Abstract
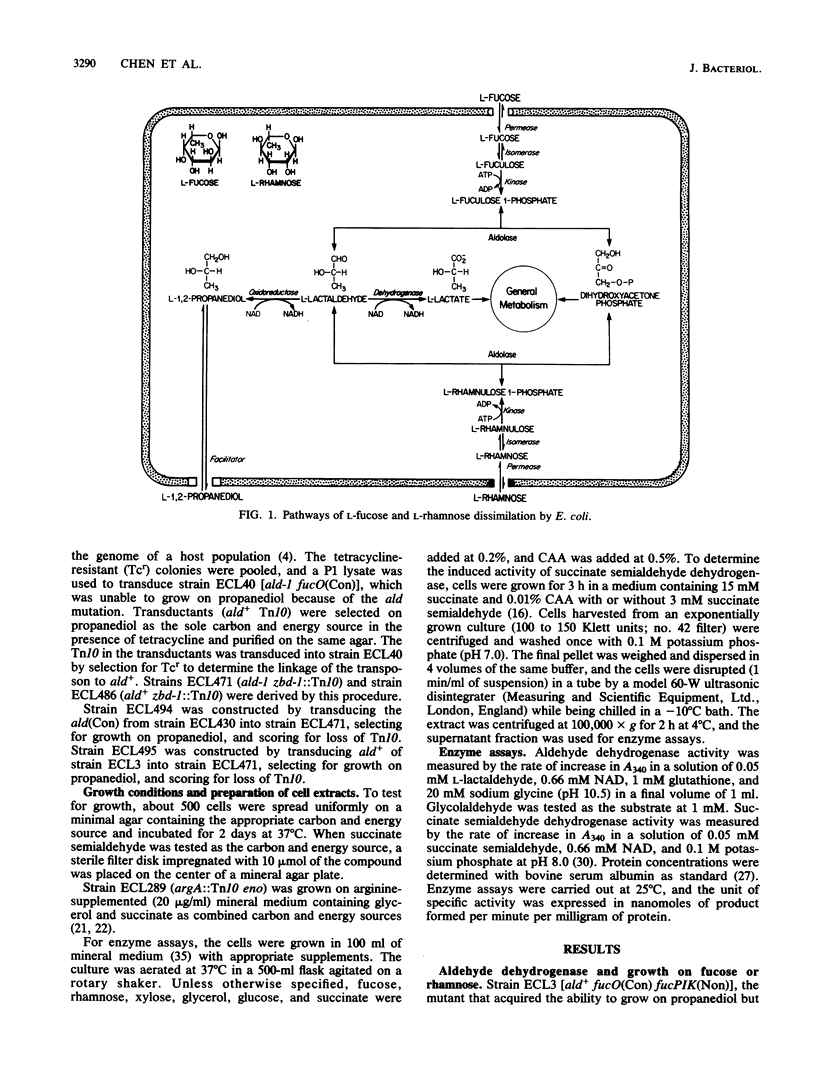
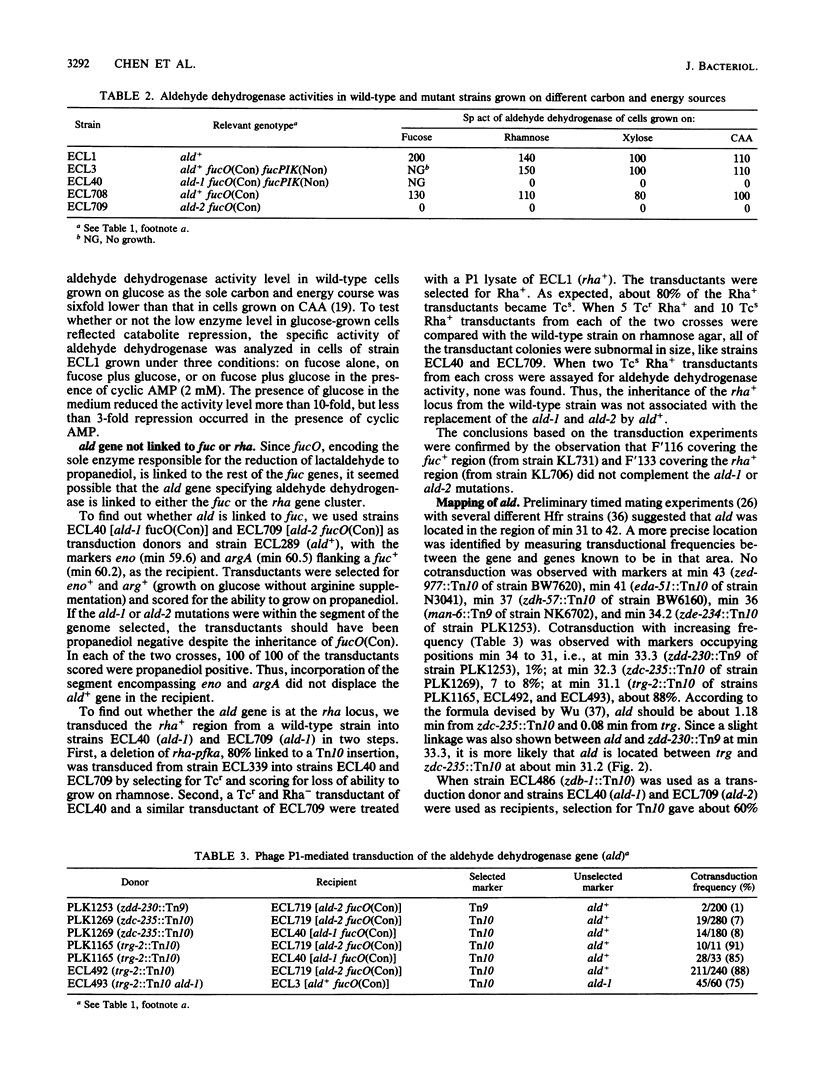
Mutant analysis revealed that complete utilization of L-fucose and L-rhamnose by Escherichia coli requires the activity of a common NAD-linked aldehyde dehydrogenase which converts L-lactaldehyde to L-lactate. Mutations affecting this activity mapped to the ald locus at min 31, well apart from the fuc genes (min 60) encoding the trunk pathway for L-fucose dissimilation (as well as L-1,2-propanediol oxidoreductase) and the rha genes (min 88) encoding the trunk pathway for L-rhamnose dissimilation. Mutants that grow on L-1,2-propanediol as a carbon and energy source also depend on the ald gene product for the conversion of L-lactaldehyde to L-lactate.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman K., Chen Y. M., Magasanik B. Physical and genetic characterization of the glnA--glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkus J. M., Mortlock R. P. Isolation of a mutation resulting in constitutive synthesis of L-fucose catabolic enzymes. J Bacteriol. 1986 Mar;165(3):710–714. doi: 10.1128/jb.165.3.710-714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitner R. M., Kuempel P. L. P1 transduction map spanning the replication terminus of Escherichia coli K12. Mol Gen Genet. 1981;184(2):208–212. doi: 10.1007/BF00272906. [DOI] [PubMed] [Google Scholar]
- Boronat A., Aguilar J. Metabolism of L-fucose and L-rhamnose in Escherichia coli: differences in induction of propanediol oxidoreductase. J Bacteriol. 1981 Jul;147(1):181–185. doi: 10.1128/jb.147.1.181-185.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronat A., Aguilar J. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J Bacteriol. 1979 Nov;140(2):320–326. doi: 10.1128/jb.140.2.320-326.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boronat A., Caballero E., Aguilar J. Experimental evolution of a metabolic pathway for ethylene glycol utilization by Escherichia coli. J Bacteriol. 1983 Jan;153(1):134–139. doi: 10.1128/jb.153.1.134-139.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero E., Baldomá L., Ros J., Boronat A., Aguilar J. Identification of lactaldehyde dehydrogenase and glycolaldehyde dehydrogenase as functions of the same protein in Escherichia coli. J Biol Chem. 1983 Jun 25;258(12):7788–7792. [PubMed] [Google Scholar]
