Abstract
Cationic antimicrobial peptides play important roles in host defense, linking innate and adaptive immunity. hCAP18, the only human antimicrobial cathelicidin, consists of a conserved N-terminal cathelin-like domain and a C-terminal peptide, LL-37. Expression is regulated during myeloid differentiation, and tightly controlled during infection and inflammation, suggesting active regulation. Using 5′ RACE (rapid amplification of cDNA ends), multiple transcription initiation sites were identified, as well as new splice variants leading to novel augmentations of hCAP18 amino acid composition in bone marrow but not peripheral blood neutrophils. Having expressed hCAP18 promoter constructs in cell lines, we found that full-length (-1739) and truncated (-978) promoter constructs had lower luciferase activities than 5′UTR deletion constructs. Transient transfection of progressively deleted constructs in the non-permissive K562 cell line led us to identify a negative regulatory element within the 53 bp immediately upstream of the ATG of hCAP18. Additionally, transient transfection of 5′ deletion constructs identified a positive regulatory element within the 101 bases 5′ of promoter sequence containing two GT-boxes. Negative and positive regulatory elements within the hCAP18 gene promoter provide new insights into the possible molecular basis of myeloid gene expression.
1. Introduction
Polymorphonuclear leukocytes play a major role in innate immunity. In addition to the generation of reactive oxygen intermediates through the respiratory burst, neutrophils secrete antimicrobial peptides into phagocytic vacuoles and the local environment. The two major antimicrobial peptide families in mammals are the defensins and the cathelicidins, with hCAP18 being the only human cathelicidin. Human CAP18 (hCAP18) maps to chromosome 3p21, with four exons and three introns. It is processed to cathelin and LL-37 components and synthesized mainly by neutrophils, myeloid bone marrow cells (Zanetti et al., 1990; Sorensen et al., 1997), lung and gastric epithelium (Frohm et al., 1999; Bals et al., 1998), and appears in sweat and airway surface fluids. Plasma has been reported to contain LL-37 bound to lipoproteins (Sorensen et al., 1999).
The full-length hCAP18 precursor consists of an N-terminal putative signal peptide, a conserved cathelin-like domain, and a C-terminal microbicidal domain, LL-37. The proform is thought to be inactive and is stored in neutrophil peroxidase-negative granules. Upon stimulation, LL-37 is liberated by the serine protease, PR3. Proteolytic processing of seminal hCAP18 occurs in the vagina by the prostate-derived protease gastricsin (Sorensen et al., 2001; Sorensen et al., 2003). In addition to direct microbial killing, hCAP18 neutralizes host responses to bacterial LPS from Gram-bacteria, lipoteichoic acid from Gram+ bacteria, and unmethylated bacterial CpG DNA. It releases histamine from mast cells and is chemoattractant for neutrophils, monocytes, and T cells (De et al., 2000). hCAP18 also has an angiogenic effect and is important for wound healing (Bals and Wilson, 2003).
hCAP18 and mouse cathelin-related antimicrobial peptide (CRAMP) are up-regulated in the skin in response to cutaneous infection or injury and in inflammatory disorders of the skin such as psoriasis, lupus erythematosus, and contact dermatitis (Frohm et al., 1997). In human keratinocytes, hCAP18 is up-regulated by insulin-like growth factor I (IGF-I) at both the protein and mRNA levels. In leukocytes, hCAP18 is regulated by certain cytokines but can be repressed by certain pathogenic microbes such as pathogenic Shigella species (Islam et al., 2001), and Nesseria gonorrheae (Bergman et al., 2005).
The promoter for the porcine cathelicidin, PR-39, contains clusters of transcription regulatory elements including those for NFκB, NF-IL-6, GM-CSF, IL-6 response elements (IL-6 RE), and the retinoic acid response element (RARE) (Zhao et al., 1995). The PR-39 promoter has inducible activity with an upstream open reading frame that represses gene expression (Wu et al., 1999; Wu et al., 2002).
CRAMP, the product of the mouse cathelicidin gene (Cnlp), maps to chromosome 9, orthologous to the location of cathelicidins in man and pig (Gudmundsson et al., 1995). The CRAMP promoter has many binding domains for NFκB, NF-IL-6, GM-CSF, IFN-γ, C/EBP, GATA, and interferon regulatory element delta (Pestonjamasp et al., 2001). Larrick et al. (1996) cloned the hCAP18 gene and characterized the promoter region and transcription initiation site being at 16 nucleotides upstream from the ATG; no activity in K562 cells was found.
We hypothesized that genetic regulation of hCAP18 was complex and important, given its broad but variable expression in myeloid and epithelial cells as well as its developmental regulation. Therefore, we developed deletion mutants to evaluate 5′UTR region as well as other promoter sequences that we thought might be relevant to gene regulation. Since hCAP18 expression is controlled by both host and pathogen, further understanding its regulation should provide insights into the pathophysiology of infection, inflammation, and wound healing.
2. Materials and methods
2.1. Neutrophil isolation and cell culture
Human neutrophils were prepared using Ficoll separation followed by dextran sedimentation and osmotic lysis of erythrocytes as described (Boyum, 1974).
U937 monocyte-like cells, K562 erythroid /myeloid cells, HL-60 promyelocytic leukemia cells, and A549 lung epithelial cells (all purchased from ATCC, Rockville, MD, USA) were grown at 37°C and 5% CO2, and maintained respectively in RPMI 1640, IMDM, and DMEM media supplemented with 10% heat inactivated fetal calf serum (Gemini Bioproducts), and passaged twice a week.
Cells were transfected with promoter constructs and then incubated with cytokines, retinoic acid (RA, 10 μM), trichostatin A (TSA, 100 ng/ml), mitromycin (100 nM), Cobalt (II) chloride hexahydrate (CoCl2, 100 μM) and Desferrioxamine mesylate salt (DFOM, 100 μM) for the indicated times.
2.2. 5′ and 3′ RACE
Human neutrophils were isolated and stimulated with or without LPS (1 μg/ml) for one hour in complete media, followed by total RNA isolation using RNAaquous for PCR Kit (Ambion Inc, TX). Human bone marrow RNA, pooled from six Caucasian males and females, was purchased from Clontech. Using the Smart Race cDNA amplification Kit (BD Biosciences, CA), 50 ng of total RNA was reverse transcribed and full length cDNA was used for both 5′ and 3′ race PCR. The amplification product was then sequenced.
2.3. Recombinant Human CAP18 reporter constructs
The 1739 bp 5′ flanking region of the hCAP18 gene was amplified from human genomic DNA using primers based on the published sequence (GenBank accession no. NM_004345). This promoter fragment was digested with XhoI and MluI and cloned into the multiple cloning site of the promoterless luciferase vector pGL3-basic (Promega).
Mutants were constructed (Stratagene Quickchange) using the cloned genomic DNA as template. Primers used to make these constructs are reported in Table 1.
Table 1.
Primers used in generating promoter constructs and in site directed mutagenesis.
