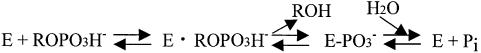Abstract
The structure of Escherichia blattae non-specific acid phosphatase (EB-NSAP) has been determined at 1.9 Å resolution with a bound sulfate marking the phosphate-binding site. The enzyme is a 150 kDa homohexamer. EB-NSAP shares a conserved sequence motif not only with several lipid phosphatases and the mammalian glucose-6-phosphatases, but also with the vanadium-containing chloroperoxidase (CPO) of Curvularia inaequalis. Comparison of the crystal structures of EB-NSAP and CPO reveals striking similarity in the active site structures. In addition, the topology of the EB-NSAP core shows considerable similarity to the fold of the active site containing part of the monomeric 67 kDa CPO, despite the lack of further sequence identity. These two enzymes are apparently related by divergent evolution. We have also determined the crystal structure of EB-NSAP complexed with the transition-state analog molybdate. Structural comparison of the native enzyme and the enzyme–molybdate complex reveals that the side-chain of His150, a putative catalytic residue, moves toward the molybdate so that it forms a hydrogen bond with the metal oxyanion when the molybdenum forms a covalent bond with NE2 of His189.
Keywords: crystal structure/Escherichia blattae/hexamer/non-specific acid phosphatase/protein crystallography
Introduction
Phosphatases constitute a diverse group of enzymes that hydrolyze phosphoesters in various kinds of substrates under different conditions (Vincent et al., 1992). Based on criteria such as specificity and optimum pH, the enzymes can be classified into several families, one of which is a family of bacterial non-specific acid phosphatases (NSAPs). The NSAPs that catalyze phosphomonoester hydrolysis are further divided into three classes, designated A, B and C, on the basis of amino acid sequence similarity (Thaller et al., 1998). Class A NSAPs have a 25–27 kDa polypeptide component (Thaller et al., 1998) and are resistant to EDTA, Pi, fluoride and tartrate (Thaller et al., 1994). The physiological function of the class A NSAPs remains to be determined. To date, this class of enzymes has been isolated from Zymomonas mobilis (Pond et al., 1989), Salmonella typhimurium (Kasahara et al., 1991), Morganella morganii (Thaller et al., 1994) and Shigella flexneri (Uchiya et al., 1996). The class A NSAPs possess a conserved sequence motif, KX6RP- (X12–54)-PSGH-(X31–54)-SRX5HX3D, which is shared by several lipid phosphatases and the mammalian glucose-6-phosphatases (Stukey and Carman, 1997). Curiously, this motif is also found in the vanadium-containing chloroperoxidase (CPO) from Curvularia inaequalis. Hemrika et al. (1997) also found the motif independently, and discovered that apo-CPO exhibits phosphatase activity. The crystal structure of CPO (Messerschmidt and Wever, 1996) revealed that the conserved residues are in close proximity so that they embrace the co-factor vanadium in a concerted manner. Consequently, the vanadate in CPO is thought to be comparable to the phosphate in the phosphatases. One of the conserved histidine residues of CPO coordinates to the vanadium, suggesting that the corresponding histidine residue in the phosphatases is transiently phosphorylated during catalysis; hence, they are histidine phosphatases.
To date, the crystal structures of acid phosphatases have been reported for kidney bean purple acid phosphatase (Sträter et al., 1995; Klabunde et al., 1996), pig purple acid phosphatase (Guddat et al., 1999), rat acid phosphatase (Lindqvist et al., 1993, 1994; Schneider et al., 1993), Aspergillus ficuum phytase (Kostrewa et al., 1997) and Aspergillus niger pH 2.5 acid phosphatase (Kostrewa et al., 1999). Purple acid phosphatases contain a binuclear iron center in their active sites, and the mechanism of phosphoester hydrolysis involves a nucleophilic attack on the phosphate group by an Fe(III)-coordinated hydroxide ion. The other acid phosphatases described above are histidine phosphatases, but they have no amino acid sequence identity to the class A NSAPs.
