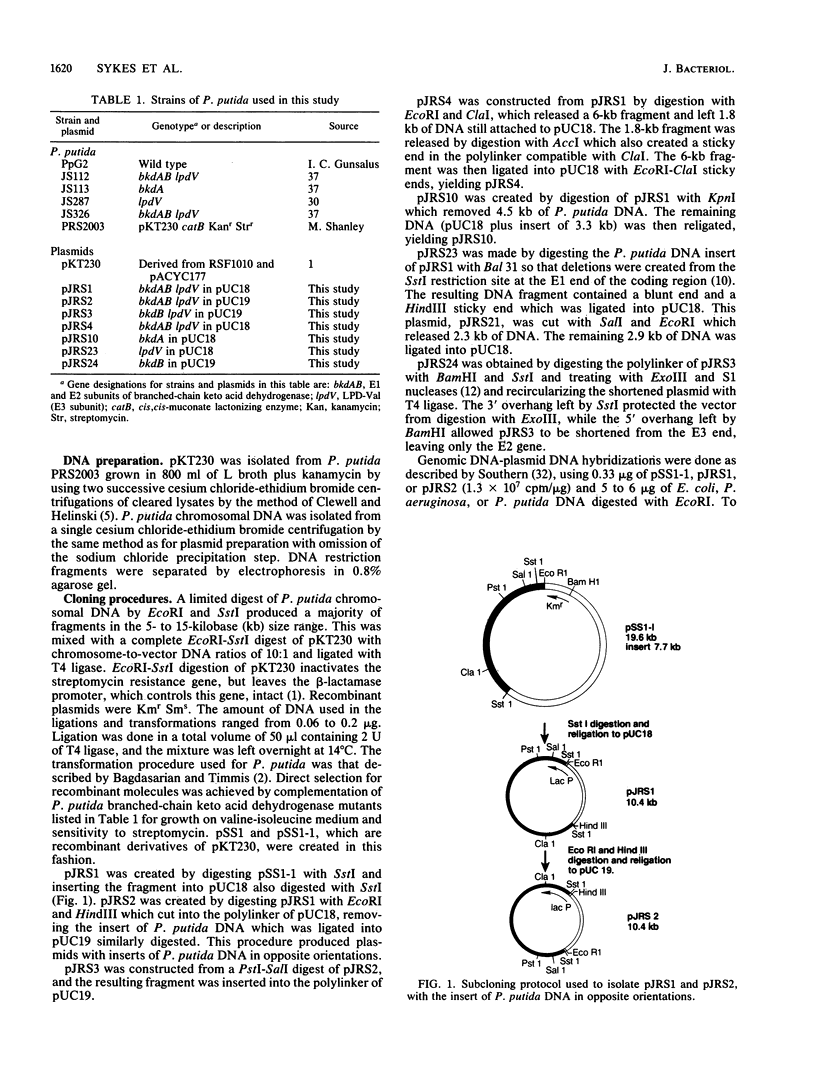
We cloned the structural genes for the individual subunits of the branched-chain keto acid dehydrogenase multienzyme complex on a 7.8-kilobase EcoRI-SstI restriction fragment of Pseudomonas putida chromosomal DNA by cloning into the broad-host-range vector pKT230. A direct selection system for growth on valine-isoleucine agar was achieved by complementation of P. putida branched-chain keto acid dehydrogenase mutants. The recombinant plasmid, pSS1-1, increased expression of branched-chain keto acid dehydrogenase up to five times in wild-type P. putida. The complex was expressed constitutively in P. putida(pSS1-1) but was inducible in Escherichia coli HB101(pSS1-1) by high valine. E. coli minicells transformed with pSS1-1 produced three polypeptides which did not match the four polypeptides of the purified complex. To resolve this problem, we inserted P. putida DNA from pSS1-1 into pUC18 and pUC19. The pUC-derived plasmids were used as DNA templates in an E. coli transcription-translation system. Four polypeptides were produced from the pUC18-derived plasmid which had the correct molecular weights, showing that the structural genes had been cloned. Since only weak bands were produced with the pUC19-derived plasmid, the direction of transcription was established. The locations and order of all the structural genes of branched-chain keto acid dehydrogenase were located by restriction enzyme mapping.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Clark-Curtiss J. E., Curtiss R., 3rd Analysis of recombinant DNA using Escherichia coli minicells. Methods Enzymol. 1983;101:347–362. doi: 10.1016/0076-6879(83)01026-5. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Cook K. G., Lawson R., Yeaman S. J. Multi-site phosphorylation of bovine kidney branched-chain 2-oxoacid dehydrogenase complex. FEBS Lett. 1983 Jun 27;157(1):59–62. doi: 10.1016/0014-5793(83)81116-8. [DOI] [PubMed] [Google Scholar]
- Darlison M. G., Spencer M. E., Guest J. R. Nucleotide sequence of the sucA gene encoding the 2-oxoglutarate dehydrogenase of Escherichia coli K12. Eur J Biochem. 1984 Jun 1;141(2):351–359. doi: 10.1111/j.1432-1033.1984.tb08199.x. [DOI] [PubMed] [Google Scholar]
- Delaney R., Burns G., Sokatch J. R. Relationship of lipoamide dehydrogenases from Pseudomonas putida to other FAD-linked dehydrogenases. FEBS Lett. 1984 Mar 26;168(2):265–270. doi: 10.1016/0014-5793(84)80259-8. [DOI] [PubMed] [Google Scholar]
- Fatania H. R., Patston P. A., Randle P. J. Dephosphorylation and reactivation of phosphorylated purified ox-kidney branched-chain dehydrogenase complex by co-purified phosphatase. FEBS Lett. 1983 Jul 25;158(2):234–238. doi: 10.1016/0014-5793(83)80585-7. [DOI] [PubMed] [Google Scholar]
- Guest J. R. Aspects of the molecular biology of lipoamide dehydrogenase. Adv Neurol. 1978;21:219–244. [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Henning U., Dennert G., Hertel R., Shipp W. S. Translation of the structural genes of the E. coli pyruvate dehydrogenase complex. Cold Spring Harb Symp Quant Biol. 1966;31:227–234. doi: 10.1101/sqb.1966.031.01.031. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Litwer S., Danner D. J. Identification of a cDNA clone in lambda gt11 for the transacylase component of branched chain ketoacid dehydrogenase. Biochem Biophys Res Commun. 1985 Sep 16;131(2):961–967. doi: 10.1016/0006-291x(85)91333-6. [DOI] [PubMed] [Google Scholar]
- Lowe P. N., Hodgson J. A., Perham R. N. Dual role of a single multienzyme complex in the oxidative decarboxylation of pyruvate and branched-chain 2-oxo acids in Bacillus subtilis. Biochem J. 1983 Oct 1;215(1):133–140. doi: 10.1042/bj2150133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B. Genetic control of nitrogen assimilation in bacteria. Annu Rev Genet. 1982;16:135–168. doi: 10.1146/annurev.ge.16.120182.001031. [DOI] [PubMed] [Google Scholar]