| Name | Sequence (5′-3′) | Position |
|---|---|---|
| Primers for cloning of the hCAP18 promoter and deletion constructs | ||
| CAP18 -1739F | ACCGACGCGTGCATACGTGACAGTCTGGCA | -1739 to -1720 |
| CAP18 -1582F | ACCGACGCGTCAACGGTGTGGAGGGTGGAC | -1582 to -1563 |
| CAP18-1200F | ACCGACGCGTTAAGAAGACAGCCCTTAGGG | -1200 to -1181 |
| CAP18 -978F | ACCGACGCGTCGTAGACAACCTTTCCTAAG | -978 to -959 |
| CAP18 -708F | ACCGACGCGTCATACTGAGTCTCACTCTGT | -708 to -689 |
| CAP18 -510F | ACCGACGCGTGCTTGTCTCGAACTCCTTAT | -510 to -491 |
| CAP18 -105R | ACCGCTCGAGGGCTGCCTGCCAGGGTGTGG | -105 to -124 |
| CAP18 -89R | ACCGCTCGAGTGATCCACCCATCCCTGGC | -89 to -107 |
| CAP18 -74R | ACCGCTCGAGAACCAGGAGCCTTCCTGATC | -74 to -93 |
| CAP18 -54R | ACCGCTCGAGTGAGCCTGATGCAAAAGCCC | -54 to -73 |
| CAP18 -42R | ACCGCTCGAGTTATGCCCAGCCTGAGCCTG | -42 to -61 |
| CAP18 -21R | ACCGCTCGAGCTAGCCCACAGGAGCCTCCT | -21 to -40 |
| CAP18 -1R | ACCGCTCGAGGGTCCCCATGTCTGCCTCCC | -1 to -20 |
| Primers for site directed mutagenesis | ||
| Mut HIF-1 F | GCATAATTCACAGTCTGGCA | -1739 to -1720 |
| Mut HIF-1 R | TGCCAGACTGTGAATTATGC | |
| Del Sp1F | GCATCAGGCTCAGCATAAAGGAGGCTCCTG | -60 to -20 |
| Del Sp1R | CAGGAGCCTCCTTTATGCTGAGCCTGATGC | |
| Del AML1F | GCATAAAGGAGGCTCCTTAGAGGGAGGCAG | -47 to -12 |
| Del AML1R | CTGCCTCCCTCTAAGGAGCCTCCTTTATGC | |
| Del GTbox1F | AGTGTGCAGGGCTAGGGCAGTGGGAACC | -1716 to -1681 |
| Del GTbox1R | GGTTCCCACTGCCCTAGCCCTGCACACTGC | |
| Del GTbox2F | CTGATGCCAGCCTAGTGTGGGCTCCTGAAG | -1665 to -1630 |
| Del GTbox2R | CTTCAGGAGCCCACACTAGGCTGGCATCAG | |
2.4. ORF constructs
The hCAP18 coding region was amplified by reverse transcriptase reaction of normal human bone marrow RNA using the Advantage RT for PCR Kit (Clontech, BD biosciences). The PCR products were then cloned in-frame 5′ to the V5 epitope into pcDNA3.1/V5-His TOPO TA expression vector (Invitrogen). Different constructs were prepared. ORF1CAP18 contained the full length open reading frame with the natural Kozak sequence (GACCATGA). ORF3CAP18 had an extra 108 bp upstream the ATG translation start, corresponding to partial hCAP18 5′UTR. The primers used were the following:ORF1F 5′-GGGACCATGAAGACCCAAAG-3′; ORF3F 5′-AGCCAGGGATGGGTGGATC-3′; and ORFR 5′-GGACTCTGTCCTGGGTACAA-3′. A shorter construct, UORF, missing 352 bp from the 3′ end of the cDNA and coding for a theoretical protein starting from the upstream ATG within the 5′UTR, using ORF3CAP18F primer and the reverse primer ORF3R 5′-GCATCCGAGGACCGCTGGTT-3′ (Fig. 1).
Fig. 1.

ORF construct. The hCAP18 mRNA sequence (GenBank accession no. NM_004345) is shown. Primers used to amplify the different inserts of ORF1CAP18, ORF3CAP18, and UORF constructs are underlined. ATG and stop codons are in bold. The initiator codon of the upstream ORF is in a favorable context and has the potential to encode a polypeptide of 86 amino acids.
The sequences and insert orientation for each construct were confirmed prior to cell transfection.
2.5. Chimeric Luciferase constructs
In order to generate the pGL3promoter 5′UTR-Luc reporter construct, the 108 bp situated 5′ upstream the ATG translation start was amplified using the sense primer 5′-ACCGAAGCTTAGCCAGGGATGGGTGGATCAGGAAGGCT-3′ (HindIII site underlined) and the antisense primer 5′-GGGTCTCCATGGTCCCCATGTCTGCCTCCCTCTAGCCC-3′ (NcoI site underlined).
The resulting product was digested and cloned into the pGL3 promoter vector (Promega) upstream of the firefly luciferase cDNA using the HindIII and the NcoI restriction sites.
The pGL3promoter 5′UTR construct was obtained by inserting the 108 bp sequence within the multiple cloning site of pGL3promoter vector using MluI and XhoI sites.
2.6. Computational analysis
In order to identify possible regulatory sites, we performed a computational sequence search for potential transcription factor binding sites using TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html), TRANSFAC (http://www.gene-regulation.com/), TESS (http://www.cbil.upenn.edu/tess/), Match (http://www.gene-regulation.com/), and MatInspector (http://www.genomatix.de).
2.7. Transient transfection and luciferase assay
Nonadherent cell lines HL-60, U937, K562 and adherent cell lines A549 and COS7 cells were transfected with 2 μg of luciferase reporter constructs and 0.2 μg of Renilla expression vector in 1 ml of medium using the Amaxa nucleofector according to the manufacturer’s protocol. The transfection efficiency was variable for each of these cells: 75-80% for K562 and A549 cells, 60% for Cos7 cells, and 40% for U937 and HL-60 cells. The pGL-3 promoter vector containing the SV40 promoter, and the pGL-3 basic promoterless vector, were used as positive and negative controls, respectively. After 18h, luciferase activity was measured in cell lysates using a luminometer. Promoter activities were expressed as relative luciferase activity normalized with respect to Renilla activity.
K562 and U937 cells were transiently transfected with 2 μg of the ORF constructs. Cotransfection was carried out in the presence of GFP, serving as a transfection efficiency control. Cell extracts were obtained 24 and 48 hours post transfection, and recombinant proteins were detected by immunoblot using a mouse anti-V5 epitope antibody (Invitrogen, CA).
2.8. Electrophoretic mobility shift assay
Nuclear extracts of U937 and K562 cells were prepared and protein concentration was determined using the Bradford assay (Sigma, Saint Louis, MO). EMSA analysis was carried out using the Panomics kit (Fremont, CA) following the manufacturer’s instructions. Complementary biotinylated DNA oligonucleotides corresponding to different regions from the hCAP18 promoter were annealed and mixed with the nuclear extract in the binding buffer and poly (dI-dC) at 15 to 20°C for 30 min. For competition reactions, a 100-fold excess of unlabeled DNA probe was incorporated into the reaction mix. The sequences of the probes used in this study are listed in Table 2. For supershift assays, 2 μg of specific Sp1 (Active Motif, CA) and Sp3 (Santa Cruz Biotechnology, CA) antibodies were incubated at 4°C for 20 minutes with the nuclear extract before adding the probes.
Table 2.
DNA sequence of oligonucleotide probes used for EMSA. Mutated nucleotides are in bold. Translation start as well as upstream ATG are underlined.
| Probe name | Sequence 5′ - 3′ | Position |
|---|---|---|
| Box -8 | CTAGAGGGAGGCAGACATGGGGACCATG | -25 to +3 |
| Box -50 | CAGGCTGGGCATAAAGGAGGCTCCTGTGGGCTAGAG | -56 to -20 |
| Box-50mut TFIID | CAGGCTGGGCTCGTAGGAGGCTCCTGTGGGCTAGAG | -56 to -20 |
| Box -74 | GGAAGGCTCCTGGTTGGGCTTTTGCATC | -88 to -61 |
| Box -99 | AGGCAGCCAGGGATGGGTGGATCAGGAAGG | -122 to -83 |
| Box HRE | CATACGTGACAGTCTGGCACTTGCAGTGTGCAGGGCTGGGG | -1738 to -1698 |
| Box RBP | AGGGGCTTGGGAACATTTTG | -919 to -900 |
| Box RBP MUT | AGGGGCTATTCAACATTTTG | -919 to -900 |
The samples were then separated on a 6% non-denaturing polyacrylamide gel. Shifted bands corresponding to the protein / DNA complexes were visualized after exposure to film.
3. Results
3.1. RACE and identification of alternative splice forms
In an effort to comprehensively characterize regulation of the hCAP18 gene, analyses of the 5′ and 3′ flanking regions were performed. Larrick et al. (1996) reported that the transcription initiation start is located 16 nucleotides upstream from the translation start. Our 5′ RACE analysis of human peripheral blood neutrophils showed that the start of transcription was heterogeneous. Among the clones sequenced, 50% had -13 as the transcription start, 25% had -17, and the remaining 25% had different transcription start sites (-14, -16, -40, -43, -73, -81, -104, and -140). Although truncated forms may arise from premature termination of cDNA synthesis by the reverse transcriptase in vitro, they may also indicate multiple transcription start sites, as is reported for TATA-less genes (Travis et al., 1991). In addition to the variability noted in the 5′ RACE, variability of 3′ RACE products was found by direct sequencing of the clones deriving from both bone marrow and neutrophils (Table 3).
