Abstract
Seed development in angiosperms initiates after double fertilization, leading to the formation of a diploid embryo and a triploid endosperm. The active repression of precocious initiation of certain aspects of seed development in the absence of fertilization requires the Polycomb group proteins MEDEA (MEA), FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) and FERTILIZATION-INDEPENDENT SEED2. Here we show that the Arabidopsis WD-40 domain protein MSI1 is present together with MEA and FIE in a 600 kDa complex and interacts directly with FIE. Mutant plants heterozygous for msi1 show a seed abortion ratio of 50% with seeds aborting when the mutant allele is maternally inherited, irrespective of a paternal wild-type or mutant MSI1 allele. Further more, msi1 mutant gametophytes initiate endosperm development in the absence of fertilization at a high penetrance. After pollination, only the egg cell becomes fertilized, the central cell starts dividing prior to fertilization, resulting in the formation of seeds containing embryos surrounded by diploid endosperm. Our results establish that MSI1 has an essential function in the correct initiation and progression of seed development.
Keywords: Arabidopsis/chromatin/MEDEA/MSI1/Polycomb group complex
Introduction
In contrast with animals, plant gametes do not differentiate directly after meiosis but are produced by multicellular gametophytes that develop from meiotically derived spores. In flowering plants the female gametophyte is formed through a well-defined programme of nuclear divisions, nuclear migration and cellularization that gives rise to a multicellular structure consisting of two synergids, three antipodals, a central cell and an egg cell (for review see Grossniklaus and Schneitz, 1998). Seed development is initiated by double fertilization in which two sperm cells fuse with the egg and the central cell of the female gametophyte, respectively. The fertilized egg cell develops into the embryo, while the fertilized central cell forms the nourishing endosperm. In addition, maternal tissue of the ovule, in which the female gametophyte is embedded, contributes to the developing seed by forming the seed coat (testa). Interestingly, in most angiosperms, including Arabidopsis, embryo and testa are diploid but the endosperm is triploid with two maternal copies and one paternal copy of the genome. Double fertilization and formation of the endosperm are key events in angiosperm evolution; however, the origin and evolutionary advantages of these two processes are not yet fully understood (for review see Chaudhury et al., 1998; Berger, 1999, 2003; Grossniklaus et al., 2001; Baroux et al, 2002).
After fertilization, development of testa, endosperm and embryo are highly coordinated. Genetic studies have shown that seed and fruit development are actively repressed in the absence of fertilization and that early phases of embryo and endosperm development are largely under maternal control (Vielle-Calzada et al., 2000; Vivian-Smith et al., 2001; Walbot and Evans, 2003). Several genes that encode regulators of early seed development are also required to repress fertilization-independent seed development. With reference to the Greek priestess Medea, who killed her own children in revenge for the unfaithfulness of her husband Jason, one of these genes was named MEDEA (MEA) because a mutant allele causes seed abortion only when inherited maternally (Grossniklaus et al., 1998). FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) and FERTILI ZATION-INDEPENDENT SEED2 (FIS2) are two other genes that are required to repress seed development in the absence of fertilization (Luo et al., 1999; Ohad et al., 1999), a phenotype also shared by mea (Grossniklaus and Vielle-Calzada, 1998; Kiyosue et al., 1999). All three fis-class mutants show a gametophytic maternal effect: the mutant phenotype can only be observed when the mutation is inherited through the female gametophyte. Early seed development of fis mutant seeds is indistinguishable from that of wild-type seeds. However, from the globular stage onwards fis embryo development is delayed and eventually arrests with oversized heart-shaped embryos and an abnormally proliferated endosperm (Grossniklaus et al., 2001).
MEA encodes a protein similar to the Drosophila Polycomb group (PcG) protein Enhancer of Zeste [E(Z)] (Grossniklaus et al., 1998). In Drosophila, E(Z) interacts with ESC, a protein sharing similarity with FIE (Jones et al., 1998; Ohad et al., 1999; Ng et al., 2000; Tie et al., 2001). MEA and FIE, as well as the mammalian homologues ENX1 and EED, interact, indicating a strong evolutionary conservation of PcG proteins and their interactions (Sewalt et al., 1998; Luo et al, 2000; Spillane et al., 2000; Yadegari et al., 2000). The E(Z)–ESC complex has a molecular mass of about 600 kDa and contains additional subunits, including p55 and the FIS2 homologue Su(Z)12 (Luo et al., 1999; Tie et al., 2001; Müller et al., 2002). Because many SET domain proteins like E(Z) have histone methyltransferase activity, it has been proposed that PcG complexes establish repressive chromatin environments at certain target loci through histone deacetylation and histone methylation (for review see Francis and Kingston, 2001). It is now increasingly appreciated that maintenance of intact chromatin states is critical for normal development and the maintenance of developmental decisions (for review see Muller and Leutz, 2001; Köhler and Grossniklaus, 2002; Reyes et al., 2002; Reyes and Grossniklaus, 2003).
WD-40 proteins similar to yeast MSI1 have key functions in the maintenance and modulation of chromatin. These proteins exist in all eukaryotes and participate in various complexes involved in chromatin dynamics, including the above mentioned PcG complexes, as well as chromatin assembly factor CAF-1, nucleosome remodelling factor NURF, histone acetyl transferase and histone deacetylase complexes, and they interact with the retinoblastoma tumor suppressor protein (Qian et al., 1993; Parthun et al., 1996; Taunton et al., 1996; Verreault et al., 1996). Members of this family include p55 in Drosophila, the retinoblastoma-associated proteins RbAp46 and RbAp48 in vertebrates and CAC3p in Saccharomyces cerevisiae. Despite their ubiquitous presence, very little is known about the in vivo function of MSI1-like proteins. In yeast, CAC3p is required for efficient gene silencing at telomeres and the mating type loci, and cac3 mutants show enhanced sensitivity to ultraviolet radiation (Kaufman et al., 1997). In Caenorhabditis elegans, dominant negative alleles of the MSI1-like LIN-53 gene cause defects in vulva differentiation (Lu and Horvitz, 1998), but no other functions of MSI1-like proteins in multicellular eukaryotes have been reported. Arabidopsis has five MSI1-like genes (MSI1–5) (Ach et al., 1997; Kenzior and Folk, 1998; Hennig et al, 2003), and reducing Arabidopsis MSI1 levels by co-suppression causes ectopic expression of floral homeotic genes and strongly interferes with cellular differentiation (Hennig et al., 2003). Because the severity of phenotypic alterations increased during development, we suggested that MSI1 is required for the inheritance of epigenetic states during mitosis.
Here we describe an Arabidopsis MSI1 insertion allele that causes seed abortion early in development when maternally inherited. Heterozygous msi1 mutants also show a high penetrance of fertilization-independent seed development. We demonstrate that Arabidopsis MSI1 physically interacts with FIE and that MSI1, FIE and MEA are part of a 600 kDa protein complex in vivo that is required for seed development.
Results
Identification of an msi1 T-DNA insertion mutant
To analyse the function of Arabidopsis MSI1 we searched several collections of T-DNA insertion mutants for a disruption of the MSI1 gene and identified one candidate in the SAIL collection (Sessions et al., 2002). Genomic DNA blots revealed a complex insertion of two T-DNAs at the same locus. PCR analysis and sequencing confirmed the insertion into the second exon of the MSI1 gene (data not shown). Heterozygous plants developed normally from the seedling stage to maturity. The T-DNA insertion allele in Arabidopsis MSI1 was termed msi1 and was used in all subsequent experiments.
