Abstract
The infectious reovirus RNA system was used to construct a mutant with two temperature-sensitive (ts) lesions in genome segments M2 and S2, respectively. The double mutant is about 300 times more ts than either of its parents, which are about 1,500 and 170 times more ts than their wild-type parent reovirus ST3 strain Dearing. At 39°C the double mutant is essentially unable to multiply. In spite of its striking temperature sensitivity, the double mutant elicits the formation of significant amounts of neutralizing antibodies in newborn mice. Possible mechanisms responsible for this are discussed, as is the significance of this double ts mutant in relation to current searches for safe and efficient vaccine strains.
The genome of mammalian reoviruses comprises 10 segments of double-stranded (ds) RNA that range in length from about 1,200 to about 3,900 bp. Upon infection these genome segments are transcribed conservatively into single-stranded (ss) RNA species that are then translated and eventually transcribed into minus strands with which they remain associated, thereby providing the progeny dsRNA genome segments.
Both the ss and the ds forms of reovirus genome segments are infectious in the sense that if complete sets of either form of, say, reovirus serotype (ST) 3 are lipofected into mouse fibroblasts, together with reaction mixtures of cell-free protein-synthesizing systems such as rabbit reticulocyte lysates in which complete sets of the ssRNA species had been translated for 60 min, and further, if these cells are also infected with a helper virus such as reovirus ST1 or ST2, infectious reovirus ST3 is formed in them (1). This system, the infectious reovirus RNA system, is extremely powerful in that it permits, on the one hand, studies of the nature of the processes involved in the generation of the segmented genome of reoviruses, and, on the other, the introduction of foreign genetic information into the reovirus genome. Using this system, it has recently been found that the introduction of heterologous genome segments into the reovirus genome, such as the introduction of ST2 genome segments into the ST3 genome, requires the presence of “acceptance” signals that are located in the S4 genome segment (2–4). These signals consist of two mutations, G74 to A and G624 to A, the presence of which is absolutely essential if any ST2 genome segment, or any truncated genome segment such as are present in the genomes of defective interfering particles, is to be incorporated into the ST3 genome. Interestingly, although the S4 genome segment variant with the G74 and G624 mutations is essential for the acceptance of heterologous (heterotypic, that is, containing foreign genetic information or truncated) genome segments into the ST3 genome, virus particles with genomes that contain it together with the other nine wild-type (wt) ST3 genome segments are noninfectious (2).
Introduction of heterologous genome segments into the reovirus genome proceeds optimally if the wt genome segments that are to be replaced are removed completely from the accepting genome segment population: eliminating such genome segments also confers the major advantage of obviating the necessity for selecting the desired product of the construction, which may be difficult, if not impossible. We describe here a technique for achieving this purpose and use it to construct a double temperature-sensitive (ts) mutant of reovirus ST3 strain Dearing, which we characterize with respect to its ability to multiply at nonpermissive temperatures and to generate neutralizing antibodies; clearly mutants with ts lesions in two genome segments could provide superior vaccine strains. The Reoviridae family includes several important pathogens, like human rotaviruses and the sheep bluetongue viruses, that could conceivably be controlled using this approach. It turns out that although the double ts mutant is extraordinarily ts, it nevertheless elicits the formation of significant amounts of neutralizing antibodies in mice.
MATERIALS AND METHODS
Virus.
The following strains of reovirus were used: the ST3 strain Dearing, the ST2 strain D/5 Jones, the group A strain Dearing mutant ts201 (tsA 201), and the group C strain Dearing mutant ts447 (tsC 447)(5–7). All were grown in mouse L929 fibroblasts in MEM or Joklik-modified MEM (GIBCO/BRL), both supplemented with 5% fetal bovine serum (HyClone).
Reovirus ssRNA.
The ss forms of the viral genome segments were transcribed by cores and isolated as described (1).
The Infectious Reovirus RNA System.
The system was used as described by Roner et al. (1). Reovirus ST2 was used as the helper virus. The system was modified with respect to incubation times and temperature to accommodate the the use of ts mutants: lipofected cells were harvested after incubation for 5 days at 30°C and plaques were isolated after incubation for 12 days at 30°C. Under these conditions ST2 formed plaques only after 16 days.