Table 3.
Frequency of the 3′ variants. 3′-RACE was performed on RNA isolated from human bone marrow and polymorphonuclear cells. Alignment of the different 3′-RACE products shows variability at the 3′UTR ends.
| 3′ RACE Sequence | Frequency (%) |
|---|---|
| AGAGC(A)n | 12.25 |
| AGAGCC(A)n | 3 |
| AGAGCAATTT(A)n | 6.1 |
| AGAGCAATTTT(A)n | 3 |
| AGAGCAATTTC(A)n | 30.35 |
| AGAGCAATTCC(A)n | 21.3 |
| AGAGCAATTAC(A)n | 3 |
| AGAGCAATTTCC(A)n | 3 |
| AGAGCAATTTCG(A)n | 3 |
| AGAGCAATTTCAT(A)n | 3 |
| AGAGCAATTTCCG(A)n | 3 |
| AGAGCAATTTCAAG(A)n | 3 |
| AGAGCAATTTCCTCAG(A)n | 3 |
| AGAGCAATTTCCTCAGG(A)n | 3 |
Two different alternatively spliced forms of hCAP18 mRNA were identified from bone marrow but not from peripheral blood neutrophils. Neither form has been previously reported in the alternative splicing databases PALSdb (http://palsdb.ym.edu.tw/) and ASG (http://statgen.ncsu.edu/asg/).
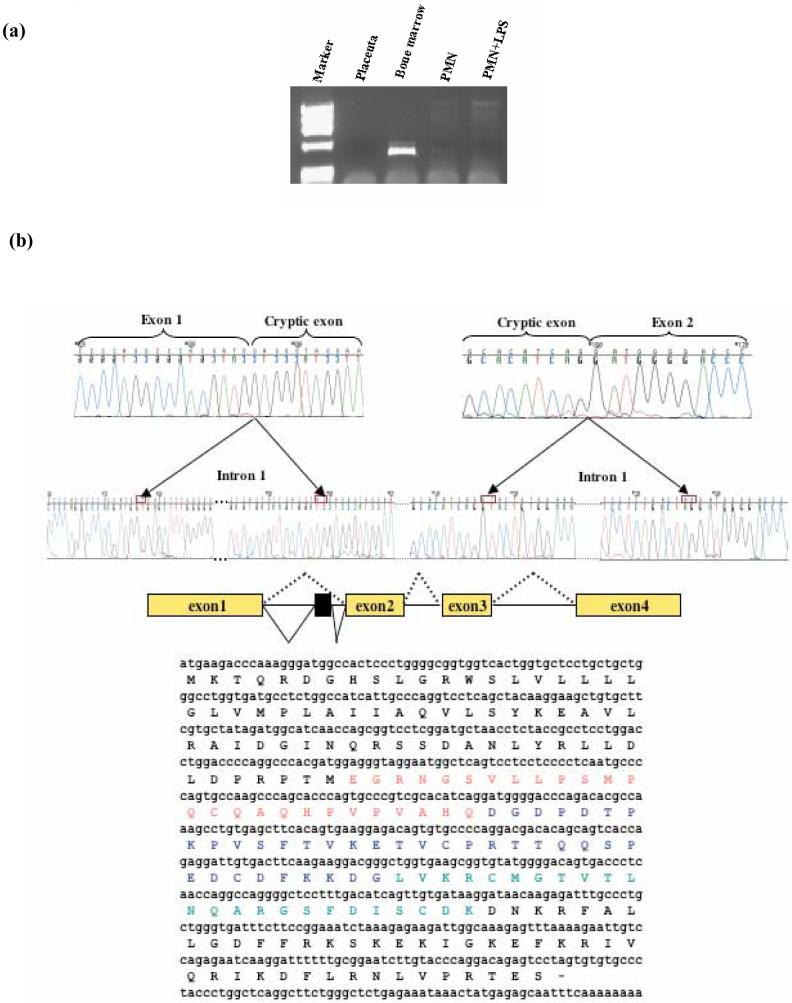
To confirm the presence of these splice variants, specific primers were designed and used in a fresh RT-PCR (Fig. 2a). Sequencing of the products showed classical splice donor and acceptor sites. One splice variant was due to a 78 bp insertion from intron 1 (exon 1b), predicting a 26 aa insertion within the cathelin part of the hCAP18 protein (Fig. 2b). Comparison of the inserted amino acid sequence with an antimicrobial protein database (http://cbcsrv.watson.ibm.com/Tamp.html) showed a 45.45% identity with bovine β defensin 13. The other alternative message resulted from the insertion of 30 bp from distal intron 3 (exon 3b) and involved a new splice acceptor site. The predicted protein encoded by this isoform has an insertion of 10 aa, also within the cathelin domain (Fig. 2c).
Fig. 2.
Novel hCAP18 splice variants are present in bone marrow. (a) By using a sense primer within the cryptic exon (exon1b) and an antisense within the 3′UTR of hCAP18 gene, a 450 bp amplimer obtained by RT-PCR. This alternative splice form was detected only from bone marrow. (b) Partial sequence chromatogram showing the junctions of exon1 and exon1b in the hCAP18 spliced mRNA. Boxes represent exons. hCAP18 gene is comprised of 4 exons; inclusion of 78 bp from intron 1 forms exon 1b. (c) Another transcript is generated by inclusion of 30 bp from intron 3 using a different acceptor splice site. The newly formed exon was designated exon 3b. The partial sequence chromatogram shows the junction of exon 4 and exon 3b in hCAP18 spliced mRNA.
3.2. Analysis of promoter activity
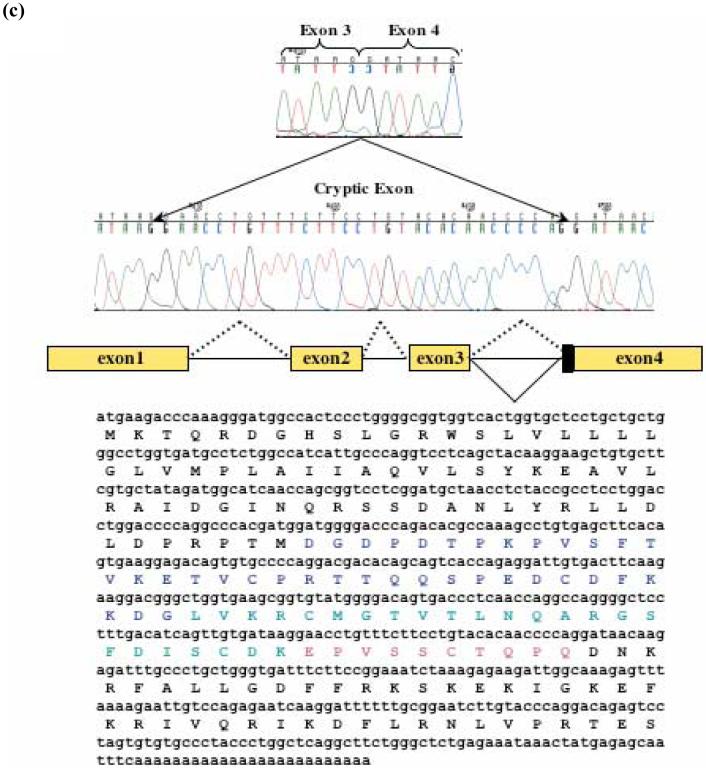
Larrick et al. (1996) characterized the hCAP18 promoter and showed the absence of canonical TATA or CAAT boxes. They analyzed 700 bp upstream of the translational start and found modest promoter activity in Cos7 cells and no activity in K562 cells. In order to further understand the regulation of hCAP18 expression, we amplified 1739 bp of 5′ flanking sequence of the hCAP18 gene from human genomic DNA by PCR using primers based on the published sequence. This 1739 bp fragment was digested with XhoI and MluI and cloned into the multiple cloning site of the promoterless luciferase vector pGL3-basic (the -1739 bp promoter).