- Marshall V. D., Sokatch J. R. Regulation of valine catabolism in Pseudomonas putida. J Bacteriol. 1972 Jun;110(3):1073–1081. doi: 10.1128/jb.110.3.1073-1081.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. R., Marshall V. D., Sokatch J. R., Unger L. Common enzymes of branched-chain amino acid catabolism in Pseudomonas putida. J Bacteriol. 1973 Jul;115(1):198–204. doi: 10.1128/jb.115.1.198-204.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCully V., Burns G., Sokatch J. R. Resolution of branched-chain oxo acid dehydrogenase complex of Pseudomonas aeruginosa PAO. Biochem J. 1986 Feb 1;233(3):737–742. doi: 10.1042/bj2330737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odessey R. Reversible ATP-induced inactivation of branched-chain 2-oxo acid dehydrogenase. Biochem J. 1980 Oct 15;192(1):155–163. doi: 10.1042/bj1920155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker P. J., Randle P. J. Inactivation of rat heart branched-chain 2-oxoacid dehydrogenase complex by adenosine triphosphate. FEBS Lett. 1978 Nov 1;95(1):153–156. doi: 10.1016/0014-5793(78)80072-6. [DOI] [PubMed] [Google Scholar]
- Paxton R., Harris R. A. Isolation of rabbit liver branched chain alpha-ketoacid dehydrogenase and regulation by phosphorylation. J Biol Chem. 1982 Dec 10;257(23):14433–14439. [PubMed] [Google Scholar]
- Pettit F. H., Yeaman S. J., Reed L. J. Purification and characterization of branched chain alpha-keto acid dehydrogenase complex of bovine kidney. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4881–4885. doi: 10.1073/pnas.75.10.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts J. R., Clarke P. H. The effect of nitrogen limitation on catabolite repression of amidase, histidase and urocanase in Pseudomonas aeruginosa. J Gen Microbiol. 1976 Apr;93(2):377–387. doi: 10.1099/00221287-93-2-377. [DOI] [PubMed] [Google Scholar]
- Sokatch J. R., Burns G. Oxidation of glycine by Pseudomonas putida requires a specific lipoamide dehydrogenase. Arch Biochem Biophys. 1984 Feb 1;228(2):660–666. doi: 10.1016/0003-9861(84)90036-5. [DOI] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Gebrosky J., Sokatch D. J. Isolation of a specific lipoamide dehydrogenase for a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):639–646. doi: 10.1128/jb.148.2.639-646.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Roberts C. M. Purification of a branched-chain keto acid dehydrogenase from Pseudomonas putida. J Bacteriol. 1981 Nov;148(2):647–652. doi: 10.1128/jb.148.2.647-652.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokatch J. R., McCully V., Sahm J. G., Reyes-Maguire M. Mutations affecting lipoamide dehydrogenases of Pseudomonas putida. J Bacteriol. 1983 Feb;153(2):969–975. doi: 10.1128/jb.153.2.969-975.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spencer M. E., Darlison M. G., Stephens P. E., Duckenfield I. K., Guest J. R. Nucleotide sequence of the sucB gene encoding the dihydrolipoamide succinyltransferase of Escherichia coli K12 and homology with the corresponding acetyltransferase. Eur J Biochem. 1984 Jun 1;141(2):361–374. doi: 10.1111/j.1432-1033.1984.tb08200.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983 Jun 1;133(1):155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Lewis H. M., Darlison M. G., Guest J. R. Nucleotide sequence of the lipoamide dehydrogenase gene of Escherichia coli K12. Eur J Biochem. 1983 Oct 3;135(3):519–527. doi: 10.1111/j.1432-1033.1983.tb07683.x. [DOI] [PubMed] [Google Scholar]
- Sykes P. J., Menard J., McCully V., Sokatch J. R. Conjugative mapping of pyruvate, 2-ketoglutarate, and branched-chain keto acid dehydrogenase genes in Pseudomonas putida mutants. J Bacteriol. 1985 Apr;162(1):203–208. doi: 10.1128/jb.162.1.203-208.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]