Loss of MSI1 causes a maternal effect seed abortion phenotype
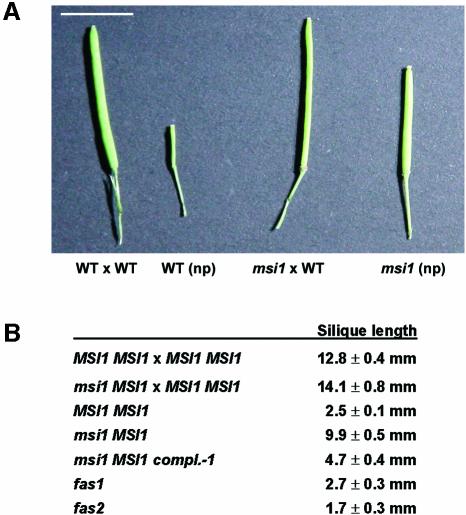
We were unable to obtain homozygous msi1 plants, suggesting that seed development is defective in MSI1/msi1 mutants. In contrast with wild-type plants that contained only normally developing seeds, MSI1/msi1 siliques had aborted seeds that appeared brown and shrunken (Figure 1). Quantification of seed abortion revealed that 50% of the seeds from heterozygous msi1 mutants aborted, in contrast with less than 1% abortion observed in the wild type (Table I). Arabidopsis MSI1 was previously reported to participate in CAF-1 (Kaya et al., 2001), but we found no striking overlapping phenotypes between the Arabidopsis CAF-1 mutants fasciata1 (fas1) and fasciata2 (fas2) and MSI1 co-suppression plants (Hennig et al., 2003). Therefore we assayed seed abortion in fas1 and fas2, but, in contrast with MSI1/msi1 plants, seed development was not impaired in the CAF-1 mutants (Table I).

Fig. 1. Siliques of MSI1/msi1 plants contain 50% normal and 50% aborted seeds. Opened siliques of (A) wild-type (WT), selfed, (B) MSI1/msi1, selfed, (C) WT × MSI1/msi1 and (D) MSI1/msi1 × WT. Scale bars, 500 µm.
Table I. Seed abortion in msi1 and fasciata mutants.
| Normal | Aborted | Expected | p value | |
|---|---|---|---|---|
| WT | 549 | 1 (0.2%) | NA | NA |
| MSI1/msi1 ×MSI1/msi1 | 268 | 261 (49.3%) | 50 | 0.76 |
| fas1 | 162 | 2 (1.2%) | NA | NA |
| fas2 | 176 | 4 (2.2%) | NA | NA |
| msi1 compl-1 | 184 | 66 (26.4%) | 25 | 0.61 |
| MSI1/msi1 ×MSI1/MSI1 | 93 | 99 (48.4%) | 50 | 0.66 |
| MSI1/MSI1 × MSI1/msi1 | 262 | 0 (0.0%) | 0 | NA |
NA, not applicable.
A single-locus recessive embryo-lethal mutant is expected to give rise to 25% aborted seeds. However, seed abortion in MSI1/msi1 occurred at a ratio of 50%. Because 50% seed abortion is a strong indication for a defect that is under female gametophytic control, we performed reciprocal crosses between MSI1/msi1 and wild-type plants. A paternally derived mutant allele (msi1 pollen) did not impair seed development, while the msi1 phenotype could not be complemented by fertilization of msi1 mutant gametophytes with wild-type pollen (Figure 1 and Table I). These observations suggest that the fate of seeds is determined only by the maternally derived MSI1 allele. Seeds derived from ovules with an msi1 gametophyte (hereafter referred to as msi1 seeds) abort regardless of the paternal genotype. This hypothesis predicts that the msi1 mutant allele can be transmitted only paternally. We tested for the presence of the msi1 allele in the progeny of selfed msi1 heterozygous plants and of reciprocal crosses between MSI1/msi1 and wild-type plants. The results shown in Table II confirm this hypothesis.
Table II. Transmission of the msi1 allele.
| Phosphinotrocin sensitive | Phosphinotrocin resistant | Expected | p value | |
|---|---|---|---|---|
| MSI1/msi1 × MSI1/msi1 | 113 | 125 (52.5%) | 50 | 0.44 |
| 75 | < 0.01 | |||
| MSI1/MSI1 × MSI1/msi1 | 42 | 44 (51.1%) | 50 | > 0.05 |
| MSI1/msi1 × MSI1/MSI1 | 51 | 0 (0.0%) | 0 | NA |
| msi1 compl-1 × MSI1/MSI1 | 48 | 26 (36.8%) | 33 | 0.696 |
NA, not applicable.
Genetic complementation of msi1
T-DNA insertion mutagenesis can also result in mutations that are not tagged by the inserted T-DNA (Azpiroz-Leehan and Feldman, 1997). Therefore we tested whether the observed msi1 phenotype is indeed caused by the insertion into the MSI1 gene. Plants heterozygous for msi1 were transformed with the MSI1 cDNA under control of the endogenous MSI1 promoter consisting of a 2 kbp DNA fragment upstream of the translation start codon. Two randomly chosen transgenic lines showed less seed abortion than the parental MSI1/msi1 mutant. We selected a transgenic line with a single insert of the transgene (msi1 compl-1, data not shown) for further analysis. As msi1 is a gametophytic maternal effect mutant, the expected ratio of aborted seeds in an MSI1/msi1 plant is 50%. An unlinked single copy MSI1 transgene will segregate randomly in the gametes such that half of the msi1 seeds are expected to contain the transgene rescuing the msi1 phenotype. This will reduce the ratio of aborted seeds to 25% (see below). As expected, the fraction of aborting seeds dropped from 49.3% in MSI1/msi1 to 26.4% in MSI1/msi1 compl-1 (Table I). Because seed abortion can be complemented by an intact MSI1 cDNA, we conclude that this phenotype of MSI1/msi1 plants is indeed caused by loss of MSI1 function.
Aborting seeds in msi1 arrest early during embryo development
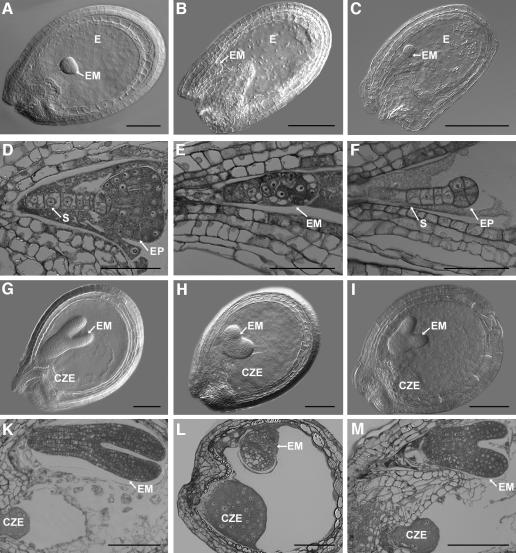
We analyzed the morphology of mutant and wild-type seeds in cleared whole-mount and semi-thin sectioned specimens to characterize the defects of msi1 mutant seeds in more detail (Figure 2). Mutant msi1 seeds were developmentally delayed compared with their wild-type siblings. The developmental arrest occurred at different developmental stages and was highly variable among different siliques. Thus, when about half of the seeds of MSI1/msi1 plants contained transition or heart-stage embryos (Figure 2A and D), about 36% (n = 361) of the seeds contained preglobular and globular stage embryos (Figures 2B and E). The remaining seeds aborted shortly after fertilization without the formation of any embryo. In contrast with the well-organized structure of preglobular embryos in wild-type seeds (Figure 2C and F), delayed preglobular seeds in MSI1/msi1 siliques often contained abnormal embryos with irregular orientation of cell division planes. In fact, there was no recognizable separation between the embryo proper and the suspensor (Figure 2B and E). Very few msi1 embryos continued development until the heart stage. However, msi1 heart-stage embryos showed overproliferation and the seeds contained an enlarged chalazal endosperm (Figure 2H and L). When developing wild-type seeds had reached the late torpedo stage (Figure 2G and K), the majority of the mutant siblings were already aborted and only very few contained overproliferated heart-stage embryos. Con sequently, seed development is initiated in msi1 female gametophytes but suffers from early defects in embryo and endosperm development, eventually leading to seed abortion.
