Abstract
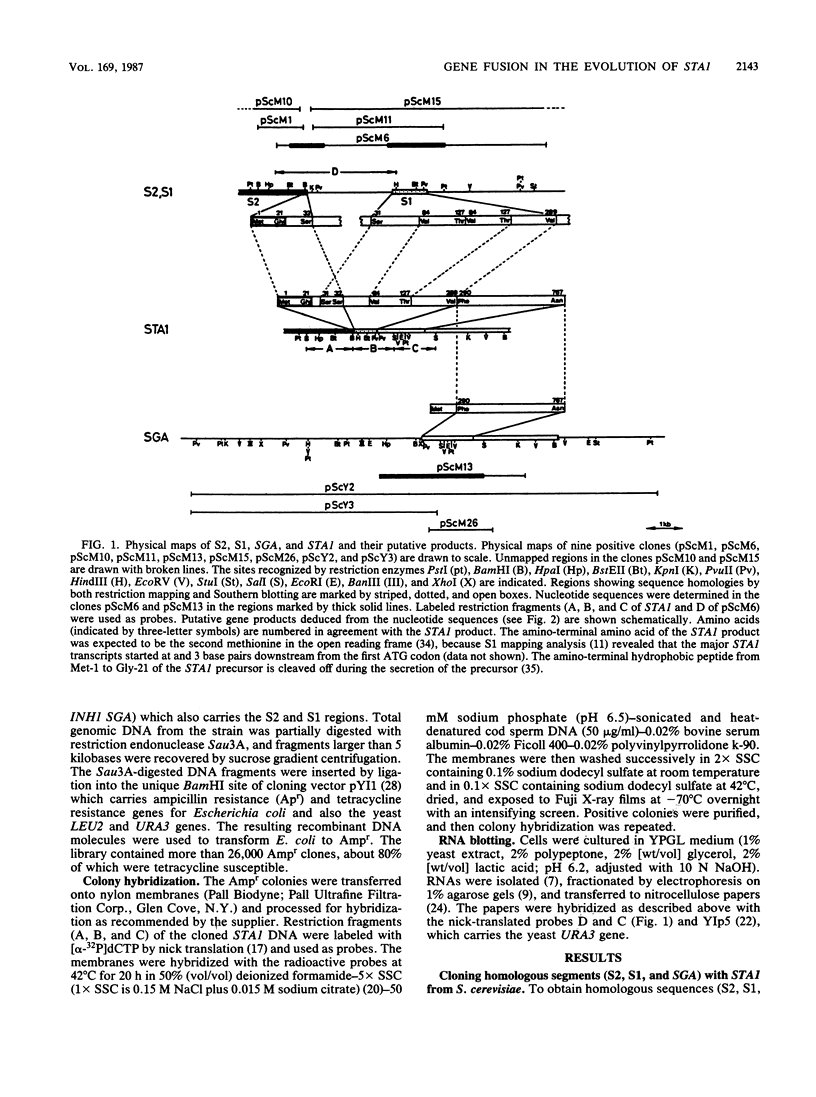
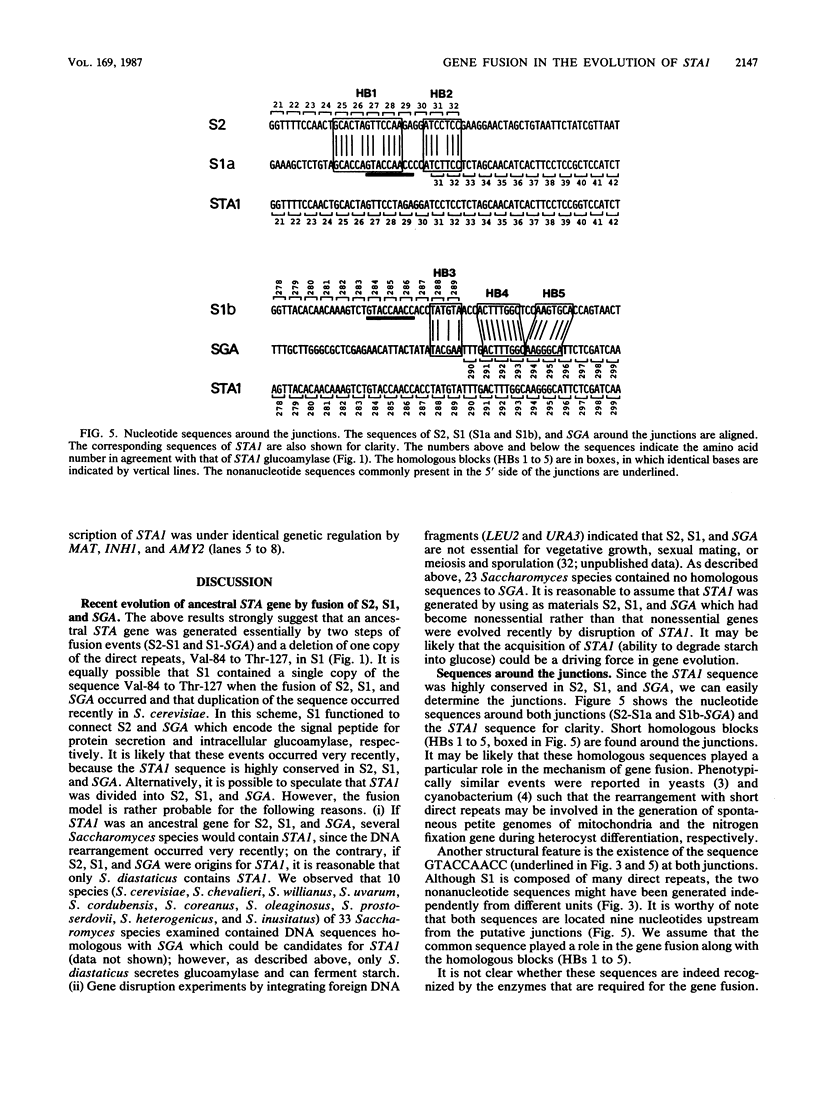
DNA from the STA1 (extracellular glucoamylase) gene of Saccharomyces diastaticus was used as a probe to enable the cloning by colony hybridization of three DNA fragments from Saccharomyces cerevisiae; these were designated S1, S2, and SGA (intracellular, sporulation-specific glucoamylase gene). To examine the evolutionary relationship among these sequences at the nucleotide level, we sequenced S2, S1, SGA and compared them with STA1. These data and RNA blot analysis revealed that the following regions of STA1 were highly conserved in S2, S1, and SGA: upstream regulatory sequences responsible for transcription, a signal sequence for protein secretion, a threonine- and serine-rich domain, and a catalytic domain for glucoamylase activity. These results suggest that an ancestral STA gene was generated relatively recently in an evolutionary time scale by the sequential fusions of S2, S1, and SGA, with S1 functioning as a connector for S2 and SGA. We describe a model for the involvement of short nucleotide sequences flanking the junctions in the gene fusions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedouelle H., Bassford P. J., Jr, Fowler A. V., Zabin I., Beckwith J., Hofnung M. Mutations which alter the function of the signal sequence of the maltose binding protein of Escherichia coli. Nature. 1980 May 8;285(5760):78–81. doi: 10.1038/285078a0. [DOI] [PubMed] [Google Scholar]
- Emr S. D., Silhavy T. J. Importance of secondary structure in the signal sequence for protein secretion. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4599–4603. doi: 10.1073/pnas.80.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard C., Strauss F., Bernardi G. Excision sequences in the mitochondrial genome of yeast. Nature. 1980 Jan 10;283(5743):218–220. doi: 10.1038/283218a0. [DOI] [PubMed] [Google Scholar]