Construction of the tsA/C Double Mutant.
The tsA/C double mutant was constructed from two populations of RNA. The first was the ssRNA population transcribed by cores of tsA 201 from which the s2 genome segment had been removed. This was accomplished by adding to 2 pmol of tsA 201 ssRNA 10 pmol of the oligodeoxyribonucleotide complementary to residues 774–760 of reovirus ST3 genome segment S2 (6), together with 5 units of RNase H. After 20 min incubation at 50°C the RNA preparation was extracted three times with phenol/chloroform and the RNA was precipitated with sodium acetate. Selective removal of only the s2 RNA species was confirmed by gel electrophoretic analysis of the RNA and analysis of the protein products formed after translation in a rabbit reticulocyte lysate. The second parent RNA preparation comprised the small (s) size class RNA species (s1–s4) of the RNA population transcribed by tsC 447 cores. These two RNA preparations were lipofected into mouse L929 fibroblasts, both before and after translation in a rabbit reticulocyte lysate, using the experimental conditions of the infectious reovirus RNA system.
Virus Titration.
Infected mouse L929 fibroblast monolayers were incubated at 30°C (the permissive temperature), 37°C (the standard temperature), or 39°C (the nonpermissive temperature). Incubation was for 12, 5, and 5 days, respectively. Neutral red was added 24 hr before counting.
Conditions for Determining Virus Growth Curves.
To examine the growth characteristics of the new mutant, L929 fibroblast monolayers (1 × 106 cells) were infected with either wt ST3 virus, tsA 201, tsC 447, or isolates of the new double ts mutant (tsA/C). After adsorption for 1 hr at 4°C, the cells were washed (time 0) and incubation was continued at either 30°C or 39°C. At the indicated times, cells in triplicate wells were harvested, sonicated, and frozen at −20°C. Virus was then titrated as described above.
Measurement of the Ability of Virus to Replicate and Generate Neutralizing Antibodies in Newborn Mice.
Three litters of mice were randomized and divided into three groups. One was inoculated with wt reovirus ST3, the second was inoculated with a sample of the same virus that had been UV inactivated, and the third was inoculated with tsA/C (see below). Each mouse was injected into the left hindlimb with 20 μl gelatin saline containing 3.2 × 108 virus particles. After 2, 7, and 28 days, two or three mice (see below) from each group were sacrificed, and their left hindlimbs and brains were excised, placed into 2 ml gelatin saline, and frozen. The tissues were then frozen and thawed three times, sonicated, and the virus in them was then titrated. Samples of blood (20 μl) were also collected at all time points, diluted into 80 μl of 1.25 x SSC (0.15 M NaCl/0.015 M sodium citrate, pH 7.5) and stored at −20°C for antibody titer measurement.
Measurement of Neutralizing Antibody Titers.
Neutralizing antibody titers were measured using a plaque reduction assay. To 25 μl of the various samples of mouse blood 200 plaque-forming units of reovirus ST3 in 50 μl of PBS (0.14 M NaCl/0.003 M KCl/0.008 M Na2HPO4/0.0015 M KH2PO4) were added. After incubation for 60 min at 37°C, an additional 425 μl of PBS was added and 250 μl of the mixtures were then added to each of two microtiter wells containing confluent L929 mouse fibroblast monolayers for standard plaque assays.
RESULTS
Construction of the ST3 tsA/C Double Mutant.
The parents chosen for construction of the double mutant were tsA 201, a group A mutant (5), and tsC 447, a group C mutant (5–7). The ts lesion in tsA 201 is in genome segment M2, which encodes protein μ1, which in cleaved form [to protein μ1C and component viii (8, 9)] is the major component of the outer capsid shell of reovirus particles (8). Protein μ1C controls susceptibility to proteolytic digestion and thus conversion to subviral particles (10), which is the first step in reovirus replication. It possesses strong affinity for protein σ3, the other major component of the reovirus outer capsid shell (11).