Fig. 2. Mutant msi1 seeds contain abnormal embryos arrested at different developmental stages: (A), (D) Wild-type seeds with transition stage embryos; (B), (E) mutant seeds from the same silique as in (A) and (D); (C), (F) preglobular wild-type embryos for comparison; (G), (K) wild-type seeds with torpedo stage embryos; (H), (L) mutant seeds from same silique as (G) and (K) containing enlarged mutant embryo at the heart stage and chalazal endosperm that is overproliferated; (I), (M) wild-type seeds with late heart-stage embryos for comparison. Abbreviations: CZE, chalazal endosperm; E, endosperm; EM, embryo; EP, embryo proper; S, suspensor. Scale bars, 100 µm in (A), (B), (C), (G), (H), (K), (I) and (M); 50 µm in (D), (E), (F) and (L).
MSI1 is strongly expressed in the female gametophyte and the embryo
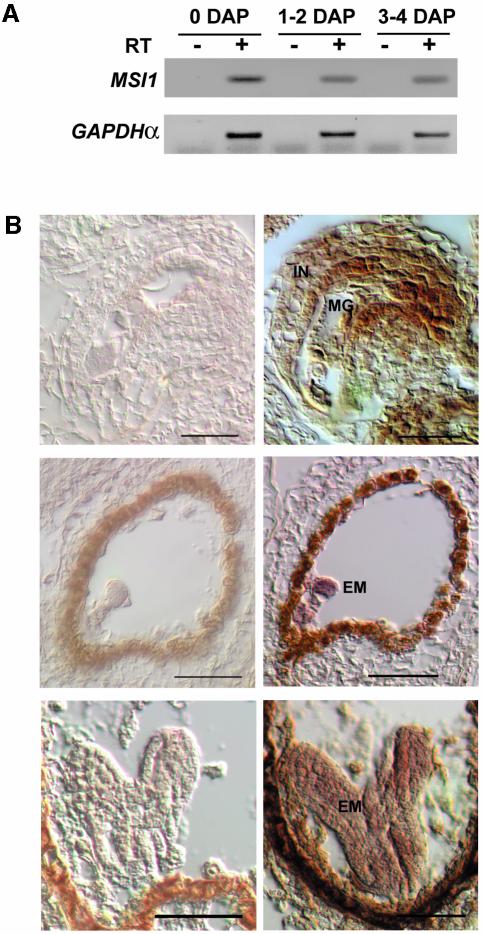
MSI1 is strongly expressed in floral buds and flowers (Hennig et al., 2003), but seed abortion in the absence of an intact maternal MSI1 allele suggested expression of MSI1 in fruits also. RT-PCR analysis demonstrated that MSI1 is strongly expressed in siliques during wild-type seed development from 0 to 4 DAP (Figure 3A). In order to characterize the tissue-specific MSI1 expression, we performed in situ hybridization experiments (Figure 3B). We obtained a strong signal with the antisense RNA probe in both the female gametophyte and the sporophytic tissue of the ovules (e.g. in integuments). After fertilization, the strongest expression was detected in developing embryos. We also observed specific hybridization signals in pollen sacs and pollen (data not shown). Together, these results support the view that MSI1 has a pivotal role in gametophyte and embryo development.
Fig. 3. MSI1 is expressed in the female gametophyte and during different stages of seed development. (A) RNA was isolated from wild-type flowers before fertilization (0 DAP), siliques containing embryos at preglobular stage (1–2 DAP) and siliques containing embryos at late globular stage (3–4 DAP). After treatment with DNase I, RNA was subjected to reverse transcription in the presence (+) or absence (–) of reverse transcriptase using oligodT primers. PCR with cDNA-specific primers was performed on aliquots of the produced cDNA, which equalled 50 ng total RNA. (B) Localization of MSI1 expression in gametophytes and developing seeds of wild-type Arabidopsis plants. Sections were hybridized with a sense (left) or antisense (right) MSI1 probe. Top, expression in female gametophytes; middle, expression in globular embryos; bottom, expression in heart-stage embryos. Abbreviations: EM, embryo; MG, megagametophyte; IN, integuments. Scale bars: top, 30 µm; middle and bottom, 50 µm.
MSI1, MEA and FIE participate in a high molecular weight complex in vivo
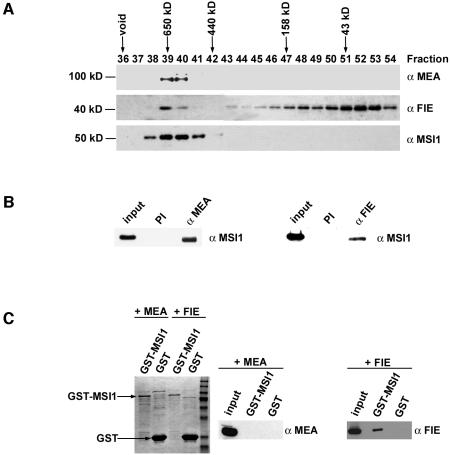
Similar to msi1, the maternal effect mutants mea, fie and fis2 also cause 50% seed abortion. Since MEA and FIE interact in vitro and in vivo (Luo et al., 2000; Spillane et al., 2000; Yadegari et al., 2000), we tested whether MSI1 is also a subunit of the MEA–FIE complex. In Drosophila, E(Z) and ESC are subunits of a large complex with a molecular mass of about 500–600 kDa (Ng et al., 2000; Tie et al., 2001). In C.elegans, however, homologues of E(Z) and ESC are found in a complex with a molecular weight of only 255 kDa (Xu et al., 2001). The molecular weight and subunit composition of the Arabidopsis MEA–FIE complex is currently unknown. Therefore we investigated the MEA–FIE complex using size exclusion chromatography (SEC) of protein extracts from Arabidopsis flowers and young siliques and tested the fractions on protein blots. The antisera used in these experiments had previously been shown to recognize the corresponding antigen and do not cross-react with other proteins of the same size (Hennig et al., 2003; Köhler et al., 2003). Figure 4A shows that MSI1, MEA and FIE co-elute at a molecular weight of about 600 kDa, suggesting that they are part of a complex similar to the PcG complex in Drosophila. Interestingly, a large amount of FIE, but not MEA and MSI1, could also be detected in a monomeric form. To confirm that MSI1 is also a subunit of the MEA–FIE complex, we performed immunoprecipitation experiments with Arabidopsis nuclear extracts. Both anti-MEA and anti-FIE, but not the preimmune sera, coprecipitated MSI1 (Figure 4B).
