Abstract
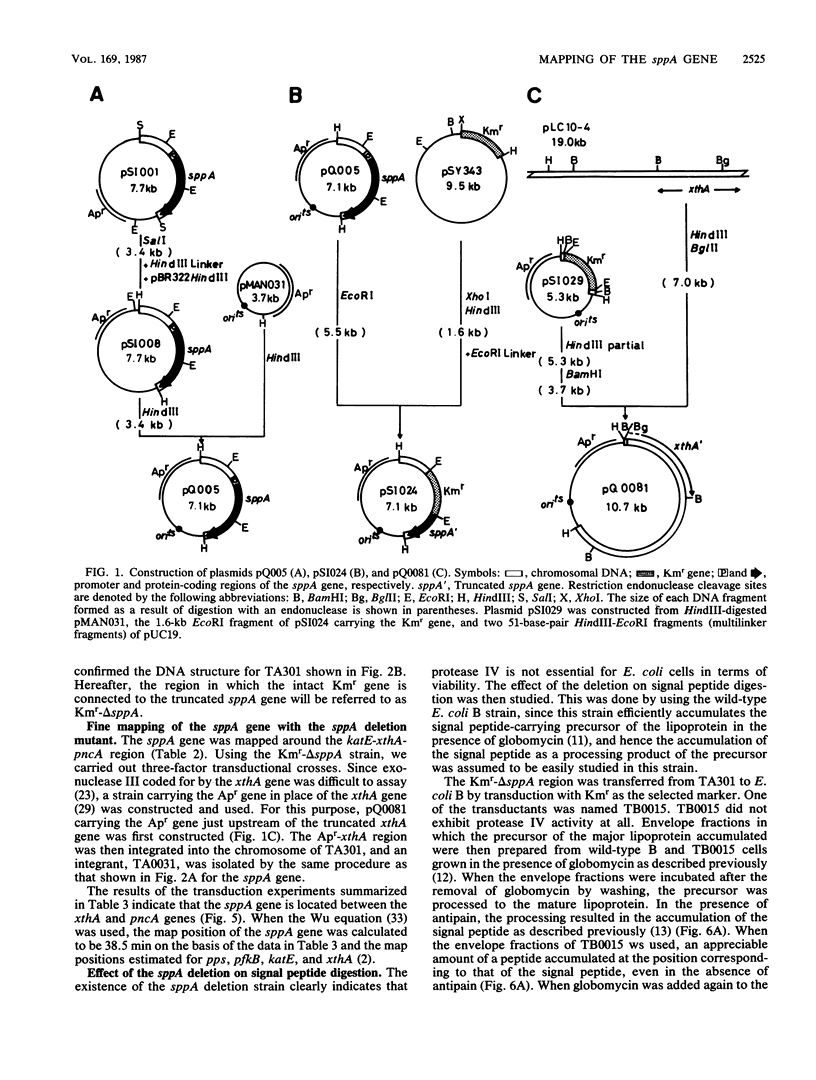
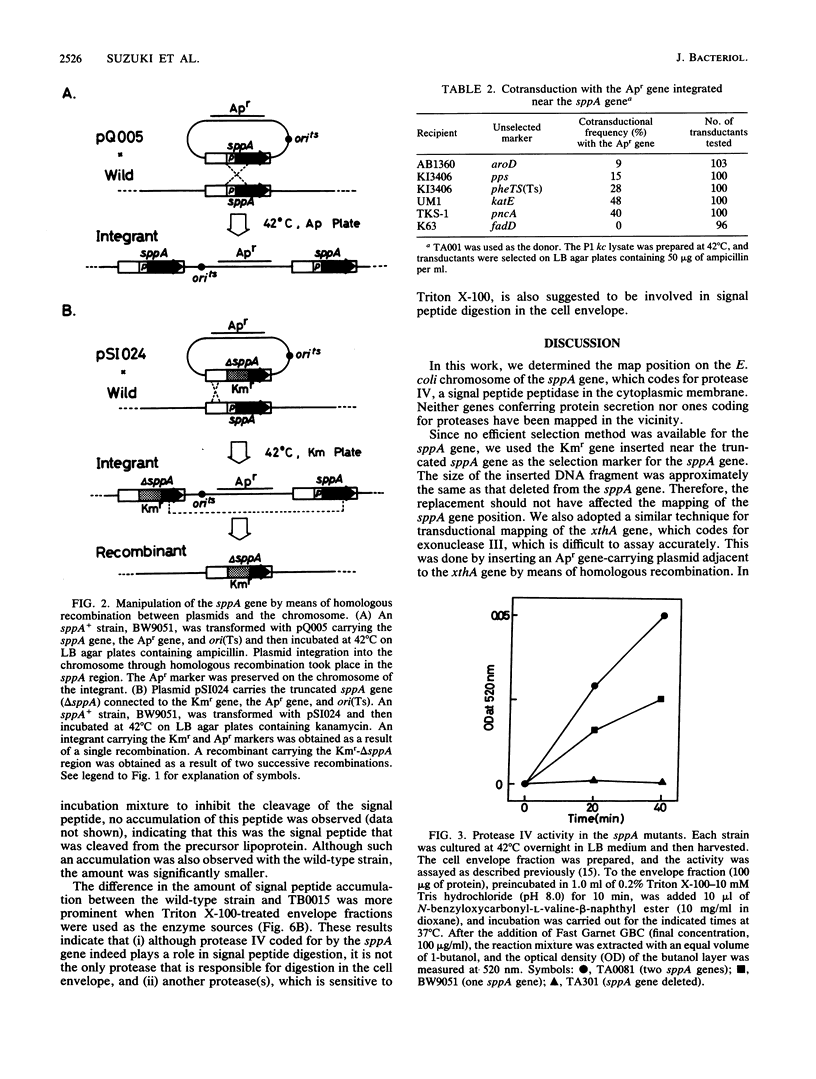
The sppA gene codes for protease IV, a signal peptide peptidase of Escherichia coli. Using the gene cloned on a plasmid, we constructed an E. coli strain carrying the ampicillin resistance gene near the chromosomal sppA gene and an sppA deletion strain in which the deleted portion was replaced by the kanamycin resistance gene. Using these strains, we mapped the sppA gene at 38.5 min on the chromosome, the gene order being katE-xthA-sppA-pncA. Although digestion of the signal peptide that accumulated in the cell envelope fraction was considerably slower in the deletion mutant than in the sppA+ strain, it was still significant, suggesting the participation of another envelope protease(s) in signal peptide digestion.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K. A., Acosta R., Ledner E., Machida Y., Pancotto M., McCormick M., Ohtsubo H., Ohtsubo E. A 37 X 10(3) molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984 May 25;175(3):331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice C. B., Kornberg H. L. Location on the chromosome of Escherichia coli of a gene specifying phosphopyruvate synthase activity. Biochim Biophys Acta. 1967 Mar 22;136(2):412–414. doi: 10.1016/0304-4165(67)90094-3. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Daldal F., Fraenkel D. G. Tn10 insertions in the pfkB region of Escherichia coli. J Bacteriol. 1981 Sep;147(3):935–943. doi: 10.1128/jb.147.3.935-943.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date T., Wickner W. Isolation of the Escherichia coli leader peptidase gene and effects of leader peptidase overproduction in vivo. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6106–6110. doi: 10.1073/pnas.78.10.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson E. S., Sundaram T. K. Chromosomal location of a gene defining nicotinamide deamidase in Escherichia coli. J Bacteriol. 1970 Mar;101(3):1090–1091. doi: 10.1128/jb.101.3.1090-1091.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N. I., Koshland D. E., Jr Replacement and amplification of bacterial genes with sequences altered in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4894–4898. doi: 10.1073/pnas.80.16.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Ichihara S., Mizushima S. Accumulation of glyceride-containing precursor of the outer membrane lipoprotein in the cytoplasmic membrane of Escherichia coli treated with globomycin. J Biol Chem. 1980 Apr 25;255(8):3707–3712. [PubMed] [Google Scholar]