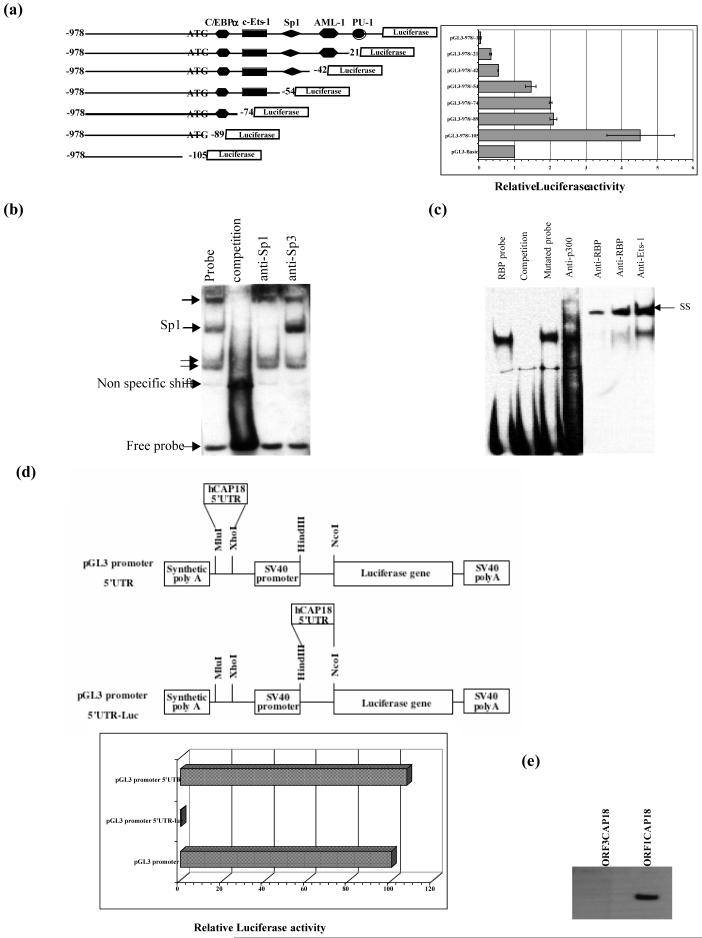
Computational analysis of the hCAP18 promoter sequence identified potential binding sites for Sp1, NF-ATp, PU-1, USF, TFIID, C/EBPα̣ and others (Fig. 3a). Transient transfection of the promoter construct in different hematopoetic cell lines revealed a low activity, mainly in K562 cells (Fig. 3b). Endogenous hCAP18 message in K562 cells was barely detected and the protein was not detected on immunoblot (data not shown).
Fig. 3.
Computational analysis of hCAP18 promoter and assessment of its activity in transiently transfected K562 cells. (a) Nucleotide sequence of the 5′ flanking region of hCAP18. Putative transcription factor binding sites are boxed. Translational start is shown in bold as well as the upstream ATG. Sequences used for EMSA oligonucleotide probes are indicated by bars. The nucleotides are numbered relative to the translation initiation codon. (b) The pGL3-1739/-1 construct was transfected into four different cell lines (U937, HL-60, A549 and K562 cells), then assayed for luciferase activity after 18 hours. Values shown are relative fold increase compared to basic and normalized to Renilla activity. (c) The 5′ deletion constructs as shown in the diagram, are transfected into K562 cells, and assayed for luciferase activity after 18 hours of culture. The pGL3-1739/-1 construct as well as the 5′ deletion constructs yielded reproducibly lower luciferase activity than the promoterless reporter alone.
To further characterize the functional properties of the hCAP18 promoter, we choose to identify and characterize the repressive elements in these non-permissive cells. We generated firefly luciferase (Luc) constructs by fusing the pGL3 basic vector with a series of 5′ promoter deletion constructs (-1739 bp to -510 bp). At 18 hours, luciferase activity of all constructs containing the immediate 5′ 100 bp was lower than that found with the promoterless control vector, indicating a repressor activity in the 5′ flanking region (Fig. 3c).
3.3. Role of upstream ATG in the promoter activity inhibition
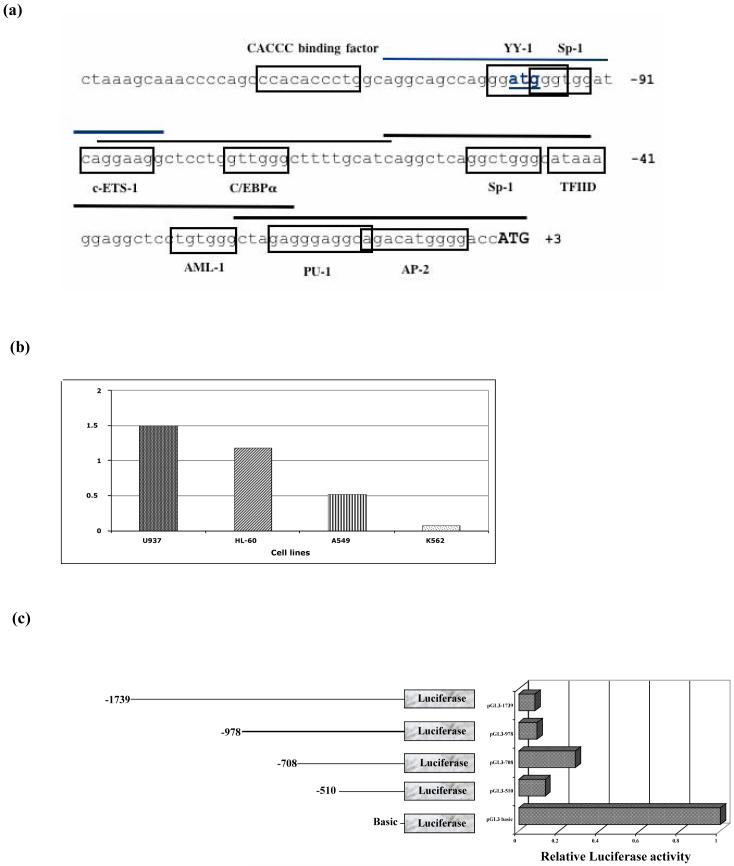
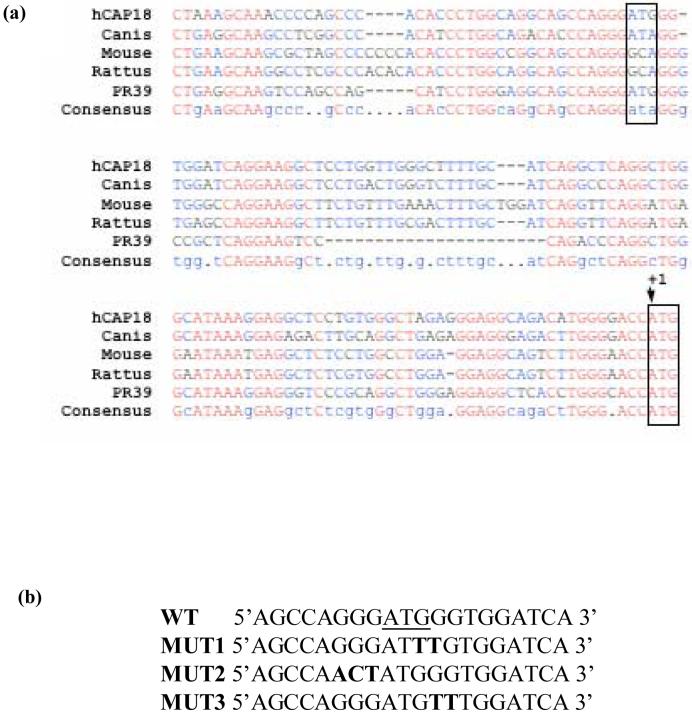
Wu et al. (2002) characterized an upstream translation start site and an upstream ORF, which acted as a repressor of the porcine cathelicidin, PR-39. Deletion analysis of the PR-39 promoter identified a negative regulatory element in the immediate 5′ flanking region of the gene. As expected, the hCAP18 promoter showed high sequence homology with the porcine PR-39 promoter, including an ATG located 100 bp upstream of the translation start site (Fig. 4a). To determine whether this upstream ATG or its immediate flanking sequences were involved in the repressive activity, three site directed mutants were created. Sequence analysis showed a potential Ying Yang1 (YY1) binding site. Since YY1 is an ubiquitous transcriptional regulator of the kruppel family of zinc finger proteins that can act as a repressor, mutants were made to alter this site and its surrounding sequence. Mutant 1 (Mut1) affects the YY1 binding site, while Mut2 and Mut3 create changes around this site (Fig. 4b).
