Abstract
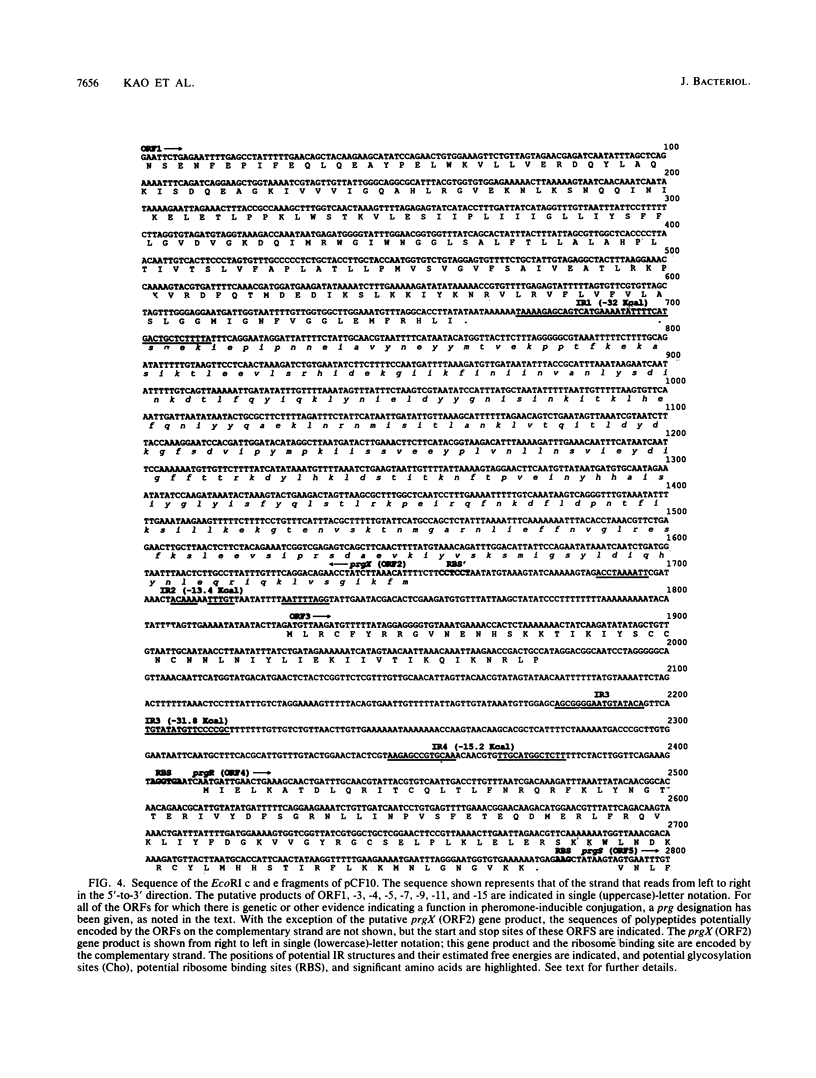
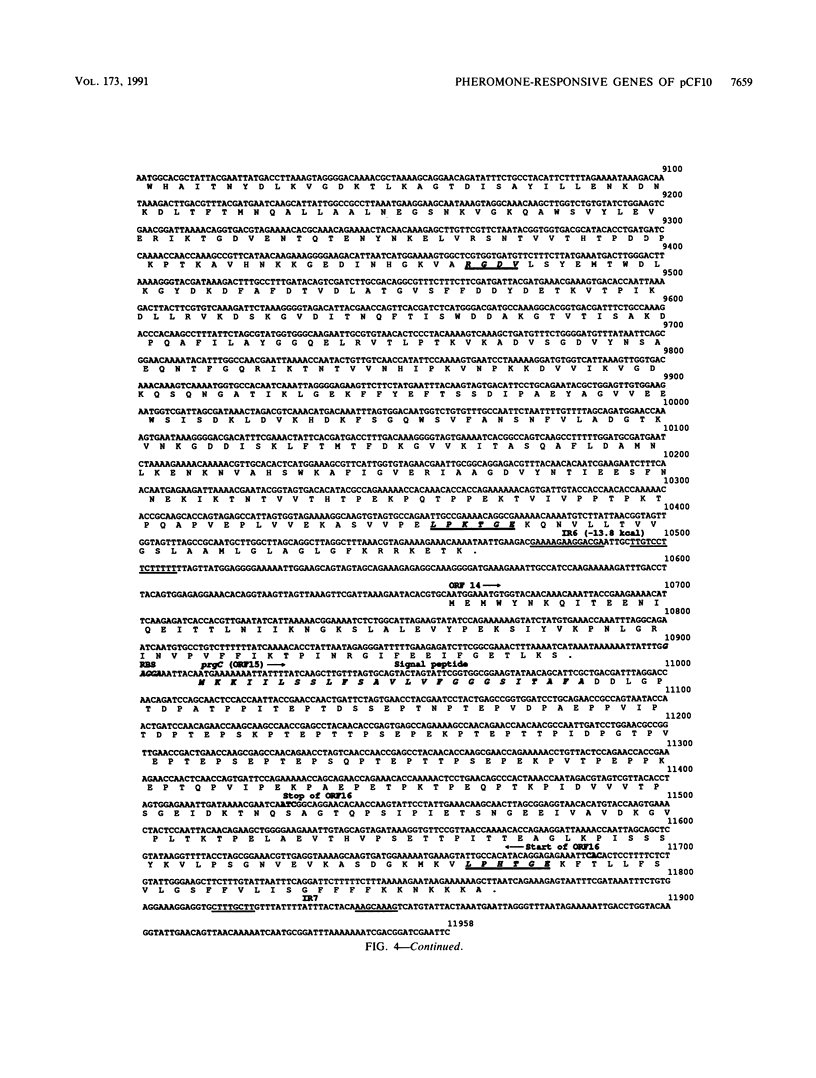
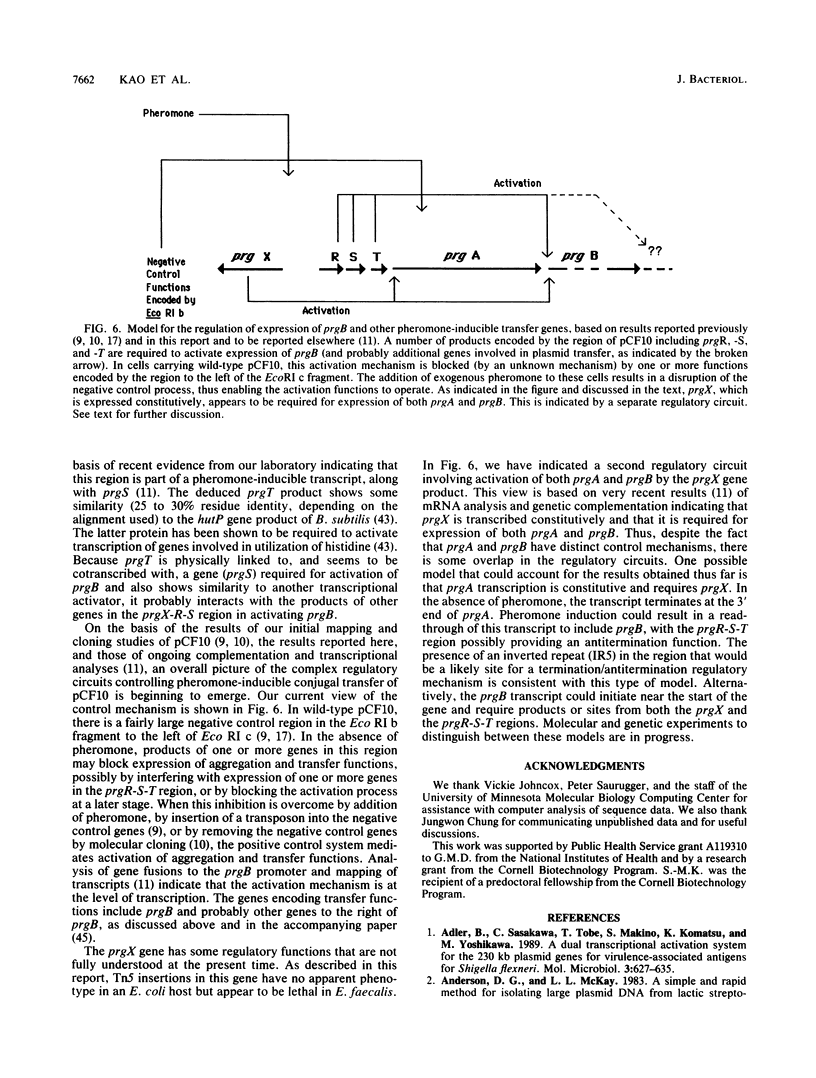
Exposure of Enterococcus faecalis cells carrying the tetracycline resistance plasmid pCF10 to the heptapeptide pheromone cCF10 results in an increase in conjugal transfer frequency by as much as 10(6)-fold. Pheromone-induced donor cells also express at least two plasmid-encoded surface proteins, the 130-kDa Sec 10 protein, which is involved in surface exclusion, and the 150-kDa Asc10 protein, which has been associated with the formation of mating aggregates. Previous subcloning and transposon mutagenesis studies indicated that the adjacent EcoRI c (7.5 kb) and e (4.5 kb) fragments of pCF10 encode the structural genes for these proteins and that the EcoRI c fragment also encodes at least two regulatory genes involved in activation of the expression of the genes encoding Asc10 and Sec10. In this paper, the results of physical and genetic analysis of this region of pCF10, along with the complete DNA sequences of the EcoRI c and e fragments, are reported. The results of the genetic studies indicate the location of the structural genes for the surface proteins and reveal important features of their transcription. In addition, we provide evidence here and in the accompanying paper (S. B. Olmsted, S.-M. Kao, L. J. van Putte, J. C. Gallo, and G. M. Dunny, J. Bacteriol. 173:7665-7672, 1991) for a role of Asc10 in mating aggregate formation. The data also reveal a complex positive control system that acts at distances of at least 3 to 6 kb to activate expression of Asc10. DNA sequence analysis presented here reveals the positions of a number of specific genes, termed prg (pheromone-responsive genes) in this region of pCF10. The genes mapped include prgA (encoding Sec10) and prgB (encoding Asc10), as well as four putative regulatory genes, prgX, -R, -S, and -T. Although the predicted amino acid sequences of Sec10 and Asc10 have some structural features in common with a number of surface proteins of gram-positive cocci, and the Asc10 sequence is highly similar to that of a similar protein encoded by the pheromone-inducible plasmid pAD1 (D. Galli, F. Lottspeich, and R. Wirth, Mol. Microbiol. 4:895-904, 1990), the regulatory genes show relatively little resemblance to any previously sequenced genes from either procaryotes or eucaryotes.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Kusećek B., Timmis K. N. Tra cistrons and proteins encoded by the Escherichia coli antibiotic resistance plasmid R6-5. Mol Gen Genet. 1978 Jul 11;163(2):169–179. doi: 10.1007/BF00267407. [DOI] [PubMed] [Google Scholar]
