Abstract
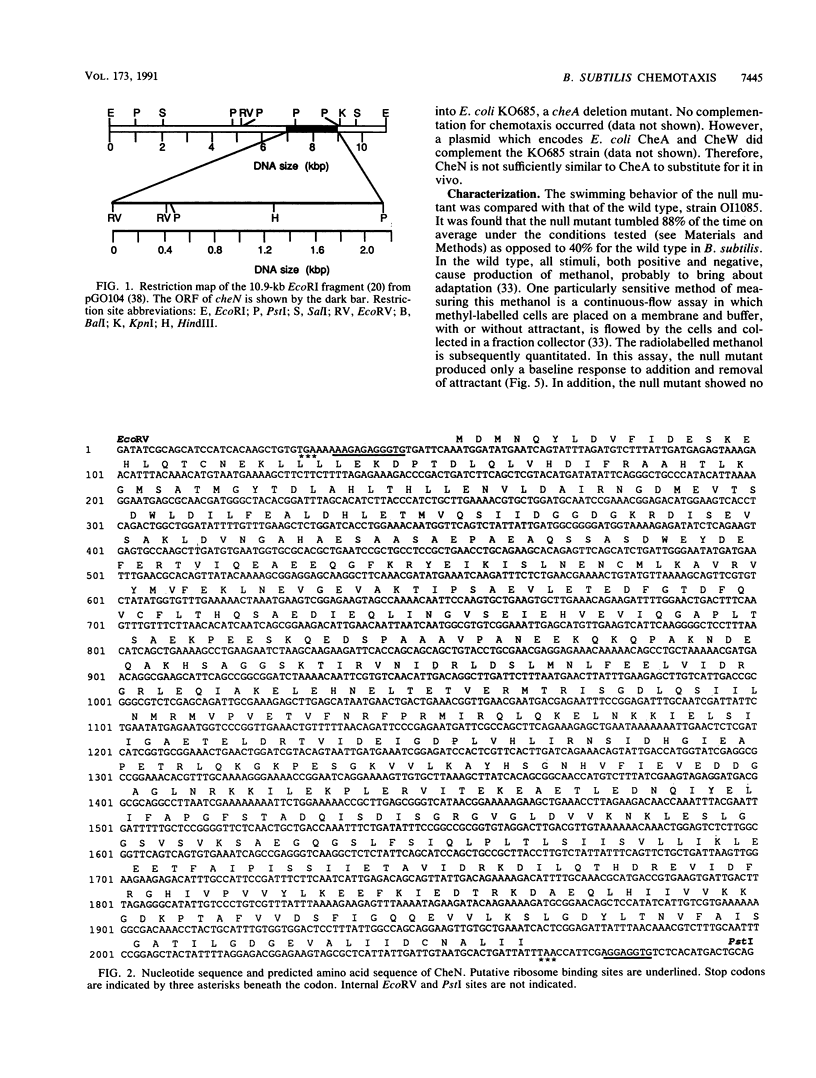
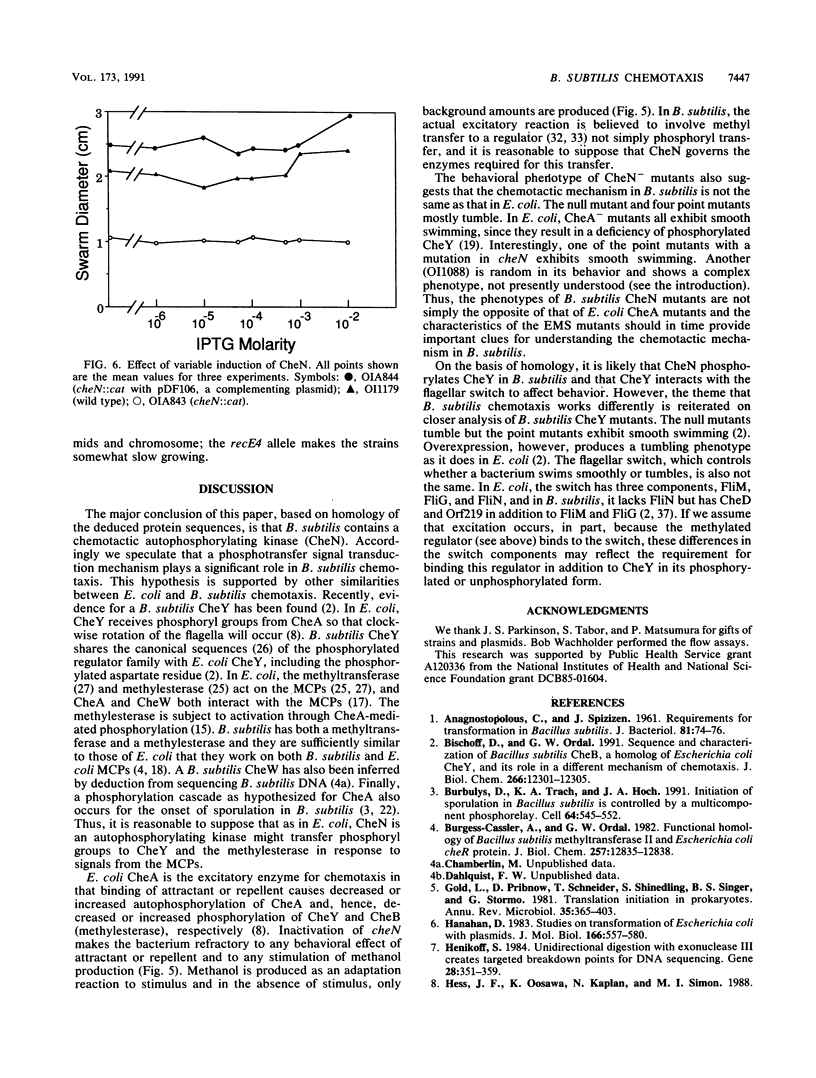
The Bacillus subtilis cheN gene was isolated, sequenced, and expressed. It encodes a large negatively charged protein with a molecular weight of approximately 74,000. The predicted protein sequence has 33 to 34% identity with the Escherichia coli and Salmonella typhimurium CheA and Myxococcus xanthus FrzE sequences. These proteins are found to autophosphorylate and are members of the same histidine kinase signal modulating family. CheN has several conserved regions (including the histidine that is phosphorylated in CheA) that coincide with other autophosphorylated signal transducers. A null mutant is defective in attractant-induced methanol formation and shows no behavioral response to chemoeffectors. These results imply that in B. subtilis the mechanism of chemotaxis involves phosphoryl transfer similar to that in E. coli. However, the CheN null mutant mostly tumbles, whereas CheA mutants swim smoothly, and only in B. subtilis does excitation lead to methyl transfer and methanol formation. Thus, the overall mechanism of chemotaxis is different in the two organisms.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bischoff D. S., Ordal G. W. Sequence and characterization of Bacillus subtilis CheB, a homolog of Escherichia coli CheY, and its role in a different mechanism of chemotaxis. J Biol Chem. 1991 Jul 5;266(19):12301–12305. [PubMed] [Google Scholar]
- Burbulys D., Trach K. A., Hoch J. A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991 Feb 8;64(3):545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Burgess-Cassler A., Ordal G. W. Functional homology of Bacillus subtilis methyltransferase II and Escherichia coli cheR protein. J Biol Chem. 1982 Nov 10;257(21):12835–12838. [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Kaplan N., Simon M. I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988 Apr 8;53(1):79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Kehry M. R., Doak T. G., Dahlquist F. W. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J Biol Chem. 1984 Oct 10;259(19):11828–11835. [PubMed] [Google Scholar]