- Golden J. W., Robinson S. J., Haselkorn R. Rearrangement of nitrogen fixation genes during heterocyst differentiation in the cyanobacterium Anabaena. Nature. 1985 Apr 4;314(6010):419–423. doi: 10.1038/314419a0. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Hereford L., Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984 Apr;36(4):1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Horwich A. L., Kalousek F., Fenton W. A., Pollock R. A., Rosenberg L. E. Targeting of pre-ornithine transcarbamylase to mitochondria: definition of critical regions and residues in the leader peptide. Cell. 1986 Feb 14;44(3):451–459. doi: 10.1016/0092-8674(86)90466-6. [DOI] [PubMed] [Google Scholar]
- Jensen R., Sprague G. F., Jr, Herskowitz I. Regulation of yeast mating-type interconversion: feedback control of HO gene expression by the mating-type locus. Proc Natl Acad Sci U S A. 1983 May;80(10):3035–3039. doi: 10.1073/pnas.80.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. M., MacKay V. L., Nasmyth K. A. Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature. 1985 Apr 18;314(6012):598–603. doi: 10.1038/314598a0. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Molecular analysis of a cell lineage. Nature. 1983 Apr 21;302(5910):670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. Analysis of the essential and excision repair functions of the RAD3 gene of Saccharomyces cerevisiae by mutagenesis. Mol Cell Biol. 1986 Apr;6(4):1218–1227. doi: 10.1128/mcb.6.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., Emori Y., Imajoh S., Kawasaki H., Kisaragi M., Suzuki K. Evolutionary origin of a calcium-dependent protease by fusion of genes for a thiol protease and a calcium-binding protein? Nature. 1984 Dec 6;312(5994):566–570. doi: 10.1038/312566a0. [DOI] [PubMed] [Google Scholar]
- Palm D., Goerl R., Burger K. J. Evolution of catalytic and regulatory sites in phosphorylases. Nature. 1985 Feb 7;313(6002):500–502. doi: 10.1038/313500a0. [DOI] [PubMed] [Google Scholar]
- Patthy L. Evolution of the proteases of blood coagulation and fibrinolysis by assembly from modules. Cell. 1985 Jul;41(3):657–663. doi: 10.1016/s0092-8674(85)80046-5. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell. 1982 Sep;30(2):517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rosenblatt M., Beaudette N. V., Fasman G. D. Conformational studies of the synthetic precursor-specific region of preproparathyroid hormone. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3983–3987. doi: 10.1073/pnas.77.7.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stone E. M., Rothblum K. N., Schwartz R. J. Intron-dependent evolution of chicken glyceraldehyde phosphate dehydrogenase gene. Nature. 1985 Feb 7;313(6002):498–500. doi: 10.1038/313498a0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A., Coussens L., Hayflick J. S., Dull T. J., Gray A., Tam A. W., Lee J., Yarden Y., Libermann T. A., Schlessinger J. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. 1984 May 31-Jun 6Nature. 309(5967):418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- Vlasuk G. P., Inouye S., Ito H., Itakura K., Inouye M. Effects of the complete removal of basic amino acid residues from the signal peptide on secretion of lipoprotein in Escherichia coli. J Biol Chem. 1983 Jun 10;258(11):7141–7148. [PubMed] [Google Scholar]
- Weber I. T., Takio K., Titani K., Steitz T. A. The cAMP-binding domains of the regulatory subunit of cAMP-dependent protein kinase and the catabolite gene activator protein are homologous. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7679–7683. doi: 10.1073/pnas.79.24.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I., Fukui S. Transcriptional control of the sporulation-specific glucoamylase gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1985 Nov;5(11):3069–3073. doi: 10.1128/mcb.5.11.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I., Maemura T., Hatano T., Fukui S. Polymorphic extracellular glucoamylase genes and their evolutionary origin in the yeast Saccharomyces diastaticus. J Bacteriol. 1985 Feb;161(2):574–582. doi: 10.1128/jb.161.2.574-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I., Suzuki K., Fukui S. Nucleotide sequence of the extracellular glucoamylase gene STA1 in the yeast Saccharomyces diastaticus. J Bacteriol. 1985 Feb;161(2):567–573. doi: 10.1128/jb.161.2.567-573.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I., Takano Y., Fukui S. Control of STA1 gene expression by the mating-type locus in yeasts. J Bacteriol. 1985 Nov;164(2):769–773. doi: 10.1128/jb.164.2.769-773.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]