Fig. 4. MEA, FIE and MSI1 are part of a large protein complex. (A) Gel filtration analysis of nuclear extracts of plant inflorescences. Nuclear extracts were loaded onto a 14 ml Bio-SEC-250 column. Fractions were separated by SDS–PAGE and tested on protein blots. Fraction numbers are indicated at the top and arrows indicate elution positions of molecular mass standards. (B) MSI1, MEA and FIE reside in one protein complex in vivo. Co-immunoprecipitations were performed on nuclear extracts prepared from plant inflorescences. Immunoprecipitated proteins were analyzed on protein blots with the antibodies indicated at top. Input contains 3% of the protein used for the co-immunoprecipitation assay. (C) MSI1 and FIE interact physically in vitro. Bacterial extracts containing GST-MSI, MEA or FIE were mixed, incubated together and bound to glutathione beads. GST alone was used as a negative control. After extensive washing, proteins were analyzed by SDS–PAGE and Coomassie staining (left panel) or protein blotting (right panel). Input contains 4% of the protein used for the pulldown assay.
To characterize the protein–protein interactions between MSI1, MEA and FIE in more detail, an MSI1–GST fusion protein was expressed in Escherichia coli and incubated with MEA or FIE proteins before binding to gluthathione beads (Figure 4C). The binding assay revealed that MSI1 can efficiently bind to FIE, but not to MEA, when it is presented as a single binding partner. These results suggest that MSI1 and FIE interact directly in vivo, but that the interaction of MEA and MSI1 requires either post-translational modifications of at least one partner or additional proteins like FIE to mediate the interaction.
Loss of MSI1 leads to fertilization-independent seed development
Both mea and fie belong to the fis class of mutants, and the current model suggests that the MEA–FIE complex prevents seed development in the absence of a fertilization signal (for review see Grossniklaus et al., 2001). If MSI1 is an integral part of the MEA–FIE complex, we would also expect autonomous endosperm or seed development in MSI1/msi1 mutants. Floral buds of wild-type and MSI1/msi1 mutant plants were emasculated and compared with manually pollinated gynoecia. Pollinated gynoecia of wild-type and MSI1/msi1 mutant plants were indistinguishable after 6 days and reached a length of about 13 mm. In contrast, after 6 days without pollination, gynoecia of wild-type plants were only 2–3 mm long, whereas in MSI1/msi1 mutant plants unpollinated gynoecia formed siliques of 10 mm (Figure 5).
Fig. 5. MSI1/msi1 plants undergo fertilization-independent silique elongation. (A) Representative siliques of wild-type and MSI1/msi1 plants at 6 DAP or non-pollinated (np). Scale bar, 5 mm. (B) Average silique length of wild-type or MSI1/msi1 plants (mean ± SE, n = 10) at 6 DAP or 6 days after emasculation (MSI1/msi1).
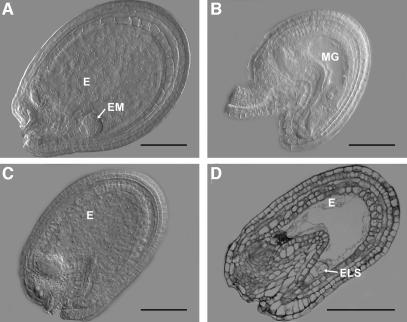
Since silique elongation in the absence of fertilization suggests autonomous endosperm and seed development we investigated the developing seeds in MSI1/msi1 plants in more detail. Figure 6 shows that unfertilized wild-type ovules did not develop (Figure 6B), whereas many unfertilized msi1 ovules initiated seed development (Figure 6C and D). Optical and semi-thin sections through such fertilization-independent seeds revealed the presence of a multinuclear endosperm but no embryo. Sometimes embryo-like structures could be observed at the micropylar end of the embryo sac (Figure 6D). However, in contrast with embryonic cells that contain small vacuoles, these cells were highly vacuolated, suggesting that they originated from the endosperm. Only 50% of the ovules carry a mutant msi1 allele and thus can be expected to initiate fertilization-independent seed development. Quantification revealed that 41.2% of the ovules displayed the FIS phenotype, demonstrating a penetrance of more than 80%. This is significantly higher than the reported penetrance of 15–20% in the mea mutants (Grossniklaus and Vielle-Calzada, 1998; Kiyosue et al., 1999).
Fig. 6. Fertilization-independent seed development in MSI1/msi1 plants. (A) Wild-type seed at 6 DAP containing a dermatogen-stage embryo. (B) Unfertilized wild-type ovule at 6 DAP. (C), (D) Unfertilized msi1 ovules that started seed development and contain multinuclear endosperm. Abbreviations: E, endosperm; EM, embryo; ELS, embryo-like structure; MG, megagametophyte. Scale bars: 100 µm in (A), (C) and (D); 50 µm in (B).
Double fertilization is impaired in msi1
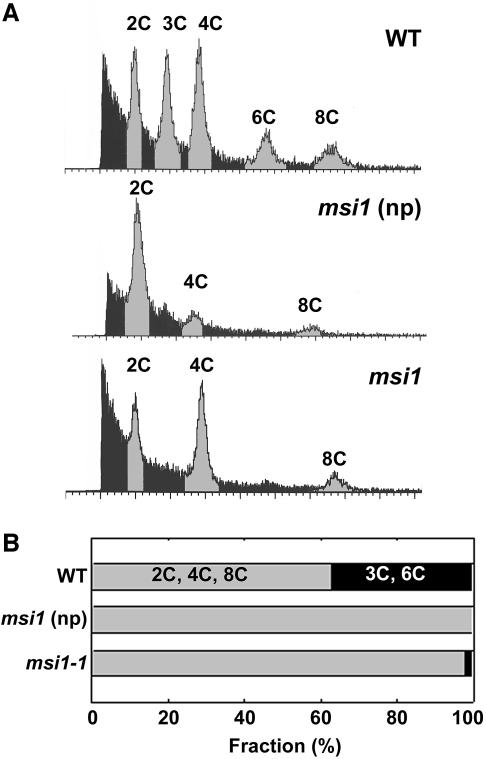
The high penetrance of the FIS phenotype in MSI1/msi1 plants suggests that endosperm development fails to arrest and starts without fertilization even after pollination. We tested this hypothesis by performing ploidy analysis using flow cytometry of nuclei from developing seeds (Figure 7). This assay was previously used to demonstrate that endoreduplication produces 6C and 12C nuclei in the endosperm and 4C, 8C and 16C nuclei in the embryos of several species (Matzk et al., 2000). We observed up to 40% 3C or 6C nuclei derived from the endosperm in wild-type seeds at 6 DAP. As expected, seeds developing in unpollinated MSI1/msi1 siliques did not yield any 3C or 6C signal, demonstrating that these seeds contain only nuclei derived from the diploid central cell. Surprisingly, 3C or 6C signals were also largely absent in msi1 mutant seeds derived from pollinated gynoecia. The small percentage of 3C and 6C nuclei detected in some preparations (compare Figure 7A, bottom, and B) suggests that double fertilization can occur in a small fraction of msi1 seeds. Because we observed embryo formation in msi1 mutant seeds only after pollination, fertilization of the egg cell appears to be required for development of the embryo but not the endosperm.