- Kofoid E. C., Parkinson J. S. Tandem translation starts in the cheA locus of Escherichia coli. J Bacteriol. 1991 Mar;173(6):2116–2119. doi: 10.1128/jb.173.6.2116-2119.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoid E. C., Parkinson J. S. Transmitter and receiver modules in bacterial signaling proteins. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4981–4985. doi: 10.1073/pnas.85.14.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lupas A., Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989 Oct 15;264(29):17337–17342. [PubMed] [Google Scholar]
- McCleary W. R., Zusman D. R. FrzE of Myxococcus xanthus is homologous to both CheA and CheY of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5898–5902. doi: 10.1073/pnas.87.15.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally D. F., Matsumura P. Bacterial chemotaxis signaling complexes: formation of a CheA/CheW complex enhances autophosphorylation and affinity for CheY. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6269–6273. doi: 10.1073/pnas.88.14.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton D. O., Ordal G. W. Functional homology of chemotactic methylesterases from Bacillus subtilis and Escherichia coli. J Bacteriol. 1989 Jan;171(1):120–123. doi: 10.1128/jb.171.1.120-123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa K., Hess J. F., Simon M. I. Mutants defective in bacterial chemotaxis show modified protein phosphorylation. Cell. 1988 Apr 8;53(1):89–96. doi: 10.1016/0092-8674(88)90490-4. [DOI] [PubMed] [Google Scholar]
- Ordal G. W., Nettleton D. O., Hoch J. A. Genetics of Bacillus subtilis chemotaxis: isolation and mapping of mutations and cloning of chemotaxis genes. J Bacteriol. 1983 Jun;154(3):1088–1097. doi: 10.1128/jb.154.3.1088-1097.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordal G. W., Parker H. M., Kirby J. R. Complementation and characterization of chemotaxis mutants of Bacillus subtilis. J Bacteriol. 1985 Nov;164(2):802–810. doi: 10.1128/jb.164.2.802-810.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Hoch J. A. Isolation and sequence of the spo0E gene: its role in initiation of sporulation in Bacillus subtilis. Mol Microbiol. 