- Adler B., Sasakawa C., Tobe T., Makino S., Komatsu K., Yoshikawa M. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol Microbiol. 1989 May;3(5):627–635. doi: 10.1111/j.1365-2958.1989.tb00210.x. [DOI] [PubMed] [Google Scholar]
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D. L., Dautricourt J. P., Maulik S., Relph J. Improved sensitivity of biological sequence database searches. Comput Appl Biosci. 1990 Jul;6(3):237–245. doi: 10.1093/bioinformatics/6.3.237. [DOI] [PubMed] [Google Scholar]
- Chen C. C., Cleary P. P. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. J Biol Chem. 1990 Feb 25;265(6):3161–3167. [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chen J. D., Morrison D. A. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong transcriptional terminators. Gene. 1987;55(2-3):179–187. doi: 10.1016/0378-1119(87)90278-2. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Dunny G. M. Identification of regions of the Streptococcus faecalis plasmid pCF-10 that encode antibiotic resistance and pheromone response functions. Plasmid. 1986 May;15(3):230–241. doi: 10.1016/0147-619x(86)90041-7. [DOI] [PubMed] [Google Scholar]
- Christie P. J., Kao S. M., Adsit J. C., Dunny G. M. Cloning and expression of genes encoding pheromone-inducible antigens of Enterococcus (Streptococcus) faecalis. J Bacteriol. 1988 Nov;170(11):5161–5168. doi: 10.1128/jb.170.11.5161-5168.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Pontius L. T., An F. Y., Ike Y., Suzuki A., Nakayama J. Nucleotide sequence of the sex pheromone inhibitor (iAD1) determinant of Enterococcus faecalis conjugative plasmid pAD1. Plasmid. 1990 Sep;24(2):156–161. doi: 10.1016/0147-619x(90)90019-9. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Weaver K. E. Sex pheromones and plasmid transfer in Enterococcus faecalis. Plasmid. 1989 May;21(3):175–184. doi: 10.1016/0147-619x(89)90041-3. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi R. H., Wang L. F. Multiple procaryotic ribonucleic acid polymerase sigma factors. Microbiol Rev. 1986 Sep;50(3):227–243. doi: 10.1128/mr.50.3.227-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G. M., Brown B. L., Clewell D. B. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G. M., Craig R. A., Carron R. L., Clewell D. B. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid. 1979 Jul;2(3):454–465. doi: 10.1016/0147-619x(79)90029-5. [DOI] [PubMed] [Google Scholar]