We sequenced the M2 genome segment of tsA 201 and found that it differed in two locations from wt M2 (12), namely G944 to U and C972 to U.
The ts lesion in tsC 447 is in genome segment S2 (6, 7), which encodes protein σ2, one of the two major components of the inner capsid shell of reovirus particles (8). It possesses strong affinity for protein λ1, the other major component of the inner capsid shell (8, 13), and weak affinity for dsRNA (14). No enzymatic activity has yet been found to be associated with either μ1 or σ2.
For constructing the double mutant, the infectious reovirus RNA system was used in its ssRNA configuration; that is, ssRNA was lipofected in free form as well as following translation for 60 min in a rabbit reticulocyte lysate. Two ssRNA preparations were used. The first comprised tsA 201 ssRNA from which the s2 RNA had been removed by hybridizing it to an excess of the oligodeoxyribonucleotide complementary to residues 774–760 of ST3 plus-stranded ST3 s2 RNA and digesting the annealed product with RNase H (see Materials and Methods). Controls showed that two such treatments reduced the amount of intact s2 RNA in tsA 201 ssRNA preparations to undetectable levels without affecting the relative amounts of the other nine RNA species. The second ssRNA preparations comprised the 4 s size class RNA species of tsC 447 isolated in density gradients. The reason for not using the large (l) and medium (m) size class RNA species of tsA 201 for construction of the double mutant rather than its specifically s2 RNA-depleted RNA complement was that density gradient fractionation does not remove s2 RNA species nearly as completely as the hybridization–digestion approach described above.
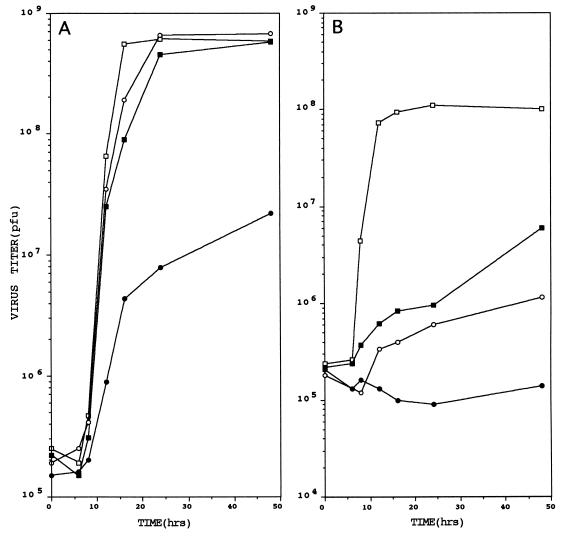
Mouse L929 fibroblasts lipofected with appropriate amounts of these two RNA preparations, in free and translated form, and also infected with reovirus ST2 as helper virus, yielded viral progeny that formed very small plaques after 12 days of incubation at 30°C. The virus in four of them (tsA/C) was isolated and the ratios of the plaquing efficiencies at 30°C to those at 39°C of the four isolates were determined. They were indistinguishable. Table 1 shows the efficiency of plaquing at 30°C/39°C for wt ST3 strain Dearing, tsA 201, tsC 447, and tsA/C; and Fig. 1 shows their one-step growth curves at 30° and 39°C. It is seen that tsA/C was very much more ts than either of its parents. For them 30°C was permissive, but for tsA/C it was very largely nonpermissive; and, at 39°C, both tsA 201 and tsC 447 were able to replicate significantly, but tsA/C was unable to replicate at all. Together with the efficiency of plaquing at 30°C/39°C data, these results indicated that the double mutant was about 300 times more ts than either of its parents, which suggested that each of its ts lesions functioned independently.
Table 1.
Plaquing efficiencies 30°/39°
| Virus | Efficiency of plaquing 30°/39° |
|---|---|
| Reovirus ST3 strain Dearing | 1.12 |
| tsA 201 | 165 |
| tsC 447 | 1,450 |
| tsA/C | 450,000 |
Figure 1.