Fig. 4.
Deletion and mutation analysis of hCAP18 promoter. (a) Sequence comparison of the upstream sequences of cathelicidin gene from different species. Translational start as well as the upstream ATG are boxed. (b) Mutations of the surrounding upstream ATG sequence were generated by site directed mutagenesis. Bold nucleotides have been changed from the wild type sequence. Constructs harboring these point mutations show no activity increase compared to the wild type.
In contrast to the PR39 promoter, neither mutations in the upstream hCAP18 ATG nor its immediate flanking regions relieved the negative regulatory activity.
Since the negative regulatory element did not appear to be in the 5′ end of the putative promoter or to surround the upstream ATG, we evaluated the 3′ most portion of the promoter. Two 3′ deletion mutants, -978/-74 (retaining the upstream ATG) and -978/-105 (removing the upstream ATG) were created. There was a two-fold increase in the activity of -978/-105 in the absence of the upstream ATG containing sequence (Fig. 5a). The deletion of the (YY1) binding site in the context of -978/-1 construct did not release the inhibitory effect.
Fig. 5.
Identification of a repressor element within the 5′UTR sequence of hCAP18. (a) A series of hCAP18 promoter 3′ deletion constructs were ligated to the luciferase gene in the pGL3 basic vector; schematic shows potential transcription factor binding sites. Following transfection of K562 cells with equal amounts of each construct, luciferase activity was measured in cell lysates and normalized to Renilla activity. The 3′ deletion construct missing the upstream ATG (pGL3-978/-105) showed at least 2-fold more promoter activity compared to pGL3-978/-74. Results are reported as the ratio of the test construct to the promoterless pGL3 basic. Data shown are means (± SD) of 3 independent experiments. (b) Binding of nuclear proteins from K562 cells to hCAP18 promoter probe -50 containing a putative repressor element. Four protein-DNA complexes are indicated by arrows. The nuclear extract is incubated with the biotinylated probe (lane 1) alone, or in the presence of excess cold competitor (lane 2). (c) EMSA analysis was performed using RBP probe incubated with nuclear extract from K562 cells. Lane 1 shows the formation of a specific protein-DNA complex, which was competed away by excess cold competitor (lane 2), but not with an unlabeled mutated probe (lane 3). A supershift was identified using antibodies specific to p300 (lane 4), RBP (lanes 5 and 6) and Ets-1 (lane 7) transcription factors. (d) hCAP18 5′UTR was amplified then inserted into pGL3 promoter vector. Schematics of two different constructs are shown. The 5′UTR was not found to inhibit downstream luciferase expression when inserted within the multiple cloning site. Insertion of the same sequence in front of the luciferase gene ORF completely abolished promoter activity. (e) Western blot analysis was performed using K562 cell lysates transfected with either ORF3CAP18 or ORF1CAP18 constructs. The blot was then probed for hCAP18 using anti-V5 antibody. 5′UTR abolished hCAP18 production as shown in the first lane.
3.4. Identification of a negative regulatory element within the hCAP18 gene promoter
Since luciferase activity from cells transfected with pGL3-978/-105 was increased 85-fold compared to K562 cells transfected with the full length promoter pGL3-978/-1, deletion mutants in the 3′ region of the hCAP18 promoter were prepared to further identify the cis-acting repressor region. Progressive truncations in this region suggested that the minimal repressor resided within the sequence between -42 and -54 (Fig. 5a), a region predicted to contain a binding site for Sp transcription factor family members. To identify this binding factor, biotinylated double stranded DNA probes derived from this region were used in electrophoretic mobility shift analyses (EMSA) using nuclear extracts from K562 and U937 cells.
EMSA was performed to determine the protein binding activity at the sequence surrounding this repressor element. Oligonucleotides corresponding to BOX-50, BOX -8, BOX-74 and BOX-99 (Table 2) were used. As shown in Fig. 5b, nuclear extracts from K562 cells formed at least 4 protein complexes that were abolished with an excess of unlabeled probe. Antibody recognizing Sp1 was able to shift one of these complexes. However, neither mutation nor deletion of the predicted Sp1 binding site abolished the repression of the hCAP18 promoter activity. Addition of Mithramycin A, a GC-rich DNA binding protein inhibitor, to K562 cells for 24h did not release the observed suppression, suggesting that Sp1 is not the repressor factor for the hCAP18 promoter (data not shown). This finding was confirmed using siRNA to knockdown Sp1 (data not shown). Since transcriptional repressors may be associated with histone deacetylase, we stimulated cells with the histone deacetylase inhibitor Trichostatin (TSA), but no release of repression was noted (data not shown).
Further deletion promoter constructs identified another repressor element, since deletion from -978 to -708 within the hCAP18 promoter resulted in a 2.5-fold increase in luciferase activity in K562 cells, compared to almost 1.5-fold in U937, HL-60 and A549 cells. Prediction programs suggested a potential binding site for the mammalian transcriptional repressor RBP (CBF1) at -912 to -907. EMSA and gel shift assays showed the binding of RBP to a probe comprised of nucleotides from -919 to -900. Cold mutant RBP probe did not compete off RBP binding (Fig. 5c). This same putative RBP sequence bound an Ets-1 transcription factor as well as the co-activator p300.
3.5. hCAP18 5′UTR dramatically effects Luciferase gene expression
To evaluate the effect of the 5′UTR sequence of the human cathelicidin gene, we prepared a chimeric construct fusing 108 bp of hCAP18 5′UTR to the luciferase ORF. Compared to the pGL3 promoter vector, we observed a dramatic decrease in Luciferase activity in the construct containing the hCAP18 5′UTR cloned upstream of the Luciferase gene (Fig. 5d). Therefore, this fragment of the hCAP18 5′UTR inhibits transcriptional activity of a downstream ORF in transiently transfected cells. Insertion of the same 108 bp of 5′UTR in front of the promoter sequence of the pGL3 promoter vector did not change luciferase activity, indicating a position-specific repressive activity.
A putative upstream ORF (UORF) is located out of frame relative to the hCAP18 coding sequence and putatively encodes an 86 amino acid polypeptide, with an upstream ATG in a favorable context for translation initiation. We transiently transfected ORF1CAP18, ORF3CAP18 and UORF constructs (described in methods) into K562 cells and examined the effect of these hCAP18 5′UTR sequences on protein expression. Cell lysates were tested for expression of the hCAP18 recombinant protein 24 and 48 hours post-transfection. As shown in Fig. 5e, hCAP18 5′UTR had a strong inhibitory effect on protein production. Furthermore, using the anti-V5 antibody, no protein corresponding to the UORF was detected by immunoblotting of cell lysates after transfection with the UORF construct. The same result was found using an antibody generated against a peptide of the UORF theoretical protein (Biogenes, Germany). No protein was detected in the media either.