- Dunny G. M. Genetic functions and cell-cell interactions in the pheromone-inducible plasmid transfer system of Enterococcus faecalis. Mol Microbiol. 1990 May;4(5):689–696. doi: 10.1111/j.1365-2958.1990.tb00639.x. [DOI] [PubMed] [Google Scholar]
- Dunny G. M., Lee L. N., LeBlanc D. J. Improved electroporation and cloning vector system for gram-positive bacteria. Appl Environ Microbiol. 1991 Apr;57(4):1194–1201. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G. M., Zimmerman D. L., Tortorello M. L. Induction of surface exclusion (entry exclusion) by Streptococcus faecalis sex pheromones: use of monoclonal antibodies to identify an inducible surface antigen involved in the exclusion process. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8582–8586. doi: 10.1073/pnas.82.24.8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny G., Funk C., Adsit J. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid. 1981 Nov;6(3):270–278. doi: 10.1016/0147-619x(81)90035-4. [DOI] [PubMed] [Google Scholar]
- Ehrenfeld E. E., Kessler R. E., Clewell D. B. Identification of pheromone-induced surface proteins in Streptococcus faecalis and evidence of a role for lipoteichoic acid in formation of mating aggregates. J Bacteriol. 1986 Oct;168(1):6–12. doi: 10.1128/jb.168.1.6-12.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freytag J. W., Noelken M. E., Hudson B. G. Physical properties of collagen--sodium dodecyl sulfate complexes. Biochemistry. 1979 Oct 16;18(21):4761–4768. doi: 10.1021/bi00588a042. [DOI] [PubMed] [Google Scholar]
- Galli D., Lottspeich F., Wirth R. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol Microbiol. 1990 Jun;4(6):895–904. doi: 10.1111/j.1365-2958.1990.tb00662.x. [DOI] [PubMed] [Google Scholar]
- Galli D., Wirth R., Wanner G. Identification of aggregation substances of Enterococcus faecalis cells after induction by sex pheromones. An immunological and ultrastructural investigation. Arch Microbiol. 1989;151(6):486–490. doi: 10.1007/BF00454863. [DOI] [PubMed] [Google Scholar]
