Abstract
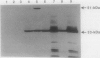
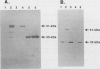
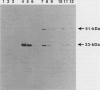
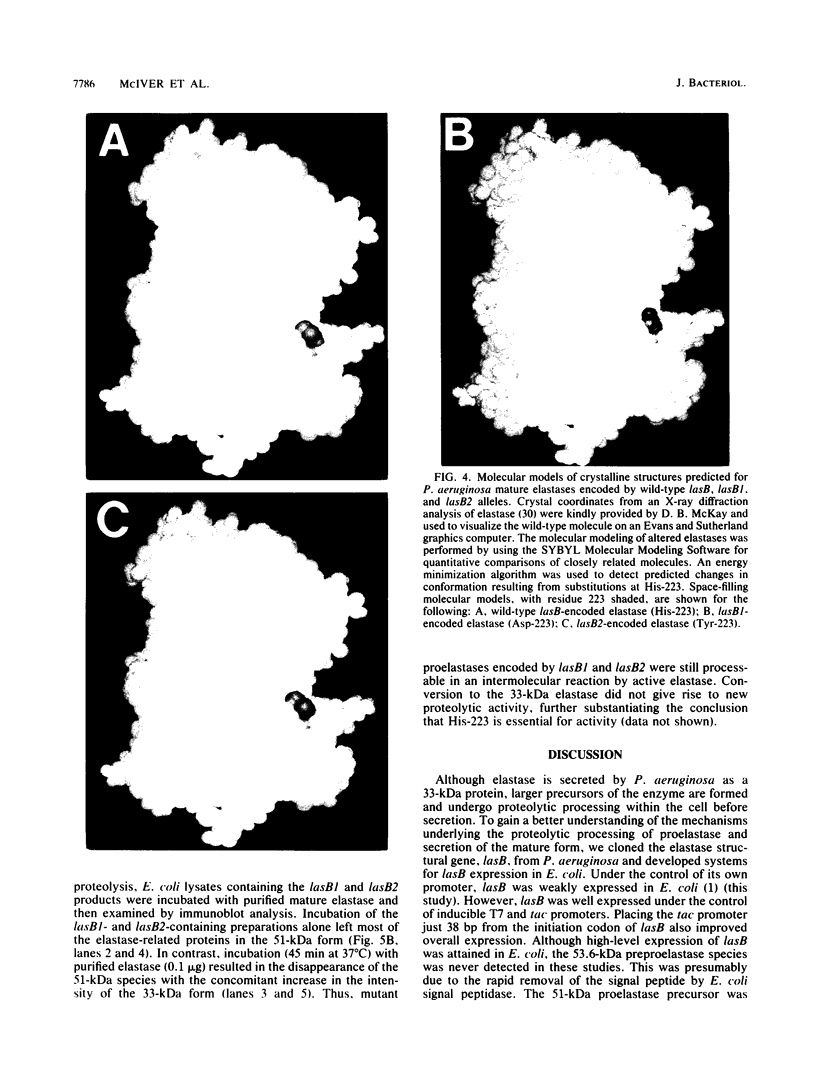
The neutral metalloprotease elastase is one of the major proteins secreted into the culture medium by many Pseudomonas aeruginosa strains. Encoded by the lasB gene, the 33-kDa elastase is initially synthesized as a 53-kDa preproenzyme which is processed to the mature form via a 51-kDa proelastase intermediate. To facilitate studies on proteolytic processing of elastase precursors and on secretion, we developed systems for overexpression of lasB in Escherichia coli under the control of the inducible T7 and tac promoters. Although the 51-kDa proelastase form was detectable in E. coli under inducible conditions, most of the elastase produced under these conditions was found in an enzymatically active 33-kDa form. The amino-terminal sequence of the first 15 amino acid residues of this 33-kDa elastase species was identical to that of the mature P. aeruginosa enzyme, suggesting that processing was autocatalytic. To test this possibility, the codon in lasB encoding His-223, a presumed active-site residue, was changed to encode Asp-223 (lasB1) and Tyr-223 (lasB2). The effects of these mutations on enzyme activity and processing were examined. No proteolytic or elastolytic activities were detected in extracts of E. coli cells containing the lasB mutant alleles. Overexpression of the mutated lasB genes in E. coli resulted in the accumulation of the corresponding 51-kDa proelastase species. These were processed in vitro to the respective 33-kDa forms by incubation with exogenous purified elastase, without an increase in proteolytic activity. Molecular modeling studies suggest that the mutations have little or no effect on the conformation of the mutant elastases. In addition, wild-type elastase and the mutant proelastases were localized to the periplasm of E. coli. The present results confirm that His-223 is essential for elastase activity and provide evidence for autoproteolytic processing of proelastase.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bever R. A., Iglewski B. H. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J Bacteriol. 1988 Sep;170(9):4309–4314. doi: 10.1128/jb.170.9.4309-4314.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Fukushima J., Yamamoto S., Morihara K., Atsumi Y., Takeuchi H., Kawamoto S., Okuda K. Structural gene and complete amino acid sequence of Pseudomonas aeruginosa IFO 3455 elastase. J Bacteriol. 1989 Mar;171(3):1698–1704. doi: 10.1128/jb.171.3.1698-1704.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Activation of an elastase precursor by the lasA gene product of Pseudomonas aeruginosa. J Bacteriol. 1987 Oct;169(10):4532–4539. doi: 10.1128/jb.169.10.4532-4539.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984 Jun;158(3):1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck L. W., Alarcon P. G., Kulhavy R. M., Morihara K., Russell M. W., Mestecky J. F. Degradation of IgA proteins by Pseudomonas aeruginosa elastase. J Immunol. 1990 Mar 15;144(6):2253–2257. [PubMed] [Google Scholar]
- Heck L. W., Morihara K., McRae W. B., Miller E. J. Specific cleavage of human type III and IV collagens by Pseudomonas aeruginosa elastase. Infect Immun. 1986 Jan;51(1):115–118. doi: 10.1128/iai.51.1.115-118.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder I. A., Neely A. N. Pseudomonas elastase acts as a virulence factor in burned hosts by Hageman factor-dependent activation of the host kinin cascade. Infect Immun. 1989 Nov;57(11):3345–3348. doi: 10.1128/iai.57.11.3345-3348.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S. E., Fecycz I. T., Stemke G. W., Campbell J. N. Demonstration of a cell-associated, inactive precursor of an exocellular protease produced by Pseudomonas aeruginosa. Can J Microbiol. 1980 Jan;26(1):87–93. doi: 10.1139/m80-013. [DOI] [PubMed] [Google Scholar]