1987 Jul;1(1):125–132. doi: 10.1111/j.1365-2958.1987.tb00536.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer W. R., Koshland D. E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci U S A. 1977 Feb;74(2):533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A., Chen T., Welsh D., Stock J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Koshland D. E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoelke M. S., Bedale W. A., Nettleton D. O., Ordal G. W. Evidence for an intermediate methyl-acceptor for chemotaxis in Bacillus subtilis. J Biol Chem. 1987 Feb 25;262(6):2811–2816. [PubMed] [Google Scholar]
- Thoelke M. S., Casper J. M., Ordal G. W. Methyl group turnover on methyl-accepting chemotaxis proteins during chemotaxis by Bacillus subtilis. J Biol Chem. 1990 Feb 5;265(4):1928–1932. [PubMed] [Google Scholar]
- Thoelke M. S., Kirby J. R., Ordal G. W. Novel methyl transfer during chemotaxis in Bacillus subtilis. Biochemistry. 1989 Jun 27;28(13):5585–5589. doi: 10.1021/bi00439a037. [DOI] [PubMed] [Google Scholar]
- Thoelke M. S., Parker H. M., Ordal E. A., Ordal G. W. Rapid attractant-induced changes in methylation of methyl-accepting chemotaxis proteins in Bacillus subtilis. Biochemistry. 1988 Nov 1;27(22):8453–8457. doi: 10.1021/bi00422a024. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Toews M. L., Goy M. F., Springer M. S., Adler J. Attractants and repellents control demethylation of methylated chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5544–5548. doi: 10.1073/pnas.76.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zuberi A. R., Bischoff D. S., Ordal G. W. Nucleotide sequence and characterization of a Bacillus subtilis gene encoding a flagellar switch protein. J Bacteriol. 1991 Jan;173(2):710–719. doi: 10.1128/jb.173.2.710-719.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuberi A. R., Ying C. W., Weinreich M. R., Ordal G. W. Transcriptional organization of a cloned chemotaxis locus of Bacillus subtilis. J Bacteriol. 1990 Apr;172(4):1870–1876. doi: 10.1128/jb.172.4.1870-1876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]