We have isolated a new member of the class A NSAPs from Escherichia blattae (EB-NSAP), cloned its gene, overexpressed it in Escherichia coli, purified its product and determined the crystal structure at 1.9 Å resolution. The first crystal structure of the bacterial acid phosphatase presented here reveals that the enzyme forms a homohexamer with a molecular mass of 150 kDa, a result in agreement with that from gel filtration chromatography, and that it has considerable structural similarity to CPO. The unprecedented structure of EB-NSAP provides insights into the reaction mechanism and has evolutionary implications. In addition, the structure can be used to improve the models of biologically important membrane-bound phosphatases that have been proposed based on the crystal structure of CPO, such as type 2 phosphatidic acid phosphatase, which plays a crucial role in signal transduction (Neuwald, 1997), and glucose-6-phosphatase, which is the key enzyme in glucose homeostasis (Hemrika and Wever, 1997; Pan et al., 1998). Thus, the structure of EB-NSAP, along with the previously determined structure of CPO, sheds light on the structure–function relationships of the enzymes that belong to the superfamily, including the mammalian glucose-6- phosphatases, several lipid phosphatases, the bacterial class A acid phosphatases and the vanadium-containing haloperoxidases.
In general, acid phosphatases hydrolyze phosphate esters via the two-step mechanism shown in Scheme 1 (Vincent et al., 1992). In the case of the histidine phosphatases, the first step of the reaction involves nucleophilic attack on the phosphate group by histidine and protonation of the leaving group by another group on the enzyme to produce a covalent phosphoenzyme intermediate and an alcohol molecule. In the next step, the phosphoenzyme intermediate is hydrolyzed, leading to the formation of inorganic phosphate. Lindqvist et al. (1994) determined the crystal structures of rat acid phosphatase complexed with the transition-state analogs vanadate and molybdate, and found the overall structure of the enzyme remains unchanged upon binding of the metal oxyanions. Messerschmidt and Wever (1998) determined the crystal structure of apo-CPO with sulfate at the vanadium-binding site, and suggested that CPO provides a rigid vanadate-binding pocket, as demonstrated by the remarkably similar structures of holo-CPO and apo-CPO. Here, the crystal structure of EB-NSAP complexed with the transition-state analog molybdate has been determined at 2.4 Å resolution, using the molecular replacement method based on the native enzyme structure. A structural comparison of the native enzyme and the enzyme–molybdate complex suggests that the formation of a covalent phosphoryl adduct induces a notable conformational change in EB-NSAP.
Scheme 1.
Results
Cloning, nucleotide sequencing and expression of the EB-NSAP-encoding gene
The E.blattae gene encoding EB-NSAP was isolated by a shotgun-cloning strategy. The nucleotide sequence of the 1225 bp PstI–HincII insert, which was thought to contain the entire EB-NSAP gene based on the expression of the enzyme activity, was determined in both directions (Figure 1). There is a single open reading frame, which encodes a polypeptide of 249 amino acid residues.
Fig. 1. Nucleotide sequence of the 1225 bp PstI–HincII fragment containing the EB-NSAP gene. Number 1 represents the first base of the start codon of the EB-NSAP open reading frame. Putative sequences involved in transcriptional control, identified on the basis of similarity to E.coli consensus sequences, are underlined (–35 and –10 regions) or boxed [ribosome-binding sites (RBS)]. The deduced amino acid sequence of EB-NSAP is shown below the nucleotide sequence. The signal sequence of EB-NSAP is doubly underlined. A potential transcriptional terminator is marked by facing arrows.
We purified EB-NSAP from E.blattae JCM1650 (the purification procedure is not shown), and sequenced its N-terminal residues, which were in good agreement with the predicted amino acid sequence. The N-terminal sequencing data revealed that the protein becomes the matured enzyme after a signal sequence cleavage by signal peptidase after the alanine residue at position 18 (see Figure 1, double underline). The calculated molecular weight of 25 004 (based on 693 bp encoding 231 amino acids, excluding the signal peptide by a posttranslational modification) is in good agreement with the value of 25 000 estimated by SDS–PAGE.
