Abstract
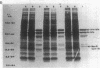
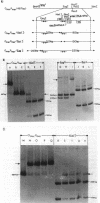
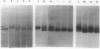
In plasmid NR1 the expression of genes involved in mercury resistance (Tn21) is regulated by the trans-acting product of the merR gene. An in vivo T7 RNA polymerase-promoter overexpression system was used to detect a protein of approximately 16,000 daltons encoded by the merR reading frame. Overexpressed MerR constituted about 5% of labeled proteins. An in vitro MerR-mer-op (mer-op is the mer operator and promoter region) gel electrophoresis binding assay established that the binding site for MerR was located between the putative -35 and -10 sequences of the promoter for the mer structural genes. A nonsense mutation in the carboxyl half of MerR resulted in the loss of biological function and the loss of in vitro mer-op binding properties.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrineau P., Gilbert P., Jackson W. J., Jones C. S., Summers A. O., Wisdom S. The DNA sequence of the mercury resistance operon of the IncFII plasmid NR1. J Mol Appl Genet. 1984;2(6):601–619. [PubMed] [Google Scholar]
- Barrineau P., Gilbert P., Jackson W. J., Jones C. S., Summers A. O., Wisdom S. The structure of the mer operon. Basic Life Sci. 1985;30:707–718. doi: 10.1007/978-1-4613-2447-8_49. [DOI] [PubMed] [Google Scholar]
- Barrineau P., Summers A. O. A second positive regulatory function in the mer (mercury resistance) operon. Gene. 1983 Nov;25(2-3):209–221. doi: 10.1016/0378-1119(83)90225-1. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., Ring J. Characterization of T7-specific ribonucleic acid polymerase. II. Inhibitors of the enzyme and their application to the study of the enzymatic reaction. J Biol Chem. 1973 Mar 25;248(6):2245–2250. [PubMed] [Google Scholar]
- Coleman J., Inouye M., Nakamura K. Mutations upstream of the ribosome-binding site affect translational efficiency. J Mol Biol. 1985 Jan 5;181(1):139–143. doi: 10.1016/0022-2836(85)90332-8. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B., McIntire S. A., Willetts N., Schottel J., Kinscherf T. G., Silver S., Shannon W. A., Jr Properties of lambda transducing bacteriophages carrying R100 plasmid DNA: mercury resistance genes. J Bacteriol. 1978 Dec;136(3):1084–1093. doi: 10.1128/jb.136.3.1084-1093.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Brown N. L. Identification of the merR gene of R100 by using mer-lac gene and operon fusions. J Bacteriol. 1985 Sep;163(3):1153–1157. doi: 10.1128/jb.163.3.1153-1157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Ginnity F. Some mercurial resistance plasmids from different incompatibility groups specify merR regulatory functions that both repress and induce the mer operon of plasmid R100. J Bacteriol. 1985 May;162(2):773–776. doi: 10.1128/jb.162.2.773-776.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Nakahara H., Weiss A. A., Silver S. Transposon A-generated mutations in the mercuric resistance genes of plasmid R100-1. J Bacteriol. 1979 Oct;140(1):167–181. doi: 10.1128/jb.140.1.167-181.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambill B. D., Summers A. O. Versatile mercury-resistant cloning and expression vectors. Gene. 1985;39(2-3):293–297. doi: 10.1016/0378-1119(85)90326-9. [DOI] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen D., Iserentant D., Derom C., Fiers W. Systematic alteration of the nucleotide sequence preceding the translation initiation codon and the effects on bacterial expression of the cloned SV40 small-t antigen gene. Gene. 1982 Jan;17(1):55–63. doi: 10.1016/0378-1119(82)90100-7. [DOI] [PubMed] [Google Scholar]
- Jackson W. J., Summers A. O. Biochemical characterization of HgCl2-inducible polypeptides encoded by the mer operon of plasmid R100. J Bacteriol. 1982 Aug;151(2):962–970. doi: 10.1128/jb.151.2.962-970.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W. J., Summers A. O. Polypeptides encoded by the mer operon. J Bacteriol. 1982 Feb;149(2):479–487. doi: 10.1128/jb.149.2.479-487.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. L., Yanofsky C. Trp aporepressor production is controlled by autogenous regulation and inefficient translation. Proc Natl Acad Sci U S A. 1982 May;79(10):3120–3124. doi: 10.1073/pnas.79.10.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lathe R., Kieny M. P., Skory S., Lecocq J. P. Linker tailing: unphosphorylated linker oligonucleotides for joining DNA termini. DNA. 1984;3(2):173–182. doi: 10.1089/dna.1984.3.173. [DOI] [PubMed] [Google Scholar]
- Lund P. A., Ford S. J., Brown N. L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986 Feb;132(2):465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Misra T. K., Brown N. L., Fritzinger D. C., Pridmore R. D., Barnes W. M., Haberstroh L., Silver S. Mercuric ion-resistance operons of plasmid R100 and transposon Tn501: the beginning of the operon including the regulatory region and the first two structural genes. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5975–5979. doi: 10.1073/pnas.81.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran T., Walsh C. Metalloregulatory DNA-binding protein encoded by the merR gene: isolation and characterization. Science. 1987 Jan 9;235(4785):211–214. doi: 10.1126/science.3798107. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Schottel J. L., Sninsky J. J., Cohen S. N. Effects of alterations in the translation control region on bacterial gene expression: use of cat gene constructs transcribed from the lac promoter as a model system. Gene. 1984 May;28(2):177–193. doi: 10.1016/0378-1119(84)90255-5. [DOI] [PubMed] [Google Scholar]
- Sherratt D., Arthur A., Burke M. Transposon-specified, site-specific recombination systems. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):275–281. doi: 10.1101/sqb.1981.045.01.040. [DOI] [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer H. A., Hui A., Comstock L. J., Wong E., Vasser M. Portable Shine-Dalgarno regions: a system for a systematic study of defined alterations of nucleotide sequences within E. coli ribosome binding sites. DNA. 1983;2(3):231–235. doi: 10.1089/dna.1983.2.231. [DOI] [PubMed] [Google Scholar]