- Kessler E., Israel M., Landshman N., Chechick A., Blumberg S. In vitro inhibition of Pseudomonas aeruginosa elastase by metal-chelating peptide derivatives. Infect Immun. 1982 Nov;38(2):716–723. doi: 10.1128/iai.38.2.716-723.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler E., Safrin M. Partial purification and characterization of an inactive precursor of Pseudomonas aeruginosa elastase. J Bacteriol. 1988 Mar;170(3):1215–1219. doi: 10.1128/jb.170.3.1215-1219.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler E., Safrin M. Synthesis, processing, and transport of Pseudomonas aeruginosa elastase. J Bacteriol. 1988 Nov;170(11):5241–5247. doi: 10.1128/jb.170.11.5241-5247.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Botstein D. Secretion of beta-lactamase requires the carboxy end of the protein. Cell. 1980 Jul;20(3):749–760. doi: 10.1016/0092-8674(80)90321-9. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Morihara K., Tsuzuki H., Oda K. Protease and elastase of Pseudomonas aeruginosa: inactivation of human plasma alpha 1-proteinase inhibitor. Infect Immun. 1979 Apr;24(1):188–193. doi: 10.1128/iai.24.1.188-193.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Rosenberg E. Y., Foulds J. Porin channels in Escherichia coli: studies with beta-lactams in intact cells. J Bacteriol. 1983 Jan;153(1):232–240. doi: 10.1128/jb.153.1.232-240.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., Chakrabarty A. M. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981 Jul;33(1):142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman D. E., Cryz S. J., Iglewski B. H. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980 Jun;142(3):836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. E., Galloway D. R. Purification and characterization of an active fragment of the LasA protein from Pseudomonas aeruginosa: enhancement of elastase activity. J Bacteriol. 1990 May;172(5):2236–2240. doi: 10.1128/jb.172.5.2236-2240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power S. D., Adams R. M., Wells J. A. Secretion and autoproteolytic maturation of subtilisin. Proc Natl Acad Sci U S A. 1986 May;83(10):3096–3100. doi: 10.1073/pnas.83.10.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schad P. A., Iglewski B. H. Nucleotide sequence and expression in Escherichia coli of the Pseudomonas aeruginosa lasA gene. J Bacteriol. 1988 Jun;170(6):2784–2789. doi: 10.1128/jb.170.6.2784-2789.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. R., Miller K. D. Elastase of Pseudomonas aeruginosa: inactivation of complement components and complement-derived chemotactic and phagocytic factors. Infect Immun. 1974 Jul;10(1):128–135. doi: 10.1128/iai.10.1.128-135.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol P. A., Ohman D. E., Iglewski B. H. A more sensitive plate assay for detection of protease production by Pseudomanas aeruginosa. J Clin Microbiol. 1979 Apr;9(4):538–540. doi: 10.1128/jcm.9.4.538-540.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Thayer M. M., Flaherty K. M., McKay D. B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-A resolution. J Biol Chem. 1991 Feb 15;266(5):2864–2871. doi: 10.2210/pdb1ezm/pdb. [DOI] [PubMed] [Google Scholar]
- Wandersman C. Secretion, processing and activation of bacterial extracellular proteases. Mol Microbiol. 1989 Dec;3(12):1825–1831. doi: 10.1111/j.1365-2958.1989.tb00169.x. [DOI] [PubMed] [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983 Nov 25;11(22):7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Fukushima J., Atsumi Y., Takeuchi H., Kawamoto S., Okuda K., Morihara K. Cloning and characterization of elastase structural gene from Pseudomonas aeruginosa IFO 3455. Biochem Biophys Res Commun. 1988 May 16;152(3):1117–1122. doi: 10.1016/s0006-291x(88)80400-5. [DOI] [PubMed] [Google Scholar]