Fig. 7. Mutant msi1 seeds contain diploid endosperm. (A) Ploidy analysis of nuclei from wild-type seeds at 6 DAP (WT), msi1 seeds developing without fertilization (msi1, np) and msi1 seeds at 6 DAP (msi1). (B) Quantified results from ploidy analysis (n ≥ 4, SD < 5%).
Discussion
MSI1-like proteins are components of various chromatin-modifying protein complexes, but their biological function is not well understood at present. In yeast, MSI1 is required for gene silencing at telomeres and mating type loci (Kaufman et al., 1997; Enomoto and Berman, 1998). In C.elegans, LIN53 is repressing genes required for the development of vulval cell fates (Lu and Horvitz, 1998). Recently, we have shown that Arabidopsis MSI1 is required for the maintenance of differentiation processes in vegetative and reproductive development (Hennig et al., 2003). In many organisms, including yeast, vertebrates and many plants, several MSI1-like proteins exist that are functionally distinct (Verreault et al., 1996; Ach et al., 1997; Kaufman et al., 1997; Hennig et al., 2003). However, insects like Drosophila and Anopheles contain only a single MSI1-like gene. Sequence similarities suggest that MSI1-like genes in different clades diverged independently during evolution. However, the advantages or costs of MSI1 diversification are not well understood. The Arabidopsis genome encodes five MSI1-like proteins (MSI1–5). MSI2 and MSI3, as well as MSI4 and MSI5, are pairs of closely related genes, while MSI1 is more distantly related and has no close homologue (Hennig et al., 2003). We isolated an insertion mutant in the MSI1 gene that is most likely a complete knockout of MSI1 function. Heterozygous mutant plants developed normally until the seed set. This observation is consistent with the fate of msi1 antisense plants, which contain about 30% of wild-type MSI1 protein levels but have a normal morphology (unpublished results). Only the reduction of MSI1 protein content to about 5% of wild-type levels in msi1 co-suppression plants caused severe defects during vegetative and reproductive development (Hennig et al., 2003). Seed abortion in heterozygous msi1 insertion mutants demonstrated a central function of MSI1 in seed development that was not uncovered in the co-suppression plants.
The maternal MSI1 allele is required for seed development
Similar to the loss-of-function alleles of MEA, FIE and FIS2, loss of MSI1 function also has a gametophytic maternal effect. Reciprocal crosses demonstrated that wild-type seed development occurs in the presence of a maternally inherited MSI1 allele, but seeds that contain a maternally derived msi1 allele abort their development regardless of the paternal contribution. Maternal effects can occur for a variety of reasons (Grossniklaus and Schneitz, 1998; Grossniklaus et al., 1998), including dosage effects in the endosperm, monoallelic expression after fertilization (genomic imprinting) or by affecting a stored gene product that is produced prior to fertilization but is required during seed development. In the triploid endosperm a single wild-type allele delivered by the pollen might not suffice to compensate two maternally derived mutant alleles. However, pollination of MSI1/msi1 plants with pollen of a tetraploid wild-type plant did not change the fraction of aborting seeds despite the presence of two wild-type MSI1 alleles (data not shown). Similarly, plants that were homozygous for the endogenous MSI1 wild-type allele and for a complementing MSI1 transgene carry two MSI1 copies in their gametes, but pollination with pollen from these plants did not affect seed abortion ratios in MSI1/msi1 plants (data not shown). Parent-of-origin effects can also be due to maternal or paternal allele-specific differential expression (genomic imprinting) (Grossniklaus et al., 2001). It was shown that the paternal alleles of the FIS genes are not expressed after fertilization, indicating that they are regulated by genomic imprinting (Vielle-Calzada et al., 1999; Luo et al., 2000). Further studies will be necessary to test whether the parent-of-origin effect of msi1 is due to a paternally silenced MSI1 allele or the deficiency in the cytoplasmic store of the MSI1 protein. The seed arrest phenotype of mea and fis2, but not fie, is alleviated by pollination with a low-methylation parent or the introduction of the ddm1 mutation, which is deficient in a putative chromatin remodelling factor and reduces DNA methylation (Jeddeloh et al., 1999; Vielle-Calzada et al., 1999; Luo et al., 2000; Yadegari et al., 2000). In contrast, we did not observe higher fractions of developing seeds in selfed MSI1/msi1; DDM1/ddm1 plants (data not shown), indicating that defects caused by the loss of MSI1 activity cannot be rescued by activating silenced genes. Together, our data show that a maternal copy of MSI1 is essential for successful seed development.
Loss of MSI1 function relaxes endosperm development in the absence or presence of fertilization
In most plants, seed and fruit development are actively repressed and usually initiate only after fertilization. Female gametophytes in MSI1/msi1 mutants initiate endosperm development without fertilization and form seed-like structures that eventually abort as reported for mea, fie and fis2. Interestingly, the penetrance of the various fis-class mutants varies considerably and is much higher in msi1 than mea mutants (Table III). Considering that many fis-class alleles are likely null alleles, the difference in penetrance could be attributed to partial redundancy of MEA and other E(Z) homologues in Arabidopsis. In contrast, the high penetrance in MSI1/msi1 suggests that none of the other four MSI1-like proteins in Arabidopsis (MSI2–5) can substitute for MSI1 function in preventing endosperm development in the absence of fertilization. In fact, in msi1 mutant gametophytes fertilization-independent endosperm formation occurred even when the gynoecia were pollinated, leading to the formation of a diploid endosperm surrounding the embryo. This suggests that the need for double fertilization is strongly relaxed. Because embryo development was never observed without pollination, we conclude that after pollination only the egg cell is fertilized while the central cell nucleus starts to divide without fertilization. Two main hypotheses attempt to rationalize the evolutionary origin of the endosperm (Friedman, 2001; Baroux et al., 2002; Berger, 2003). One hypothesis assumes a sporophytic nature of the endosperm, which would have developed from an altruistic second embryo. The other hypothesis suggests that the endosperm is derived from the female gametophyte and the second fertilization event developed only later to prevent accumulation of nourishing endosperm tissue in the absence of an embryo. The unique phenotype of MSI1/msi1 provides some support for the idea of a gametophytic endosperm origin.
Table III. Penetrance of fertilization-independent seed development (6 DAP).
| Seed-like | Ovules | Percentage seed-like | Penetrance (%) | Reference | |
|---|---|---|---|---|---|
| MSI1/MSI1 | 0 | 146 | 0.0 | NA | This work |
| MSI1/msi1 | 147 | 210 | 41.2 | 82.4 | This work |
| MEA/mea3 (Ler) | 7.5 | 15.0 | Kiyosue et al. (1999) | ||
| MEA/mea3 (Col) | 10.3 | 20.6 | Kiyosue et al. (1999) | ||
| MEA/mea4 (WS) | 1.8 | 3.6 | Kiyosue et al. (1999) |
NA, not applicable.
Our results do not explain directly why msi1 mutant seeds contain abnormal embryos and abort. Since MSI1 is expressed in the developing embryo, it is most likely also functionally required during embryo development. However, it is also possible that the defective endosperm perturbs embryo development. Distinguishing between these two possibilities will be subject of future investigations. Several forward genetic screens for fis-class mutants have been reported in which several alleles of mea, fie and fis2 were isolated. Considering the high penetrance of the fis phenotype in MSI1/msi1 mutants, it is striking that no mutant allele of MSI1 was previously identified. It is possible that the screens were not yet saturated, suggesting that still other fis-class mutants can be expected.