One-step growth curves of wt reovirus ST3, tsA 201, tsC 447, and tsA/C at 30°C (A) and 39°C (B) in mouse fibroblast monolayers. For experimental details see Materials and Methods and text. □, wt virus; ▪, tsA 201; ○, tsC 447; •, tsA/C.
To prove that tsA/C possessed both parental mutant genome segments, its M2 and S2 RNA species were sequenced. In M2, wt virus and tsC 447 possessed G at position 944, whereas tsA 201 and tsA/C possessed U; and at position 972 wt virus and tsC 447 possessed C, whereas tsA 201 and tsA/C possessed U. In S2, wt virus and tsA 201 possessed A at position 1166, whereas tsC 447 and tsA/C possessed G. Thus, there is no doubt that in tsA/C the M2 and S2 genome segments were derived from tsA 201 and tsC 447, respectively.
Ability of tsA/C to Multiply and Generate Neutralizing Antibodies in Mice.
The greatly diminished capacity of tsA/C to replicate at body temperature, coupled with the fact that its temperature sensitivity is caused by mutations in two genome segments, make it a potentially very attractive model vaccine strain. The question is, is it so ts, that is, does it replicate so poorly at body temperature, that it does not elicit the formation of significant amounts of neutralizing antibodies?
To answer this question tsA/C was injected into the left hindlimbs of newborn mice. On days 2, 5, 7, and 28 after injection some of the mice were sacrificed and the amount of virus present at the site of inoculation and in the brain, as well as the amount of neutralizing antibodies in the blood, were determined. Two controls were used: wt reovirus ST3 strain Dearing, the original parent of tsA/C, and UV-inactivated reovirus ST3 strain Dearing (so as to determine the immunogenicity of nonreplicating wt virus). One-third of three randomized 2-day old mouse litters were injected with each virus preparation, with each animal receiving 20 μl containing 3 × 108 virus particles. Two animals were sacrificed on days 2 and 5, and three on days 7 and 28.
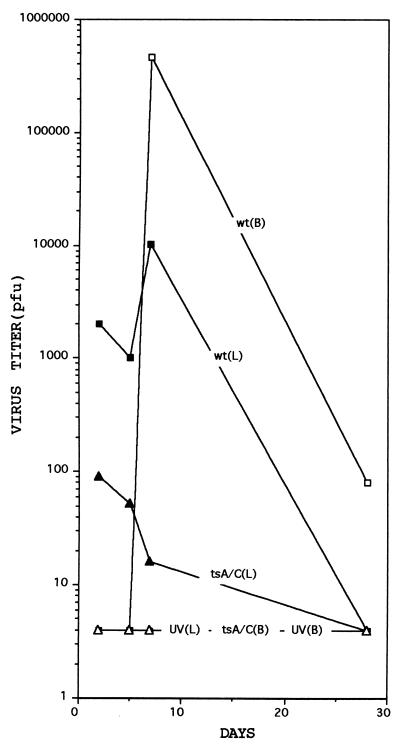
The amounts of virus at the site of inoculation and in the brain are shown in Fig. 2. The amount of wt virus in the hindlimb increased about 10-fold between days 5 and 7, and then decreased to undetectable levels by day 28. No virus could be detected in the brain on days 2 and 5; the amount of virus then increased rapidly during the next 48 hr and then declined. Small amounts of the double mutant could be demonstrated at the site of inoculation until day 7, but could never be detected in the brain. Thus, tsA/C was essentially unable to multiply in mice (Fig. 2).
Figure 2.
Growth of reovirus ST3 and tsA/C in newborn mice at the site of inoculation (L) and in the brain (B). UV-inactivated ST3 virus was used as the negative control. For experimental details see Materials and Methods and text. Virus titers are the total amounts of virus present in the hindlimb or in the brain.
The relative amounts of neutralizing antibodies that were generated are shown in Fig. 3. No significant amounts of antibody were present before day 7 when the amounts of antibody elicited by all three virus preparations were about the same. Three weeks later both wt virus and tsA/C had elicited the formation of amounts of neutralizing antibodies that greatly exceeded that elicited by UV-inactivated virus. This result was confirmed by a 50% plaque reduction assay. The neutralization titers of the day 28 wt reovirus ST3, tsA/C, and UV-inactivated reovirus ST3 blood samples were 1,024, 512, and 128, respectively.