Escherichia coli transformants were cultured for 16 h at 37°C with shaking in LB medium containing ampicillin, and the enzyme activities of the crude extracts were measured. Acid phosphatase activity was hardly detected in E.coli JM109 harboring pUC19, whereas the specific activity of acid phosphatase in E.coli JM109 harboring pEAP320 was 31.3 U/mg, and the value was ∼120-fold higher than that in E.blattae (0.26 U/mg). The transformant showed almost the same activity with or without isopropyl-β-d-thiogalactopyranoside (IPTG) induction. The EB-NSAP gene of E.blattae, therefore, appeared to be expressed under the control of its own promoter.
Purification and characterization of EB-NSAP
EB-NSAP was purified from the crude extract of E.coli JM109 harboring pEAP320, by SP-Toyopearl column chromatography and Butyl-Toyopearl column chromatography. The N-terminal sequence of the enzyme purified from the E.coli JM109 transformant was determined as NH2-LALVATGNDTTTKPD, and was identical to that of the enzyme purified from E.blattae. The specific activity of the purified enzyme was 129 U/mg.
The optimum reaction conditions were investigated by measurements at 30°C and various pH values in sodium acetate buffer (pH 3.0–5.5), MES–NaOH buffer (pH 5.0–7.0) and potassium phosphate buffer (pH 6.0–8.0), and the optimum pH was found to be 6.0. The enzyme activity was also measured at pH 6.0 and various temperatures from 4 to 70°C, and it was maximal at ∼35°C. The enzyme activity was partially inhibited by metal ions such as Hg2+ and Ag+. Chelating reagents, such as EDTA, o-phenanthroline, 2,2′-dipyridyl-8-hydroxyquinoline and NaF did not inhibit the activity, indicating that metal ions are not required for the activity. The enzyme activity was fully retained in the presence of up to 10 mM phosphate, but was slightly inhibited in the presence of 100 mM phosphate.
The enzyme was able to dephosphorylate various phosphoesters. In addition to p-nitrophenylphosphate (pNPP), phenylphosphate (89% relative to pNPP), carbamylphosphate (326%), pyrophosphate (36%), glucose-6-phosphate (33%) and ATP (23%) acted as good substrates. The activity for glucose-1-phosphate was lower (4.5%).
The purified enzyme exhibited not only phosphatase activity but also PPi-glucose phosphotransferase activity. The optimum pH for the phosphotransferase activity was found to be 5.2. The specific activity of the purified enzyme in the transphosphorylation reaction was 7.7 U/mg.
Subunit structure