The function of the MSI1–FIS–MEA PcG-like complex is separate from other MSI1 functions
FIE and MEA interact, suggesting that they function together in a complex similar to PcG complexes in animals. We find that the Arabidopsis MEA–FIE complex has a molecular mass of about 600 kDa and therefore is similar to the Drosophila 600 kDa ESC/E(Z) complex and the homologous complex from mammals (Ng et al., 2000; Tie et al., 2001; Czermin et al., 2002; Kuzmichev, 2002; Müller et al., 2002). MSI1 co-migrates with MEA and FIE and can be co-immunoprecipitated with both proteins in vivo. These results show that MSI1 is an additional member of the FIE–MEA complex. In vitro binding experiments revealed that MSI1 binds efficiently to FIE but not to MEA, suggesting that the topology of the FIS complex involves FIE as a bridge between MEA and MSI1. While MEA function is specific for seed development, MSI1 and FIE were also shown to act during later stages of development. In particular, MSI1 is part of chromatin assembly factor complex CAF 1 that is required for the maintenance of shoot and root apical meristems. However, fas1 and fas2, which lack the two other CAF-1 subunits (Kaya et al., 2001), do not show seed abortion and fertilization-independent endosperm or seed development. The normal reproductive development of fas mutants demonstrates that MSI1 acts independently of CAF-1 in seed development. Furthermore, MSI1 is the only MSI1-like protein that is strictly required for seed development in Arabidopsis as mutants in any of the other four MSI1-like genes had normal seed set (data not shown). However, additional functions of MSI1 could account for the more severe phenotype of msi1 compared with mea or fie mutants.
The combined molecular mass of MEA, FIE and MSI1 is only 169 kDa, suggesting that additional proteins are present in the complex. One likely candidate is FIS2, although no direct interaction has been observed so far (unpublished results). The MEA–FIE–MSI1 complex is most likely conserved between monocots and dicots, because all members of this complex have homologues in monocots like maize (Rossi et al., 2001; Springer et al., 2002; Danilevskaya et al., 2003).
Together, we demonstrate the presence of a 600 kDa PcG-like complex in Arabidopsis that consists of MEA, FIE and MSI1. This complex is of ancient origin and has been evolutionarily conserved between plants and animals. Similar to MEA and FIE, MSI1 is a gametophytic maternal effect gene because the paternal copy of MSI1 has no effect on the fate of the offspring. In msi1, endosperm development of mutant seeds initiates independently of fertilization even in pollinated gynoecia, leading to the formation of embryos surrounded by diploid endosperm. This most likely leads to an earlier arrest of embryo development compared with mea and fie mutants. Overall, our reverse genetics approach has facilitated the identification of a new subunit of the MEA–FIE complex and revealed the role of MSI1 in the correct initiation and progress of seed development.
Materials and methods
Plant material and growth conditions
Plants were grown in Conviron growth chambers with mixed cold inflorescent and incandescent light (150 µmol/m2/s, 23°C) under long days (16 h of light). Seeds of Columbia and Landsberg erecta wild-type accessions and of fas1-1 and fas2-1 (Reinholz, 1966; Leyser and Furner, 1992) mutants were obtained from the Nottingham Arabidopsis Stock Centre (NASC On-Line Catalogue at http://nasc.nott.ac.uk/home.html).
To complement the MSI1/msi1 mutant, a 2000 bp fragment upstream of the MSI1 start codon was amplified by PCR using primers CGGGATCCAGGTTTGGAATCGACCAAGA and TGTCGACCGAT GTCTTTGTTATTCCCG. The MSI1 cDNA was amplified using primers CGGTCGACATGGGGAAAGACGAAGAGGAA and GACCATGGA AAGAAGCTTTTGATGGTTCTTC, and MSI1 promoter and cDNA were inserted into a modified pCAMBIA1380 vector. Heterozygous msi1 plants were transformed by floral dip and transgenic plants were selected on hygromycin. T1 plants were assayed for complementation of seed abortion and several complementing lines were obtained. After performing DNA blots with an HPT probe, we chose a line with a single insert for further analysis (msi1 compl.-1)
RNA isolation and RT-PCR
RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. For RT-PCR analysis, 1 µg total RNA was treated with DNase I. Half of the DNA-free RNA (0.5 µg) was reverse transcribed using an oligodT primer and MMLV reverse transcriptase (Clontech, Palo Alto, CA), while the remaining RNA was incubated without reverse transcriptase. Aliquots of the generated cDNA, which equalled 50 ng total RNA, were used as template for PCR with gene-specific primers AtMSI1-F (GCACCGCTCTTCACACATTTG) and AtMSI1-R (TCGATCCTGCTAAGGTCCCAA) or GAPDH-F (TTCTTGGCACCAGCTTCAAT) and GAPDH-R (CTCCCTTGGAA GGAGCTAGG).
In situ hybridization
In situ hybridization was performed as described (Vielle-Calzada et al., 1999) with modifications. For sense and antisense 11-digoxigenin-UTP-labelled probes, a plasmid pKS (Stratagene) containing a 450 bp fragment (5′ end of the MSI1 cDNA) was linearized with restriction enzymes cutting in the polylinker (BamHI and HindIII, respectively) and 1 µg was used for probe synthesis. Mature flowers and siliques of Arabidopsis thaliana wild-type plants were fixed in 4% paraformaldehyde and embedded in Paraplast Plus (Sigma). Sections 10 µm thick were cut using a Leica microtome (Leica RM 2145) and mounted on ProbeOnPlus slides (Fischer Biotech). Sections were digested with proteinase K for 30 min at 37°C, treated with acetic acid anhydride, dried in ethanol and then hybridized with about 40 ng of DIG-labelled probes per slide overnight at 55°C. After washing with 0.2× SSC at 55°C, the slides were processed for detection of the DIG antigen. This involved blocking with DIG-blocking reagent and bovine serum albumin, followed by incubation with an anti-DIG antibody conjugated to alkaline phosphatase (Roche). After washing with blocking reagent, the immunological detection was performed by incubation in NBT and X-phosphate for periods of 16 to 18 h. Reactions were stopped with TE buffer 10 mM (pH 8.0) and mounted in TE/glycerol (v/v) for microscope analysis.
Histological analysis
Material was fixed in FAA (3.7% formaldehyde, 5% acetic acid, 50% ethanol) overnight at 4°C and embedded in Technovit 7100 resin (Kulzer, Wahrheim, Germany). Sections 5 µm thick were stained with Toluidine blue and observed using an Axioplan 2 microscope (Zeiss, Jena, Germany). Alternatively, tissues were cleared with chloralhydrate after fixation in ethanol–acetic acid (9:1) and observed under differential interference contrast (DIC) optics. Images were recorded with an Axiocam HRC CCD camera (Zeiss, Jena, Germany) and edited with ZeissVision software.