- Guo L. H., Yang R. C., Wu R. An improved strategy for rapid direct sequencing of both strands of long DNA molecules cloned in a plasmid. Nucleic Acids Res. 1983 Aug 25;11(16):5521–5540. doi: 10.1093/nar/11.16.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss B., Uhlén M., Nilsson B., Lindberg M., Sjöquist J., Sjödahl J. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur J Biochem. 1984 Jan 16;138(2):413–420. doi: 10.1111/j.1432-1033.1984.tb07931.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hollingshead S. K., Fischetti V. A., Scott J. R. Complete nucleotide sequence of type 6 M protein of the group A Streptococcus. Repetitive structure and membrane anchor. J Biol Chem. 1986 Feb 5;261(4):1677–1686. [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hynes R. O. Integrins: a family of cell surface receptors. Cell. 1987 Feb 27;48(4):549–554. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- Kessler R. E., Yagi Y. Identification and partial characterization of a pheromone-induced adhesive surface antigen of Streptococcus faecalis. J Bacteriol. 1983 Aug;155(2):714–721. doi: 10.1128/jb.155.2.714-721.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornitzer D., Teff D., Altuvia S., Oppenheim A. B. Genetic analysis of bacteriophage lambda cIII gene: mRNA structural requirements for translation initiation. J Bacteriol. 1989 May;171(5):2563–2572. doi: 10.1128/jb.171.5.2563-2572.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci E., Garzino V., Mary C., Bennani N., Pradel J. Modulo, a new maternally expressed Drosophila gene encodes a DNA-binding protein with distinct acidic and basic regions. Nucleic Acids Res. 1989 Oct 25;17(20):8101–8115. doi: 10.1093/nar/17.20.8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mori M., Sakagami Y., Ishii Y., Isogai A., Kitada C., Fujino M., Adsit J. C., Dunny G. M., Suzuki A. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J Biol Chem. 1988 Oct 5;263(28):14574–14578. [PubMed] [Google Scholar]
- Nakayama J., Nagasawa H., Isogai A., Clewell D. B., Suzuki A. Amino acid sequence of pheromone-inducible surface protein in Enterococcus faecalis, that is encoded on the conjugative plasmid pPD1. FEBS Lett. 1990 Jul 2;267(1):81–84. doi: 10.1016/0014-5793(90)80293-r. [DOI] [PubMed] [Google Scholar]