- Hussain M., Ichihara S., Mizushima S. Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J Biol Chem. 1982 May 10;257(9):5177–5182. [PubMed] [Google Scholar]
- Hussain M., Ozawa Y., Ichihara S., Mizushima S. Signal peptide digestion in Escherichia coli. Effect of protease inhibitors on hydrolysis of the cleaved signal peptide of the major outer-membrane lipoprotein. Eur J Biochem. 1982 Dec;129(1):233–239. doi: 10.1111/j.1432-1033.1982.tb07044.x. [DOI] [PubMed] [Google Scholar]
- Ichihara S., Beppu N., Mizushima S. Protease IV, a cytoplasmic membrane protein of Escherichia coli, has signal peptide peptidase activity. J Biol Chem. 1984 Aug 10;259(15):9853–9857. [PubMed] [Google Scholar]
- Ichihara S., Suzuki T., Suzuki M., Mizushima S. Molecular cloning and sequencing of the sppA gene and characterization of the encoded protease IV, a signal peptide peptidase, of Escherichia coli. J Biol Chem. 1986 Jul 15;261(20):9405–9411. [PubMed] [Google Scholar]
- Innis M. A., Tokunaga M., Williams M. E., Loranger J. M., Chang S. Y., Chang S., Wu H. C. Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3708–3712. doi: 10.1073/pnas.81.12.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen P. C. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol. 1984 Feb;157(2):622–626. doi: 10.1128/jb.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen P. C., Triggs B. L., George C. S., Hrabarchuk B. E. Genetic mapping of katG, a locus that affects synthesis of the bifunctional catalase-peroxidase hydroperoxidase I in Escherichia coli. J Bacteriol. 1985 May;162(2):661–667. doi: 10.1128/jb.162.2.661-667.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Inokuchi K., Mizushima S. Promoter exchange between ompF and ompC, genes for osmoregulated major outer membrane proteins of Escherichia coli K-12. J Bacteriol. 1984 Jun;158(3):1041–1047. doi: 10.1128/jb.158.3.1041-1047.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Mizushima S. Construction and characterization of a deletion mutant lacking micF, a proposed regulatory gene for OmpF synthesis in Escherichia coli. J Bacteriol. 1985 Jun;162(3):1196–1202. doi: 10.1128/jb.162.3.1196-1202.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milcarek C., Weiss B. Mutants of Escherichia coli with altered deoxyribonucleases. I. Isolation and characterization of mutants for exonuclease 3. J Mol Biol. 1972 Jul 21;68(2):303–318. doi: 10.1016/0022-2836(72)90215-x. [DOI] [PubMed] [Google Scholar]