- Chakrabarti T., Chen Y. M., Lin E. C. Clustering of genes for L-fucose dissimilation by Escherichia coli. J Bacteriol. 1984 Mar;157(3):984–986. doi: 10.1128/jb.157.3.984-986.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Chakrabarti T., Lin E. C. Constitutive activation of L-fucose genes by an unlinked mutation in Escherichia coli. J Bacteriol. 1984 Aug;159(2):725–729. doi: 10.1128/jb.159.2.725-729.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Lin E. C. Dual control of a common L-1,2-propanediol oxidoreductase by L-fucose and L-rhamnose in Escherichia coli. J Bacteriol. 1984 Mar;157(3):828–832. doi: 10.1128/jb.157.3.828-832.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Lin E. C. Post-transcriptional control of L-1,2-propanediol oxidoreductase in the L-fucose pathway of Escherichia coli K-12. J Bacteriol. 1984 Jan;157(1):341–344. doi: 10.1128/jb.157.1.341-344.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Lin E. C., Ros J., Aguilar J. Use of operon fusions to examine the regulation of the L-1,2-propanediol oxidoreductase gene of the fucose system in Escherichia coli K12. J Gen Microbiol. 1983 Nov;129(11):3355–3362. doi: 10.1099/00221287-129-11-3355. [DOI] [PubMed] [Google Scholar]
- Cocks G. T., Aguilar T., Lin E. C. Evolution of L-1, 2-propanediol catabolism in Escherichia coli by recruitment of enzymes for L-fucose and L-lactate metabolism. J Bacteriol. 1974 Apr;118(1):83–88. doi: 10.1128/jb.118.1.83-88.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly M. I., Cooper R. A. Two succinic semialdehyde dehydrogenases are induced when Escherichia coli K-12 Is grown on gamma-aminobutyrate. J Bacteriol. 1981 Mar;145(3):1425–1427. doi: 10.1128/jb.145.3.1425-1427.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FALKOW S., SCHNEIDER H., BARON L. S., FORMAL S. B. VIRULENCE OF ESCHERICHIA-SHIGELLA GENETIC HYBRIDS FOR THE GUINEA PIG. J Bacteriol. 1963 Dec;86:1251–1258. doi: 10.1128/jb.86.6.1251-1258.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacking A. J., Aguilar J., Lin E. C. Evolution of propanediol utilization in Escherichia coli: mutant with improved substrate-scavenging power. J Bacteriol. 1978 Nov;136(2):522–530. doi: 10.1128/jb.136.2.522-530.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacking A. J., Lin E. C. Disruption of the fucose pathway as a consequence of genetic adaptation to propanediol as a carbon source in Escherichia coli. J Bacteriol. 1976 Jun;126(3):1166–1172. doi: 10.1128/jb.126.3.1166-1172.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacking A. J., Lin E. C. Regulatory changes in the fucose system associated with the evolution of a catabolic pathway for propanediol in Escherichia coli. J Bacteriol. 1977 May;130(2):832–838. doi: 10.1128/jb.130.2.832-838.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman J. D., Fraenkel D. G. Glyceraldehyde 3-phosphate dehydrogenase mutants of Escherichia coli. J Bacteriol. 1975 Jun;122(3):1175–1179. doi: 10.1128/jb.122.3.1175-1179.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani M., Maitra P. K. Isolation and characterization of Escherichia coli mutants defective in enzymes of glycolysis. Biochem Biophys Res Commun. 