- Oda M., Sugishita A., Furukawa K. Cloning and nucleotide sequences of histidase and regulatory genes in the Bacillus subtilis hut operon and positive regulation of the operon. J Bacteriol. 1988 Jul;170(7):3199–3205. doi: 10.1128/jb.170.7.3199-3205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- Olmsted S. B., Kao S. M., van Putte L. J., Gallo J. C., Dunny G. M. Role of the pheromone-inducible surface protein Asc10 in mating aggregate formation and conjugal transfer of the Enterococcus faecalis plasmid pCF10. J Bacteriol. 1991 Dec;173(23):7665–7672. doi: 10.1128/jb.173.23.7665-7672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V., Fischetti V. A. Isolation and characterization of the cell-associated region of group A streptococcal M6 protein. J Bacteriol. 1988 Jun;170(6):2618–2624. doi: 10.1128/jb.170.6.2618-2624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riede I., Eschbach M. L. Evidence that TraT interacts with OmpA of Escherichia coli. FEBS Lett. 1986 Sep 15;205(2):241–245. doi: 10.1016/0014-5793(86)80905-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Swinfield T. J., Oultram J. D., Thompson D. E., Brehm J. K., Minton N. P. Physical characterisation of the replication region of the Streptococcus faecalis plasmid pAM beta 1. Gene. 1990 Mar 1;87(1):79–90. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tortorello M. L., Dunny G. M. Identification of multiple cell surface antigens associated with the sex pheromone response of Streptococcus faecalis. J Bacteriol. 1985 Apr;162(1):131–137. doi: 10.1128/jb.162.1.131-137.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura T., Lau P. C. Nucleotide sequences from the colicin E8 operon: homology with plasmid ColE2-P9. Mol Gen Genet. 1987 Oct;209(3):489–493. doi: 10.1007/BF00331154. [DOI] [PubMed] [Google Scholar]
- Wanner G., Formanek H., Galli D., Wirth R. Localization of aggregation substances of Enterococcus faecalis after induction by sex pheromones. An ultrastructural comparison using immuno labelling, transmission and high resolution scanning electron microscopic techniques. Arch Microbiol. 1989;151(6):491–497. doi: 10.1007/BF00454864. [DOI] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth R., An F. Y., Clewell D. B. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J Bacteriol. 1986 Mar;165(3):831–836. doi: 10.1128/jb.165.3.831-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Kessler R. E., Shaw J. H., Lopatin D. E., An F., Clewell D. B. Plasmid content of Streptococcus faecalis strain 39-5 and identification of a pheromone (cPD1)-induced surface antigen. J Gen Microbiol. 1983 Apr;129(4):1207–1215. doi: 10.1099/00221287-129-4-1207. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]