Gel filtration analysis
Inflorescences (3 g) were harvested and immersed in 30 ml of Buffer 1 [1 M hexylene glycol, 10 mM PIPES pH 7.0, 10 mM MgCl2, 0.2% Triton X100, 5 mM β-mercaptoethanol, 1 µM ZnSO4, 0.8 mM phenyl-methylsulfonyl fluoride (PMSF), 1× complete Protease Inhibitors (PI) (Roche)]. The tissue was homogenized using a Polytron and then filtered through 500 µm and 50 µm meshes. The solution was centrifuged (2000g) for 10 min and the pellet was resuspended in 15 ml of Buffer 2 (0.5 M hexylene glycol, 10 mM PIPES pH 7.0, 10 mM MgCl2, 0.2% Triton X100, 5 mM β-mercaptoethanol, 0.8 mM PMSF, 1× PI). After centrifugation (3000g) for 10 min, the pellet was resuspended in 500 µl of Buffer 3 (110 mM KCl, 15 mM HEPES pH 7.5, 5 mM MgCl2, 1 µM ZnSO4, 1 mM dithiothreitol, 1× PI). Then, 50 µl of 4 M ammonium sulfate solution was added slowly and the solution was rocked on ice for 30 min. The solution was dialysed against NLB, and dialysed extracts (200 µl) were loaded onto a 14 ml Bio-SEC-250 column (Bio-Rad) and eluted with Buffer 3 at a flow rate of 0.5 ml/min. Fractions (200 µl) were collected and precipitated with acetone. The precipitate was resuspended in SDS sample buffer and analyzed on protein blots using anti-MEA, anti-FIE (Köhler et al., 2003) and anti-MSI1 (Ach et al., 1997; Hennig et al., 2003) antisera.
Immunoprecipitations
Anti-MEA and anti-FIE antibodies were cross-linked to protein A agarose as described previously (Weigel and Glazebrook, 2002). Nuclei were isolated as described above and washed with Buffer 2. Nuclei were dissolved in 300 µl of Buffer 4 (50 mM Tris pH 7.5, 10 mM EDTA, 1% SDS, 1× PI) and the extract was diluted 10-fold with Buffer 5 (1.1% Triton X100, 1.2 mM EDTA, 16.7 mM Tris pH 7.5, 167 mM NaCl, 1× PI). Extracts were precleared for 1 h with protein A beads. Antibodies coupled to protein A beads (20 µl) were added to 700 µl nuclear extract and incubated under rotation for 4 h at 4°C. Immunocomplexes were washed four times with 1 ml of Buffer 6 (10 mM Tris pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% Triton X100, 0.5% NP 40, 1× PI) and eluted by boiling with sample buffer.
In vitro pulldown assays
The full-length MSI1 coding sequence was cloned into the pGEX-4T expression vector (Amersham). Escherischia coli strains harbouring the pGEX-MSI1 or pGEX plasmids were grown overnight in 2× YT medium at 37°C. Cultures were diluted 1:100 into fresh medium and grown until OD600 = 0.8 followed by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 1 mM. After induction for 6 h at 28°C the cells were pelleted and resuspended in BB (20 mM Tris pH 7.5, 150 mM NaCl, 0.1% Triton X100, 1 µM ZnSO4, 1 mM PMSF, 1× PI). Lysozyme was added to 2 mg/ml and cells were lysed by incubation for 20 min on ice. After a brief sonication the solution was centrifuged (20 000g) for 10 min, and aliquots of the supernatant were frozen in liquid nitrogen. Expression of FIE protein using the pRSET-FIE plasmid was as previously described (Spillane et al., 2000). pASK-MEA contains the full-length MEA cDNA in pASK-IBA3 (Institut für Bioanalytik GmbH). Bacterial cells containing pASK-MEA were grown under similar conditions to those described for pGEX-MSI1. Expression was induced by adding anhydrotetracyclin to 0.2 µg/ml. After 6 h of induction, cells were pelleted and lysed as described above. Equal volumes of extract containing FIE or MEA were combined with extracts containing GST-MSI1 or GST and incubated under rotation for 2 h. Then, 100 µl of glutathione beads were added and incubation continued for 40 min. After six washes with BB, bound proteins were eluted with SDS sample buffer and analyzed on protein blots using anti-Xpress antibodies (Invitrogen) for detection of Xpress-FIE or anti-MEA antibodies.
Ploidy analysis
Ploidy analysis was performed as described (Matzk et al., 2000). About 50 seeds were isolated from siliques, crushed with a pestle in microfuge tubes containing 400 µl nuclear extraction buffer (Partec, Münster, Germany), incubated for 30 min on ice, filtered through 30 µm mesh, mixed with 1 ml nuclear staining buffer (Partec), incubated on ice for a further 10 min and analyzed with a Partec Ploidy Analyser. For quantification, results of four to six independent preparations were averaged.
Acknowledgments
Acknowledgements
We are grateful to Michael Federer for his help with the histological preparations. We thank Celia Baroux for helpful discussions. The msi1 T-DNA insertion mutant was generously provided by Syngenta Torrey Mesa Research Institute. C.K is supported by EMBO and HFSP Fellowships, and L.H. is supported by the Deutsche Forschungsgemeinschaft. This research was supported by E.U. Project QLG2-1999-00454 (ECCO) and funds from the Swiss Ministry for Science and Education (BBW-No. 00.0223) to W.G, and by the Kanton of Zürich and grant 31-64061.00 of the Swiss National Science Foundation to U.G.
References
- Ach R.A., Taranto,P. and Gruissem,W. (1997) A conserved family of WD-40 proteins binds to the retinoblastoma protein in both plants and animals. Plant Cell, 9, 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz-Leehan R. and Feldmann,K.A. (1997) T-DNA insertion mutagenesis in Arabidopsis: going back and forth. Trends Genet., 13, 152–156. [DOI] [PubMed] [Google Scholar]
- Baroux C., Spillane,C. and Grossniklaus,U. (2002) Evolutionary origins of the endosperm in flowering plants. Genome Biol., 3, reviews1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F. (1999) Endosperm development. Curr. Opin. Plant Biol., 2, 28–32. [DOI] [PubMed] [Google Scholar]
- Berger F. (2003) Endosperm: the crossroad of seed development. Curr. Opin. Plant Biol., 6, 42–50. [DOI] [PubMed] [Google Scholar]
- Chaudhury A.M., Craig,S., Dennis,E. and Peacock,W. (1998) Ovule and embryo development, apomixis and fertilization. Curr. Opin. Plant Biol., 1, 26–31. [DOI] [PubMed] [Google Scholar]
- Czermin B., Melfi,R., McCabe,D., Seitz,V., Imhof,A. and Pirrotta,V. (2002) Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell, 111, 185–196. [DOI] [PubMed] [Google Scholar]
- Danilevskaya O.N., Hermon,P., Hantke,S., Muszynski,M.G., Kollipara,K. and Ananiev,E.V. (2003) Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell, 15, 425–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto S. and Berman,J. (1998) Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev., 12, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N.J. and Kingston,R.E. (2001) Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell. Biol., 2, 409–421. [DOI] [PubMed] [Google Scholar]
- Friedman W.E. (2001) Developmental and evolutionary hypotheses for the origin of double fertilization and endosperm. C. R. Acad. Sci. III, 324, 559–567. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U. and Schneitz,K. (1998) The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol., 9, 227–238. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U. and Vielle-Calzada,J.-P. (1998). Parental conflict and infanticide during embryogenesis. Trends Plant Sci., 3: 328. [Google Scholar]
- Grossniklaus U., Vielle-Calzada,J.P., Hoeppner,M.A. and Gagliano,W.B. (1998) Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science, 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U., Spillane,C., Page,D.R. and Köhler,C. (2001) Genomic imprinting and seed development: endosperm formation with and without sex. Curr. Opin. Plant Biol., 4, 21–27. [DOI] [PubMed] [Google Scholar]