Figure 3.
Generation of neutralizing antibodies against reovirus ST3 in newborn mice following inoculation with wt ST3 virus (solid bars), UV-inactivated ST3 virus (open bars), or with tsA/C (shaded bars). For experimental details see Materials and Methods and text.
Thus, in spite of its greatly reduced capacity to replicate in the mouse, the double mutant tsA/C was capable of generating highly significant amounts of neutralizing antibodies.
DISCUSSION
The results reported in this paper bear on three important areas. First, they demonstrate that the infectious reovirus RNA system permits the construction of any desired reovirus genome. The example addressed here, namely the construction of the double ts mutant tsA/C, did not involve the introduction of heterotypic genome segments into the ST3 genome and therefore did not require the presence of acceptance signals G74 to A and G624 to A in genome segment S4 (2). The significant feature of the double ts mutant construction reported here is that it eliminated the necessity for selecting for the desired progeny because the genome segment that was to be replaced was removed completely from the accepting genome segment population. This was achieved via an approach that employed cleavage by RNase H of short ds regions generated by annealed complementary oligodeoxyribonucleotides. Using this very efficient removal procedure and the variant S4 genome segment that possesses the mutations that constitute the signals required for accepting heterotypic/foreign genome segments, any desired reovirus ST3 genome can now be constructed, even genomes that cannot be selected.
Second, the double ts mutant constructed here is about 300 times more ts than either of its parents: clearly, the temperature sensitivity of each of its mutations functions independently, which, although it appeared likely, could not be taken for granted. This makes it a potentially very attractive vaccine strain, because it differs minimally from the wt strain, yet, being a double mutant, would be extremely safe.
Third, in spite of its greatly reduced ability to replicate in mice relative to wt reovirus ST3, the double mutant tsA/C elicits the formation of very significant amounts of neutralizing antibodies. The reason may be as follows. Reovirus replication no doubt initiates normally in cells infected with the double ts mutant; although nonfunctional forms of proteins μ1 and σ2 would be produced, normal amounts of fragments of wt proteins σ1, μ1, σ3, and λ2, the proteins that elicit the formation of neutralizing antibodies (15–17), are very likely displayed on cell surfaces. Cells infected with reovirus are probably eliminated by apoptosis, in the induction of which proteins σ1 and μ1 play a role (18). Protein μ1 is formed in large amounts in infected cells and appears to be very cytotoxic; in particular, it causes chromium release from cell membranes (19), interferes with virus multiplication in special situations (20), and blocks transmembrane transport (21). Interestingly, ST3 μ1 is significantly more active in these functions than ST1 μ1. We have also found that when cloned into the thymidine kinase gene of vaccinia virus, 9 of the 10 reovirus genome segments are expressed very efficiently [to the extent of 0.1–1% of total cellular protein (4)]; the one genome segment that is expressed very poorly, not only via the vaccinia, but also via the Sindbis virus expression system (22), is genome segment M2, which encodes protein μ1. Since ST1 μ1 is already known to be less cytotoxic than ST3 μ1, protein μ1 of tsA 201 and tsA/C may similarly be significantly less cytotoxic than wt ST3 μ1, thus causing cells in which it is formed to survive much longer than cells infected with wt serotype 3 virus, with consequent expression of the fragments of proteins that elicit the formation of neutralizing antibodies for greatly extended periods of time without the necessity for multiple cycles of reinfection. Whatever the explanation, the double mutant tsA/C elicits the formation of significant amounts of neutralizing antibodies and would be an excellent vaccine strain. The significance of the results reported here lies in the potential of the infectious RNA technology being transferred to the rotavirus and orbivirus genera, which include several medically and economically very important viruses.
Acknowledgments
This work was supported by National Institutes of Health Grant 5R01 AI 08909 and by Grants 3233 and 3233A from the Council for Tobacco Research.