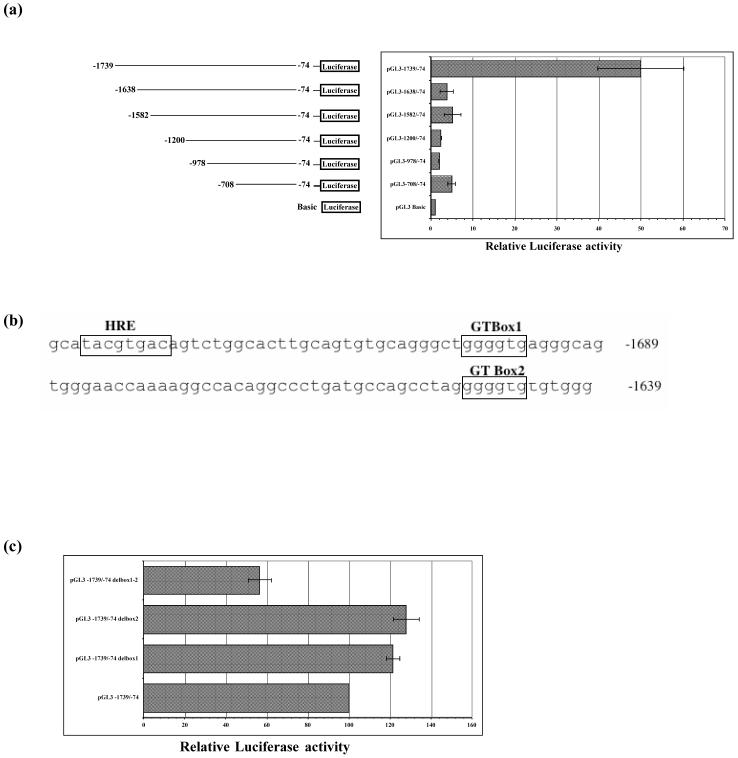
3.6. Identification of an upstream enhancer
Transient transfection of the construct pGL3-1739/-74 in K562 cells showed a 25-fold greater activity than pGL3-978/-74. To narrow down the sequence essential for this transcriptional activation, further 5′ deletion mutants were created. A 101 bp region within the 5′ promoter sequence of hCAP18 gene enhanced expression of the reporter plasmid in K562 cells (Fig. 6a). The activity of the reporter plasmid containing the enhancer pGL3-1739/-74 in HL-60, U937, and A549 was not as high as in the K562 cells. No enhancer activity was detected in Cos7 cell transfection.
Fig. 6.
Identification of an upstream positive regulatory element. (a) Promoter 5′-deletion constructs in pGL3-Basic were transiently expressed in K562 cells and assayed for luciferase reporter gene assays. Deletion of the most 5′ 101 bp dramatically reduced promoter activity. Data are reported as relative firefly luciferase, normalized to Renilla luciferase activity. The results are expressed relative to pGL3 basic vector set at 1 and represent the means ± SD of three experiments. (b) Partial sequence of hCAP18 promoter, open boxes indicated putative binding sites for transcription factors HIF-1 (HRE), and Sp1/Kruppel like factors (GT-boxes) within the enhancer element. (c) BoxGT deletion single and double mutant constructs of hCAP18 promoter were transiently transfected into K562 cells. Luciferase activity was normalized and results are expressed relative to the construct pGL3-1739/-74. Only deletion of both cis-elements affected the promoter activity.
Potential cis-elements were identified computationally, including hypoxic response element (HRE), and two GT boxes (Fig. 6b). Deletion of one GT box at a time did not affect the promoter activity. However, deletion of both GT boxes resulted in lowering luciferase activity by 35% compared to pGL3-1739/-74 (Fig. 6c).
In the murine model, hypoxia inducible transcription factor 1 (HIF-1α̣ deletion led to a dramatic reduction in the active mature form of cathelicidin (Peyssonnaux et al., 2005). To determine whether HIF-1α is playing a role in hCAP18 promoter activation, we treated K562, U937, and A549 cells, already transfected with pGL3-1739/-74 or pGL3-1739/-1, with pharmacologic inducers of HIF-1 [desferrioxamine (DFO) and CoCl2] for 5 hours prior to cell lysis. No increase in the luciferase activity was seen. No change in the luciferase activity was noted when we transfected the cells with a construct mutated for the HIF-1α binding site (HRE).
3.7. Stimulation of hCAP18 promoter by cytokines and agonists of transcription
Luciferase production following transient transfection in K562 cells was assayed in response to IL-1, LPS, IL-6, TNFα, and ATRA. Using either pGL3-978/-1 or the 3′ deleted construct pGL3-978/-74, IL-6, IL-1, TNFα̣ and LPS did not increase luciferase activity after 6 hours of stimulation in transfected K562 cells.
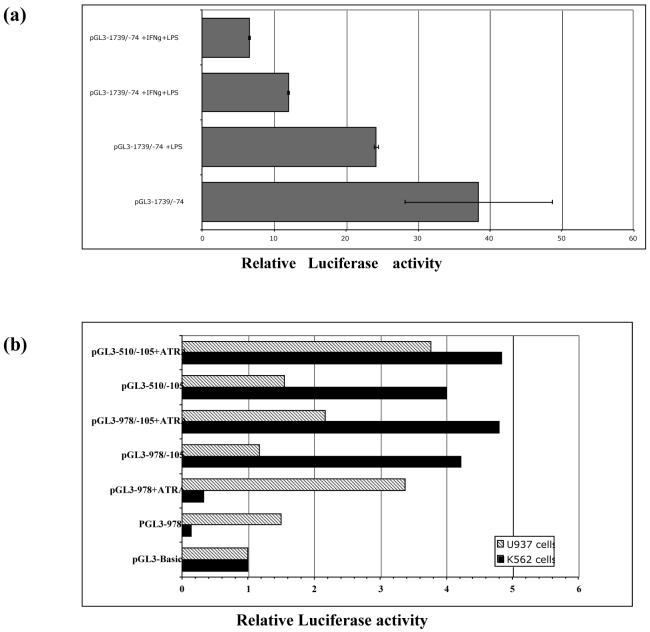
To investigate whether the response elements to these cytokines were located further in the promoter sequence, pGL3-1739/-1 was transfected into both K562 and A549 cells. Overnight stimulation using LPS alone or in conjunction with IFN-γ and TNFα did not abrogate the effect of the repressor element.
Interestingly, IFN-γ stimulation of A549 cells transfected with pGL3-1739/-74 induced less promoter activity, while adding both IFN-γ and LPS further decreased promoter activity (Fig. 7a). No effects were noted in K562 and U937 cells.
Fig. 7.
Effect of all-trans-retinoic acid, IFN-γ, and LPS on hCAP18 promoter activity. (a) A549 cells were transiently transfected with pGL3-1739/-74 construct, then stimulated with IFN-γ and/or LPS. Luciferase activity was measured after 18h. Stimulation with either IFN-γ or LPS alone decreased promoter activity. An additive repressive effect was noted using IFN-γ and LPS simultaneously. (b) U937 and K562 cells were transiently transfected with the indicated 5′ deletion promoter constructs. ATRA were added and luciferase activity was determined 18h after transfection. ATRA induces hCAP18 promoter in U937 cells but not in K562 cells.
In U937 cells, ATRA caused a 2-fold increase in hCAP18 promoter activity, even for the whole promoter sequence construct. No effect was noted in K562 cells. The silencing effect was not abolished in K562 cells upon stimulation with ATRA (Fig. 7b).
4. Discussion
Cathelicidins are multifunctional proteins found in a variety of mammalian species, as well as in hagfish and in salmonids. These potent ancient antiseptic agents are highly expressed in myeloid and epithelial cells and share a conserved preproregion containing the cathelin like domain.
The presence of alternative hCAP18 splice variants in human bone marrow as reported in the present study is novel. These variants encode augmented cathelin domains, with either 10 or 26 more amino acids with well-conserved cathelin signatures (Tomasinsig and Zanetti, 2005). Inclusion of either exon 1b or exon 3b is predicted to introduce a fifth cysteine, potentially disturbing the other disulfide bonds. The splice variant resulting in exon1b inclusion encodes a protein that harbors the potential N-glycosylation site NGSV, whereas the canonical hCAP18 does not have a glycosylation site. Alternative splicing by itself may attenuate cathelicidin gene expression. Since LL-37 is cytotoxic to the eukaryotic cells at a high concentration (Johansson et al., 1998), mechanisms of control may be critical.
After being processed and released, the cathelin domain exerts an antimicrobial activity against bacterial strains that are resistant to LL-37 (Zaiou et al., 2003). The roles of hCAP18, LL-37, and the cathelin domain outside of antimicrobial activity are complex and not well known. Issues of transcriptional activity, processing activity, and overall charge may impact on chemotaxis, cell-cell binding, and interaction with plasma proteins. Therefore, tissue and stage specific expression of the cathelin domain splice variant may have effects on maturation, emigration, and microbial killing. It will be of interest to test the antimicrobial activity of the two predicted isoforms, as well as their ability to inhibit cysteine proteinases. Along these lines, the lactoferrin gene, encoding a secondary granule protein, is alternatively spliced (Siebert and Huang, 1997).
