Abstract
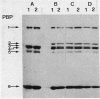
The penicillin-binding proteins (PBPs) of 209 cell division (or growth) temperature-sensitive mutants of Streptococcus faecium were analyzed in this study. A total of nine strains showed either constitutive or temperature-sensitive conditional damage in the PBPs. Analysis of these nine strains yielded the following results: one carried a PBP 1 constitutively showing a lower molecular weight; one constitutively lacked PBP 2; two lacked PBP 3 at 42 degrees C, but not at 30 degrees C; one was normal at 30 degrees C but at 42 degrees C lacked PBP 3 and overproduced PBP 5; two were normal at 42 degrees C and lacked PBP 5 at 30 degrees C; one constitutively lacked PBP 5; and one carried a PBP 6 constitutively split in two bands. The mutant lacking PBP 3 and overproducing PBP 5 continued to grow at 42 degrees C for 150 min and then lysed. Revertants selected for growth capability at 42 degrees C from the mutants altered in PBPs 5 and 6 maintained the same PBP alterations, while those isolated from the strains with altered PBP 1 or lacking PBP 2 or PBP 3 showed a normal PBP pattern. Penicillin-resistant derivatives were isolated at 30 degrees C from the mutants lacking PBP 2 and from that lacking PBP 3. All these derivatives continued to show the same PBP damage as the parents, but overproduced PBP 5 and grew at 42 degrees C. These findings indicate that high-molecular-weight, but not low-molecular-weight, PBPs are essential for cell growth in S. faecium. This is in complete agreement with previous findings obtained with a different experimental system. On the basis of both previous and present data it is suggested that PBPs 1, 2, and 3 appear necessary for cell growth at optimal temperature (and at maximal rate), but not for cell growth at a submaximal one (or at a reduced rate), and an overproduced PBP 5 is capable of taking over the function of PBPs 1, 2, and 3.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boothby D., Daneo-Moore L., Shockman G. D. A rapid, guantitative, and selective estimation of radioactively labeled peptidoglycan in gram-positive bacteria. Anal Biochem. 1971 Dec;44(2):645–653. doi: 10.1016/0003-2697(71)90255-7. [DOI] [PubMed] [Google Scholar]
- Canepari P., Lleò M. M., Cornaglia G., Fontana R., Satta G. In Streptococcus faecium penicillin-binding protein 5 alone is sufficient for growth at sub-maximal but not at maximal rate. J Gen Microbiol. 1986 Mar;132(3):625–631. doi: 10.1099/00221287-132-3-625. [DOI] [PubMed] [Google Scholar]
- Canepari P., Lléo M. M., Fontana R., Satta G., Shockman G. D., Daneo-Moore L. Division of temperature-sensitive Streptococcus faecium mutants after return to the permissive temperature. J Bacteriol. 1984 Oct;160(1):427–429. doi: 10.1128/jb.160.1.427-429.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canepari P., Lléo M. M., Satta G., Fontana R., Shockman G. D., Daneo-Moore L. Division blocks in temperature-sensitive mutants of Streptococcus faecium (S. faecalis ATCC 9790). J Bacteriol. 1983 Dec;156(3):1046–1051. doi: 10.1128/jb.156.3.1046-1051.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyette J., Ghuysen J. M., Fontana R. The penicillin-binding proteins in Streptococcus faecalis ATCC 9790. Eur J Biochem. 1980 Sep;110(2):445–456. doi: 10.1111/j.1432-1033.1980.tb04886.x. [DOI] [PubMed] [Google Scholar]
- Fontana R., Canepari P., Satta G., Coyette J. Identification of the lethal target of benzylpenicillin in Streptococcus faecalis by in vivo penicillin binding studies. Nature. 1980 Sep 4;287(5777):70–72. doi: 10.1038/287070a0. [DOI] [PubMed] [Google Scholar]
- Fontana R., Canepari P., Satta G., Coyette J. Streptococcus faecium ATCC 9790 penicillin-binding proteins and penicillin sensitivity are heavily influenced by growth conditions: proposal for an indirect mechanism of growth inhibition by beta-lactams. J Bacteriol. 1983 May;154(2):916–923. doi: 10.1128/jb.154.2.916-923.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana R., Cerini R., Longoni P., Grossato A., Canepari P. Identification of a streptococcal penicillin-binding protein that reacts very slowly with penicillin. J Bacteriol. 1983 Sep;155(3):1343–1350. doi: 10.1128/jb.155.3.1343-1350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana R., Grossato A., Rossi L., Cheng Y. R., Satta G. Transition from resistance to hypersusceptibility to beta-lactam antibiotics associated with loss of a low-affinity penicillin-binding protein in a Streptococcus faecium mutant highly resistant to penicillin. Antimicrob Agents Chemother. 1985 Nov;28(5):678–683. doi: 10.1128/aac.28.5.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles A. F., Reynolds R. E. Bacillus megaterium resistance to cloxacillin accompanied by a compensatory change in penicillin binding proteins. Nature. 1979 Jul 12;280(5718):167–168. doi: 10.1038/280167a0. [DOI] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984 May;158(2):513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaya M., Goldman R., Tipper D. J., Feingold B., Strominger J. L. Morphology of an Escherichia coli mutant with a temperature-dependent round cell shape. J Bacteriol. 1978 Dec;136(3):1143–1158. doi: 10.1128/jb.136.3.1143-1158.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Suzuki H., Hirota Y. Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol Gen Genet. 1985;200(2):272–277. doi: 10.1007/BF00425435. [DOI] [PubMed] [Google Scholar]
- Kleppe G., Yu W., Strominger J. L. Penicillin-binding proteins in Bacillus subtilis mutants. Antimicrob Agents Chemother. 1982 Jun;21(6):979–983. doi: 10.1128/aac.21.6.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci M. J., Hinks E. T., Dicker D. T., Higgins M. L., Daneo-Moore L. Inhibition of beta-lactam antibiotics at two different times in the cell cycle of Streptococcus faecium ATCC 9790. J Bacteriol. 1986 Mar;165(3):682–688. doi: 10.1128/jb.165.3.682-688.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi L., Tonin E., Cheng Y. R., Fontana R. Regulation of penicillin-binding protein activity: description of a methicillin-inducible penicillin-binding protein in Staphylococcus aureus. Antimicrob Agents Chemother. 1985 May;27(5):828–831. doi: 10.1128/aac.27.5.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater M., Schaechter M. Control of cell division in bacteria. Bacteriol Rev. 1974 Jun;38(2):199–221. doi: 10.1128/br.38.2.199-221.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Deletion of the penicillin-binding protein 5 gene of Escherichia coli. J Bacteriol. 1980 Dec;144(3):1190–1192. doi: 10.1128/jb.144.3.1190-1192.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977 Jul;131(1):293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Nishimura Y., Hirota Y. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci U S A. 1978 Feb;75(2):664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Nakajima S., Matsuhashi M. Thermosensitive mutation in Escherichia coli simultaneously causing defects in penicillin-binding protein-1Bs and in enzyme activity for peptidoglycan synthesis in vitro. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5472–5476. doi: 10.1073/pnas.74.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A. Penicillin-binding proteins in bacteria. Ann Intern Med. 1982 Apr;96(4):502–504. doi: 10.7326/0003-4819-96-4-502. [DOI] [PubMed] [Google Scholar]
- Waxman D. J., Strominger J. L. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu Rev Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]