- Novak P., Ray P. H., Dev I. K. Localization and purification of two enzymes from Escherichia coli capable of hydrolyzing a signal peptide. J Biol Chem. 1986 Jan 5;261(1):420–427. [PubMed] [Google Scholar]
- Overath P., Pauli G., Schairer H. U. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur J Biochem. 1969 Feb;7(4):559–574. [PubMed] [Google Scholar]
- Pacaud M. Identification and localization of two membrane-bound esterases from Escherichia coli. J Bacteriol. 1982 Jan;149(1):6–14. doi: 10.1128/jb.149.1.6-14.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacaud M. Purification and characterization of two novel proteolytic enzymes in membranes of Escherichia coli. Protease IV and protease V. J Biol Chem. 1982 Apr 25;257(8):4333–4339. [PubMed] [Google Scholar]
- Rogers S. G., Weiss B. Cloning of the exonuclease III gene of Escherichia coli. Gene. 1980 Nov;11(3-4):187–195. doi: 10.1016/0378-1119(80)90059-1. [DOI] [PubMed] [Google Scholar]
- Silver P., Wickner W. Genetic mapping of the Escherichia coli leader (signal) peptidase gene (lep): a new approach for determining the map position of a cloned gene. J Bacteriol. 1983 May;154(2):569–572. doi: 10.1128/jb.154.2.569-572.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Sato T., Matsuhashi M. Role of lipopolysaccharides in antibiotic resistance and bacteriophage adsorption of Escherichia coli K-12. J Bacteriol. 1971 Mar;105(3):968–975. doi: 10.1128/jb.105.3.968-975.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White B. J., Hochhauser S. J., Cintron N. M., Weiss B. Genetic mapping of xthA, the structural gene for exonuclease III in Escherichia coli K-12. J Bacteriol. 1976 Jun;126(3):1082–1088. doi: 10.1128/jb.126.3.1082-1088.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H., Taguchi N., Daishima K., Mizushima S. Genetic characterization of a gene for prolipoprotein signal peptidase in Escherichia coli. Mol Gen Genet. 1983;192(1-2):10–14. doi: 10.1007/BF00327640. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yasuda S., Takagi T. Overproduction of Escherichia coli replication proteins by the use of runaway-replication plasmids. J Bacteriol. 1983 Jun;154(3):1153–1161. doi: 10.1128/jb.154.3.1153-1161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Yamada H., Daishima K., Mizushima S. Nucleotide sequence of the lspA gene, the structural gene for lipoprotein signal peptidase of Escherichia coli. FEBS Lett. 1984 Jul 23;173(1):264–268. doi: 10.1016/0014-5793(84)81060-1. [DOI] [PubMed] [Google Scholar]
- Zwizinski C., Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980 Aug 25;255(16):7973–7977. [PubMed] [Google Scholar]