- Hennig L., Taranto,P., Walser,M., Schonrock,N. and Gruissem,W. (2003) Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development, 130, 2555–2565. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes,T.L. and Richards,E.J. (1999) Maintenance of genomic methylation requires a SWI/SNF-like protein. Nature Genet., 22, 94–97. [DOI] [PubMed] [Google Scholar]
- Jones C.A., Ng,J., Peterson,A.J., Morgan,K., Simon,J. and Jones,R.S. (1998) The Drosophila ESC and E(z) proteins are direct partners in Polycomb group-mediated repression. Mol. Cell. Biol., 18, 2825–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman P.D., Kobayashi,R. and Stillman,B. (1997) Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev., 11, 345–357. [DOI] [PubMed] [Google Scholar]
- Kaya H., Shibahara,K., Taoka,K., Iwabuchi,M., Stillman,B. and Araki,T. (2001) FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell, 104, 131–142. [DOI] [PubMed] [Google Scholar]
- Kenzior A.L. and Folk,W.R. (1998) AtMSI4 and RbAp48 WD-40 repeat proteins bind metal ions. FEBS Lett., 440, 425–429. [DOI] [PubMed] [Google Scholar]
- Kiyosue T. et al. (1999) Control of fertilization-independent endosperm development by the MEDEA Polycomb gene in Arabidopsis. Proc. Natl Acad. Sci. USA, 96, 4186–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C. and Grossniklaus,U. (2002) Epigenetic inheritance of expression states in plant development: the role of Polycomb group proteins. Curr. Opin. Cell Biol., 14, 773–779. [DOI] [PubMed] [Google Scholar]
- Köhler C., Hennig,L., Spillane,C., Pien,S., Gruissem,W. and Grossniklaus,U. (2003) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev., 17, 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka,K., Erdjument-Bromage,H., Tempst,P. and Reinberg,D. (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev., 16, 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser H.M.O. and Furner,I.J. (1992) Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development, 116, 397–403. [Google Scholar]
- Lu X.W. and Horvitz,H.R. (1998) lin-35 and lin-53, two genes that antagonize a C.elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell, 95, 981–991. [DOI] [PubMed] [Google Scholar]
- Luo M., Bilodeau,P., Koltunow,A., Dennis,E.S., Peacock,W.J. and Chaudhury,A.M. (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 96, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Bilodeau,P., Dennis,E.S., Peacock,W.J. and Chaudhury,A. (2000) Expression and parent-of-origin effects for FIS2, MEA and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl Acad. Sci. USA, 97, 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzk F., Meister,A. and Schubert,I. (2000) An efficient screen for reproductive pathways using mature seeds of monocots and dicots. Plant J., 21, 97–108. [DOI] [PubMed] [Google Scholar]
- Muller C. and Leutz,A. (2001) Chromatin remodeling in development and differentiation. Curr. Opin. Genet. Dev., 11, 167–174. [DOI] [PubMed] [Google Scholar]
- Müller J. et al. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell, 111, 197–208. [DOI] [PubMed] [Google Scholar]
- Ng J., Hart,C.M., Morgan,K. and Simon,J.A. (2000) A Drosophila ESC–E(Z) protein complex is distinct from other Polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol., 20, 3069–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N., Yadegari,R., Margossian,L., Hannon,M., Michaeli,D., Harada,J.J., Goldberg,R.B. and Fischer,R.L. (1999) Mutations in FIE, a WD Polycomb group gene, allow endosperm development without fertilization. Plant Cell, 11, 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthun M.R., Widom,J. and Gottschling,D.E. (1996) The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell, 87, 85–94. [DOI] [PubMed] [Google Scholar]
- Qian Y.W., Wang,Y.C., Hollingsworth,R.E., Jr., Jones,D., Ling,N. and Lee,E.Y. (1993) A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature, 364, 648–652. [DOI] [PubMed] [Google Scholar]
- Reinholz E. (1966) Radiation induced mutants showing changed inflorescence characteristics. Arabid. Inf. Serv., 3, 19–20. [Google Scholar]
- Reyes J.C. and Grossniklaus,U. (2003) Diverse functions of Polycomb group proteins during plant development. Semin. Cell Dev. Biol., 14, 77–84. [DOI] [PubMed] [Google Scholar]
- Reyes J.C., Hennig,L. and Gruissem,W. (2002) Chromatin-remodeling and memory factors. New regulators of plant development. Plant. Physiol., 130, 1090–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V., Varotto,S., Locatelli,S., Lanzanova,C., Lauria,M., Zanotti,E., Hartings,H. and Motto,M. (2001) The maize WD-repeat gene ZmRbAp1 encodes a member of the MSI/RbAp sub-family and is differentially expressed during endosperm development. Mol. Genet. Genom., 265, 576–584. [DOI] [PubMed] [Google Scholar]
- Sessions A. et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell, 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt R.G., van der Vlag,J., Gunster,M.J., Hamer,K.M., den Blaauwen,J.L., Satijn,D.P., Hendrix,T., van Driel,R. and Otte,A.P. (1998) Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol. Cell. Biol., 18, 3586–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane C., MacDougall,C., Stock,C., Köhler,C., Vielle-Calzada,J., Nunes,S.M., Grossniklaus,U. and Goodrich,J. (2000) Interaction of the Arabidopsis Polycomb group proteins FIE and MEA mediates their common phenotypes. Curr. Biol., 10, 1535–1538. [DOI] [PubMed] [Google Scholar]
- Springer N.M., Danilevskaya,O.N., Hermon,P., Helentjaris,T.G., Phillips,R.L., Kaeppler,H.F. and Kaeppler,S.M. (2002) Sequence relationships, conserved domains and expression patterns for maize homologs of the polycomb group genes E(z), esc and E(Pc). Plant Physiol., 128, 1332–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J., Hassig,C.A. and Schreiber,S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Tie F., Furuyama,T., Prasad-Sinha,J., Jane,E. and Harte,P.J. (2001) The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development, 128, 275–286. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman,P.D., Kobayashi,R. and Stillman,B. (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell, 87, 95–104. [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada J.P., Thomas,J., Spillane,C., Coluccio,A., Hoeppner,M.A. and Grossniklaus,U. (1999) Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev., 13, 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada J.P., Baskar,R. and Grossniklaus,U. (2000) Delayed activation of the paternal genome during seed development. Nature, 404, 91–94. [DOI] [PubMed] [Google Scholar]
- Vivian-Smith A., Luo,M., Chaudhury,A. and Koltunow,A. (2001) Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development, 128, 2321–2331. [DOI] [PubMed] [Google Scholar]
- Walbot V. and Evans,M.M. (2003) Unique features of the plant life cycle and their consequences. Nat. Rev. Genet., 4, 369–379. [DOI] [PubMed] [Google Scholar]
- Weigel D. and Glazebrook,J. Arabidopsis—a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, p. 2002. [Google Scholar]
- Xu L., Fong, T and Strome,S. (2001) The Caenorhabditis elegans maternal-effect sterile proteins, MES-1, MES-3 and MES-6, are associated in a complex in embryos. Proc. Natl Acad. Sci. USA, 98, 5061–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R. et al. (2000) Mutations in the FIE and MEA genes that encode interacting Polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell, 12, 2367–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]