1974 Jan;56(1):127–133. doi: 10.1016/s0006-291x(74)80324-4. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- LURIA S. E., ADAMS J. N., TING R. C. Transduction of lactose-utilizing ability among strains of E. coli and S. dysenteriae and the properties of the transducing phage particles. Virology. 1960 Nov;12:348–390. doi: 10.1016/0042-6822(60)90161-6. [DOI] [PubMed] [Google Scholar]
- Lin E. C. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J. The L-rhamnose genetic system in Escherichia coli K-12. Genetics. 1967 Mar;55(3):557–568. doi: 10.1093/genetics/55.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. A., Cooper R. A. An Escherichia coli mutant defective in the NAD-dependent succinate semialdehyde dehydrogenase. Arch Microbiol. 1982 Sep;132(3):270–275. doi: 10.1007/BF00407964. [DOI] [PubMed] [Google Scholar]
- Skjold A. C., Ezekiel D. H. Analysis of lambda insertions in the fucose utilization region of Escherichia coli K-12: use of lambda fuc and lambda argA transducing bacteriophages to partially order the fucose utilization genes. J Bacteriol. 1982 Oct;152(1):120–125. doi: 10.1128/jb.152.1.120-125.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skjold A. C., Ezekiel D. H. Regulation of D-arabinose utilization in Escherichia coli K-12. J Bacteriol. 1982 Oct;152(1):521–523. doi: 10.1128/jb.152.1.521-523.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhara S., Wu T. T., Chused T. M., Lin E. C. Ferrous-activated nicotinamide adenine dinucleotide-linked dehydrogenase from a mutant of Escherichia coli capable of growth on 1, 2-propanediol. J Bacteriol. 1969 Apr;98(1):87–95. doi: 10.1128/jb.98.1.87-95.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhara S., Wu T. T. Purification and properties of lactaldehyde dehydrogenase from Escherichia coli. J Biol Chem. 1969 Oct 10;244(19):5233–5238. [PubMed] [Google Scholar]
- Tanaka S., Lerner S. A., Lin E. C. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J Bacteriol. 1967 Feb;93(2):642–648. doi: 10.1128/jb.93.2.642-648.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1986 Sep 5;191(1):39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagalak B., Frey P. A., Karabatsos G. L., Abeles R. H. The stereochemistry of the conversion of D and L 1,2-propanediols to propionaldehyde. J Biol Chem. 1966 Jul 10;241(13):3028–3035. [PubMed] [Google Scholar]
- Zhu Y., Lin E. C. Loss of aldehyde dehydrogenase in an Escherichia coli mutant selected for growth on the rare sugar L-galactose. J Bacteriol. 1987 Feb;169(2):785–789. doi: 10.1128/jb.169.2.785-789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]