We have identified multiple transcription start sites by sequence analysis of 5′ RACE clones derived from bone marrow and neutrophils. The hCAP18 promoter has a high GC content and no canonical TATA box, consistent with this finding. Furthermore, heterogeneity in the 3′UTR sequence was found by analyzing 3′RACE products. This 5′ and 3′ transcriptional variability could also affect the stability of the message and translation efficiency.
Cathelicidin regulation is poorly characterized and not well understood. It is primarily transcribed in bone marrow. Neutrophils as well as the majority of hematopoietic cell lines have little, if any, protein synthesis. We identified DNA regions that mediate either gene repression or activation within the hCAP18 gene promoter. Deletion of the immediate 5′UTR results in enhanced transcriptional activity in the non-permissive K562 cells as well as in other cells tested, suggesting that this sequence negatively regulates the hCAP18 promoter. Further promoter deletions and point mutations identified 13 bp (-42 to -54) as necessary for the repressor effect. The ATG located 100 bp upstream of the translation start ATG is in a favorable Kozak context and could be recognized by the ribosome. Its deletion or mutation in the context of the pGL3-978/-1 construct did not release the repression; however, deletion in the context of the 3′ deletion promoter construct increased promoter activity 2-fold. These results suggest that repression of hCAP18 gene requires recruitment of other transcription factors to the 3′ end of the promoter sequence, interacting with factors binding the ATG element such as YY1. Further studies will be conducted to reveal the molecular basis of hCAP18 repression.
In order to identify transcriptional repressors recruited by the negative cis-acting elements, EMSA experiments were conducted using nuclear extracts from K562 cells allowing detection of Sp1 binding moiety. The Sp/kruppel like transcription factor family belongs to the conserved zinc finger DNA-binding domain proteins that recognize GC- and GT-box motifs. These proteins are mainly important for the expression of genes that do not contain TATA or CAAT-boxes in their proximal promoters, as well as certain tissue specific genes. Neither mutation nor deletion of the Sp1 binding site at the position (-53/-47) abolished repression of promoter activity. Mithramycin A, an aureolic acid antibiotic that selectively inhibits gene expression by displacing GC-rich binding transcription factors such as Sp1, also showed no effect on promoter activity in K562 cells using pGL3-978/-1.
Repressor proteins are usually linked directly or through co-repressors to the histone deacetylase complex (HDAC) (Wolffe, 1997). Recently, it has been shown that the expression of human cathelicidin is modulated by HDAC in gastrointestinal cells (Schauber et al., 2004). However, trichostatin A (TSA), a specific inhibitor of histone deacetylase, which could have abolished promoter repression, had no effect on hCAP18 promoter activity in K562 cells.
These findings rule out direct involvement of Sp1 in the repression of hCAP18 expression, as well as the presence of histone deacetylase in the repressor complex. The effects of Sp/KLH family members could be indirect, through activation of other inhibitory pathways. The basal transcription element binding protein (BTEB3) represses some genes by binding to the co-repressor mSin3A (Martin et al., 2003). However, using K562 cell nuclear extract and anti-mSin3A antiboby, no supershift was detected by EMSA (data not shown).
A DNA affinity column was used to identify factors binding the repressor element. A hypothetical zinc-finger protein, polyADP-ribose polymerase (PARP), and BCL2 associated transcription factor were identified by mass spectrometry (Unpublished data). Future experiments targeting these factors are needed to clarify their roles in hCAP18 gene regulation. Transient transfection studies using hCAP18 expression vectors with or without the upstream sequence showed a strong inhibitory effect of the hCAP18 5′UTR on protein production. High GC content and an upstream ORF (UORF) may repress gene expression (Lee et al., 2002). Analysis of the 5′UTR using Mfold program predicted a free energy of -40 Kcal/mol. A free energy of -30Kcal/mol is sufficient for inhibiting translation in vitro (Zuker, 2000). 5′UTR sequences may form stable secondary structures capable of inhibiting the translation of the main open reading frame (Kozak, 1987), and this may be position specific (Gray and Hentze, 1994). Immunoblotting with a specific antibody raised against the theoretical protein encoded by the UORF ruled out the translation of such a protein, and the chimeric luciferase constructs showed higher promoter activity when inserting more than one copy of the repressor element (data not shown). These results suggest that the 5′UTR of the hCAP18 gene can behave as a weak internal ribosome entry site (IRES), as reported for the homeodomain protein Gtx (Chappell et al., 2000). Therefore, in hCAP18 permissive cells, there must be some initiation factors, such as eIF4A, which are able to unwind this highly structured 5′UTR through an ATP dependent RNA helicase activity, as has been reported for other genes (Van der Velden and Thomas, 1999).
Transient transfection of further deletion constructs identified a potential binding site for the transcriptional repressor RBP. We showed that other factors such as Ets-1 and coactiavtor p300 could also bind to the same promoter sequence, suggesting a competition between a repressor and an activator, regulating hCAP18 expression in different cells.
Along with this potent repression, we identified other regulatory elements within the hCAP18 promoter, including an inducer in the -1739 to -1638 region. This DNA segment contains a hypoxic response element (HRE) as well as two GT-boxes, which are potential binding sites for Sp1/kruppel like transcription factors (Lania et al., 1997). Our Data suggested that either HIF-1α is not a critical factor enhancing human cathelicidin production, or it is acting in conjunction with other factors not present in K562 cells. Deletion of either GT-box caused no decrease in the promoter activity, whereas double deletion reduced activity by 35 % in K562 cells. This suggests that the transcriptional activation of hCAP18 gene is under the combined action of several transcriptional activators.
Most cathelicidins are constitutively expressed in myeloid progenitor cells and stored as propeptides in mature neutrophil granules. Cathelicidins are also expressed in other tissues and are inducible, for example by inflammation in skin (Agerberth et al., 2000). However, IL-1 stimulation of the hematopoietic cell lines K562 and U937 did not induce the hCAP18 promoter. Similarly, in colon epithelial cells, inflammatory mediators do not affect hCAP18 expression (Schauber et al., 2003).
We found the human cathelicidin gene to be inducible by retinoic acids, as have other laboratories (Wu et al., 2000). Retinoic acid is a strong inducer of defensins and induces granulocytic cell differentiation (Herwig et al., 1996). RARα binds to retinoic acid response elements consisting of direct repeat (A/G)G(G/T)TCA separated by 2 or 5 nucleotides as a heterodimer with the retinoid X receptor RXR and stimulates transcription in response to all trans-retinoic acid ATRA.
These studies demonstrate complex regulation of hCAP18, with a potent repressor within the 5′UTR sequence, an upstream enhancer and two GT boxes that positively regulate hCAP18 promoter activity. The identification of novel hCAP18 splice variants only in bone marrow-derived mRNA indicates compartmentalization of hCAP18 regulation in myeloid as well as epithelial cells. These regulatory elements will increase our understanding of myeloid development and regulation.
Acknowledgements
We thank Dr Kol Zarember (Laboratory of Host Defenses, NIAID, NIH) for his helpful discussions, Dr Li Ding for her technical help, and Dr Glenn Nardone (RTB, NIAID) for making the DNA column. This work was supported by Intramural program of the NIAID.