ABBREVIATIONS
- ss
single stranded
- ds
double stranded
- l
m, and s, large, medium, and small size class ssRNA genome segments
- L
M, and S, large, medium, and small size class dsRNA genome segments
- ST
serotype
- wt
wild type
- ts
temperature sensitive
References
- 1.Roner M R, Sutphin L A, Joklik W K. Virology. 1990;179:845–852. doi: 10.1016/0042-6822(90)90153-i. [DOI] [PubMed] [Google Scholar]
- 2.Roner M R, Lin P-N, Nepluev I, Kong L-J, Joklik W K. Proc Natl Acad Sci USA. 1995;92:12362–12366. doi: 10.1073/pnas.92.26.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joklik W K, Roner M R. J Biol Chem. 1995;270:4181–4184. doi: 10.1074/jbc.270.9.4181. [DOI] [PubMed] [Google Scholar]
- 4.Joklik W K, Roner M R. Prog Nucleic Acids Res Mol Biol. 1996;53:249–281. doi: 10.1016/s0079-6603(08)60147-6. [DOI] [PubMed] [Google Scholar]
- 5.Fields B N, Joklik W K. Virology. 1969;37:335–342. doi: 10.1016/0042-6822(69)90217-7. [DOI] [PubMed] [Google Scholar]
- 6.Cashdollar L W, Esparza J, Hudson G R, Chmelo R, Lee P W K, Joklik W K. Proc Natl Acad Sci USA. 1982;79:7644–7648. doi: 10.1073/pnas.79.24.7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiener J R, Joklik W K. Virology. 1989;170:340–341. doi: 10.1016/0042-6822(89)90392-9. [DOI] [PubMed] [Google Scholar]
- 8.Smith R E, Zweerink H J, Joklik W K. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 9.Zweerink H J, Joklik W K. Virology. 1970;41:501–522. doi: 10.1016/0042-6822(70)90171-6. [DOI] [PubMed] [Google Scholar]
- 10.Drayna D, Fields B N. J Virol. 1982;41:110–118. doi: 10.1128/jvi.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huismans H J, Joklik W K. Virology. 1976;70:411–424. doi: 10.1016/0042-6822(76)90282-8. [DOI] [PubMed] [Google Scholar]
- 12.Jayasuriya A K, Nibert M L, Fields B N. Virology. 1988;163:591–602. doi: 10.1016/0042-6822(88)90300-5. [DOI] [PubMed] [Google Scholar]
- 13.Xu P, Miller S E, Joklik W K. Virology. 1993;197:726–731. doi: 10.1006/viro.1993.1648. [DOI] [PubMed] [Google Scholar]
- 14.Dermody T S, Stiff L A, Nibert M L, Coombs K M, Fields B N. J Virol. 1991;65:5721–5731. doi: 10.1128/jvi.65.11.5721-5731.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner H L, Fields B N. J Exp Med. 1977;146:1305–1310. doi: 10.1084/jem.146.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyler K L, Mann M A, Fields B N, Virgin H W, IV. J Virol. 1993;67:3446–3453. doi: 10.1128/jvi.67.6.3446-3453.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes E C, Lee P W K, Miller S E, Joklik W K. Virology. 1981;108:147–155. doi: 10.1016/0042-6822(81)90534-1. [DOI] [PubMed] [Google Scholar]
- 18.Tyler K L, Squire M K T, Brown A L, Pike B, Willis D, Oberhaus S M, Dermody T S, Cohen J J. J Virol. 1995;70:7984–7951. doi: 10.1128/jvi.70.11.7984-7991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucia-Jandris P, Hooper J W, Fields B N. J Virol. 1993;67:5339–5345. doi: 10.1128/jvi.67.9.5339-5345.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rozinoff M N, Fields B N. J Virol. 1994;68:6667– 6671. doi: 10.1128/jvi.68.10.6667-6671.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazelton P R, Coombs K M. Virology. 1995;207:46–58. doi: 10.1006/viro.1995.1050. [DOI] [PubMed] [Google Scholar]
- 22.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]