Appendix
The abbreviations used are
- RACE
rapid amplification of cDNA ends
- PCR
polymerase chain reaction
- RT
reverse transcriptase
- ORF
open reading frame
- UTR
untranslated region
- IL-
interleukin
- LPS
lipopolysaccharide
- IFN
interferon
- TNF
tumor necrosis factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agerberth B, Charo J, Werr J, Olsson B, Idali F, Lindbom L, Kiessling R, Jomvall H, Wigzell H, Gudmundsson GH. The human antimicrobial and chemotactic peptides LL-37 and -defensins are expressed by specific lymphocyte and monocyte populations. Blood. 2000;96:3086–3093. [PubMed] [Google Scholar]
- Bals R, Wang X, Zasloff M, Wilson JM. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9541–9546. doi: 10.1073/pnas.95.16.9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals R, Wilson JM. Cathelicidins- a family of multifunctional antimicrobial peptides. Cell. Mol. Life. Sci. 2003;60:711–720. doi: 10.1007/s00018-003-2186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman P, Johansson L, Asp V, Plant L, Gudmundsson GH, Jonsson AB, Agerberth B. Neisseria gonorrhoeae downregulates expression of the human antimicrobial peptide LL-37. Cell. Microbiol. 2005;7:1009–1017. doi: 10.1111/j.1462-5822.2005.00530.x. [DOI] [PubMed] [Google Scholar]
- Boyum A. Separation of blood leucocytes, granulocytes and lymphocytes. Tissue Antigens. Tissue Antigens. 1974;4:269–274. [PubMed] [Google Scholar]
- Chappello SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl. Acad. Sci. U. S. A. 2000;397:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Y, Chen Q, Schmidt AP, Anderson GM, Wang JM, Wooters J, Oppenheim JJ, Chertov O. LL-37, the neutrophil granule- and epithelial cell-derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J. Exp. Med. 2000;192:1069–1074. doi: 10.1084/jem.192.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohm M, Agerberth B, Ahangari G, Stahle-Backdahl M, Liden S, Wigzell H, Gudmundsson GH. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J. Biol. Chem. 1997;272:15258–15263. doi: 10.1074/jbc.272.24.15258. [DOI] [PubMed] [Google Scholar]
- Frohm NM, Sandstedt B, Sorensen O, Weber G, Borregaard N, Stahle-Backdahl M. The human cationic antimicrobial protein (hCAP18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6. Infect. Immun. 1999;67:2561–2566. doi: 10.1128/iai.67.5.2561-2566.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NK, Hentze MW. Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- Gudmundsson GH, Magnusson KP, Chowdhary BP, Johansson M, Andersson L, Boman HG. Structure of the gene for porcine peptide antibiotic PR-39, a cathelin gene family member: comparative mapping of the locus for the human peptide antibiotic FALL-39. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7085–7089. doi: 10.1073/pnas.92.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwig S, Su Q, Zhang W, Ma Y, Tempst P. Distinct temporal patterns of defensin mRNA regulation during drug-induced differentiation of human myeloid leukemia cells. Blood. 1996;87:350–364. [PubMed] [Google Scholar]
- Islam D, Bandholtz L, Nilsson J, Wigzell H, Christensson B, Agerberth B, Gudmundsson G. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nat. Med. 2001;7:180–185. doi: 10.1038/84627. [DOI] [PubMed] [Google Scholar]
- Johansson J, Gudmundsson GH, Rottenberg ME, Berndt KD, Agerberth B. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J. Biol. Chem. 1998;273:3718–3724. doi: 10.1074/jbc.273.6.3718. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lania L, Majello B, De Luca P. Transcriptional regulation by the Sp family proteins. Int. J. Biochem. Cell. Biol. 1997;29:1313–1323. doi: 10.1016/s1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- Larrick JW, Lee J, Ma S, Li X, Francke U, Wright SC, Balint RF. Structural, functional analysis and localization of the human CAP18 gene. FEBS. Lett. 1996;398:74–80. doi: 10.1016/s0014-5793(96)01199-4. [DOI] [PubMed] [Google Scholar]
- Lee J, Park EH, Couture G, Harvey I, Garneau P, Pelletier J. An upstream open reading frame impedes translation of the Huntington gene. Nucleic Acids Res. 2002;30:5110–5119. doi: 10.1093/nar/gkf664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Thomas T, Laure M, Abderrahmani A, Condorelli DF, Waeber G, Haefliger J-A. Critical Role of the Transcriptional Repressor Neuron-restrictive Silencer Factor in the Specific Control of Connexin36 in Insulin-producing Cell Lines. J. Biol. Chem. 2003;278:53082–53089. doi: 10.1074/jbc.M306861200. [DOI] [PubMed] [Google Scholar]
- Pestonjamasp VK, Huttner KH, Gallo RL. Processing site and gene structure for the murine antimicrobial peptide CRAMP. Peptides. 2001;22:1643–1650. doi: 10.1016/s0196-9781(01)00499-5. [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J. Clin. Invest. 2005;115:1806–1815. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauber J, Svanholm C, Termén S, Iffland K, Menzel T, Scheppach R, Melcher R, Agerberth B, Luhrs H, Gudmundsson GH. The expression of the cathelicidin LL-37 is modulated by short-chain fatty acids in colonocytes: Relevance of signalling pathways. Gut. 2003;52:735–741. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauber J, Iffland K, Frisch S, Kudlich T, Schmausser B, Eck M, Menzel T, Gostner A, Luhrs H, Scheppach W. Histone-deacetylase inhibitors induce the cathelicidin LL-37 in gastrointestinal cells. Mol. Immunol. 2004;41:847–854. doi: 10.1016/j.molimm.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Siebert PD, Huang BCB. Identification of an alternative form of human lactoferrin mRNA that is expressed differentially in normal tissues and tumor-derived cell lines. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2198–2203. doi: 10.1073/pnas.94.6.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen O, Arnljots K, Cowland JB, Bainton DF, Borregaard N. The human antibacterial cathelicidin, hCAP-18, is synthesized in myelocytes and metamyelocytes and localized to specific granules in neutrophils. Blood. 1997;90:2796–2803. [PubMed] [Google Scholar]
- Sorensen O, Bratt T, Johnsen AH, Madsen MT, Borregaard N. The human antibacterial cathelicidin, hCAP-18, is bound to lipoproteins in plasma. J. Biol. Chem. 1999;274:22445–22451. doi: 10.1074/jbc.274.32.22445. [DOI] [PubMed] [Google Scholar]
- Sorensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, Borregaard N. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97:3951–3959. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- Sorensen OE, Cowland JB, Theilgaard-Monch K, Liu L, Ganz T, Borregaard N. Wound healing and expression of antimicrobial peptides/polypeptides in human keratinocytes, a consequence of common growth factors. J. Immunol. 2003;170:5583–5589. doi: 10.4049/jimmunol.170.11.5583. [DOI] [PubMed] [Google Scholar]
- Tomasinsig L, Zanetti M. The cathelicidins--structure and evolution. Curr Protein Pept Sci. Curr. Protein Pept. Sci. 2005;15:23–34. doi: 10.2174/1389203053027520. [DOI] [PubMed] [Google Scholar]
- Travis A, Hagman J, Grosschedl R. Heterogeneously initiated transcription from the pre-B- and B-cell-specific mb-1 promoter: analysis of the requirement for upstream factor-binding sites and initiation site sequences. Mol. Cell. Biol. 1991;11:5756–5766. doi: 10.1128/mcb.11.11.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Velden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int. J. Biochem. Cell. Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang G, Ross CR, Blecha F. Cathelicidin gene expression in porcine tissues: roles in ontogeny and tissue specificity. Infect. Immun. 1999;67:439–442. doi: 10.1128/iai.67.1.439-442.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang G, Minton JE, Ross CR, Blecha F. Regulation of cathelicidin gene expression: induction by lipopolysaccharide, interleukin-6, retinoic acid, and Salmonella enterica serovar Typhimurium infection. Infect. Immun. 2000;68:5552–5558. doi: 10.1128/iai.68.10.5552-5558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ross CR, Blecha F. Characterization of an upstream open reading frame in the 5′ untranslated region of PR-39, a cathelicidin antimicrobial peptide. Mol. Immunol. 2002;39:9–18. doi: 10.1016/s0161-5890(02)00093-7. [DOI] [PubMed] [Google Scholar]
- Zaiou M, Nizet V, Gallo RL. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J. Invest. Dermatol. 2003;120:810–816. doi: 10.1046/j.1523-1747.2003.12132.x. [DOI] [PubMed] [Google Scholar]
- Zanetti M, Litteri L, Gennaro R, Horstmann H, Romeo D. Bactenecins, defense polypeptides of bovine neutrophils, are generated from precursor molecules stored in the large granules. J. Cell. Biol. 1990;111:1363–1371. doi: 10.1083/jcb.111.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Ganz T, Lehrer RI. Structures of genes for two cathelin-associated antimicrobial peptides: prophenin-2 and PR-39. FEBS. Lett. 1995;376:130–134. doi: 10.1016/0014-5793(95)01237-3. [DOI] [PubMed] [Google Scholar]
- Zuker M. Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol. 2000;10:303–310. doi: 10.1016/s0959-440x(00)00088-9. [DOI] [PubMed] [Google Scholar]