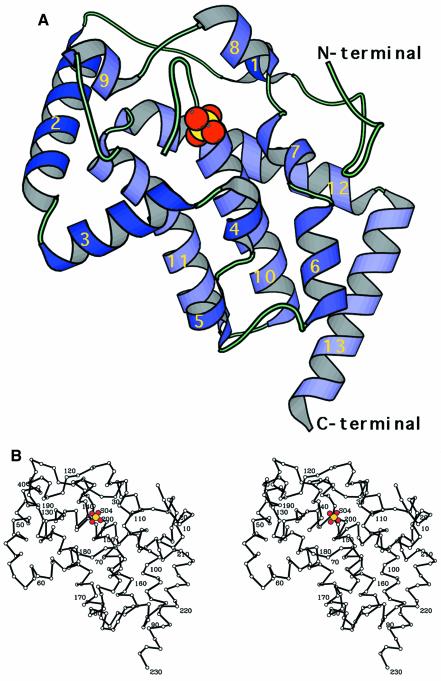
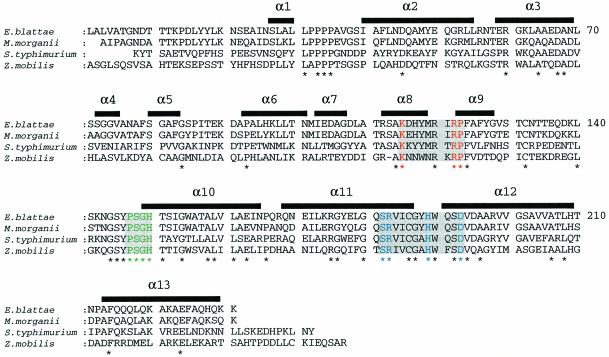
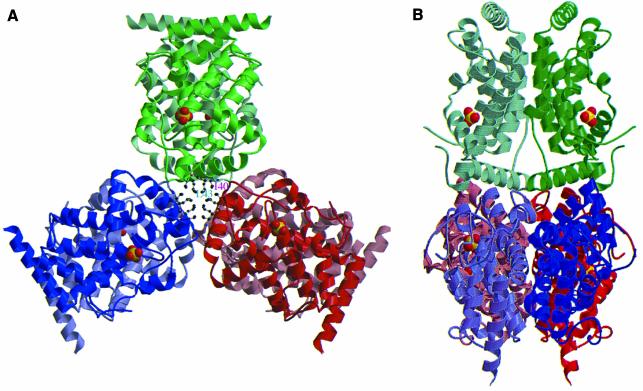
The crystal structure of EB-NSAP was solved by the multiple isomorphous replacement (MIR) method using two heavy atom derivatives at pH 8 (Figure 2). Despite the rather high pH for an acid phosphatase, the enzymatic activity is retained, since soaking the crystal with pNPP turns it yellow, meaning that the substrate is hydrolyzed to p-nitrophenol. An example of the final electron density map is shown in Figure 3. The final model of EB-NSAP includes residues 7–230 with a gap consisting of residues 135 and 136. The subunit, which consists of 13 α-helices (58% of the total residues), has the shape of a flat disk, with dimensions of 45 × 50 × 29 Å. The all α structure of EB-NSAP is comparable to the α/β structures of the other acid phosphatases with solved crystal structures, such as purple acid phosphatase (Sträter et al., 1995), rat acid phosphatase (Schneider et al., 1993) and phytase (Kostrewa et al., 1997). Sequence alignment of the class A NSAPs, together with the secondary structural elements of EB-NSAP, is shown in Figure 4. The only disulfide bridge, between residues 132–186, connects the loop between helix 9 and helix 10 to the C-terminus of helix 11. Except for the N- and C-termini of the subunit, the loop Asn133–Asn143 is the only highly flexible region in the subunit; the average B-value is 69.8 Å2 for the Cα atoms.
Fig. 2. Polypeptide chain trace in EB-NSAP from residue 7 to 230 with a gap consisting of residues 135 and 136. A bound sulfate ion is also shown. Sulfur and oxygen are colored yellow and red, respectively. (A) Schematic ribbon drawing. This figure was prepared using MOLSCRIPT (Kraulis, 1993). (B) Stereo drawing of the Cα backbone with every tenth residue labeled.
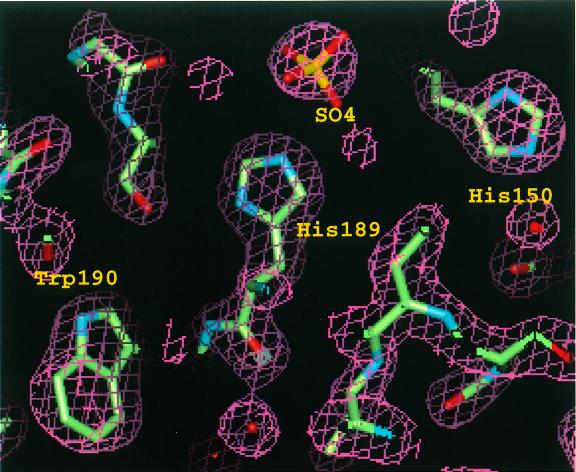
Fig. 3. Final 2Fo – Fc map in the vicinity of the active site, calculated at 1.9 Å resolution. The map was contoured at 2.0σ. Sulfur, oxygen and nitrogen are colored yellow, red and blue, respectively.
Fig. 4. Sequence alignment of EB-NSAP with the class A NSAPs from M.morganii (Thaller et al., 1994), S.typhimurium (Kasahara et al., 1991) and Z.mobilis (Pond et al., 1989). Signal sequences are not shown for the E.blattae, M.morganii and S.typhimurium NSAPs. Residues 1–18 are omitted for the Z.mobilis NSAP, since its signal sequence is unknown. The addition of the other known class A NSAPs from S.flexneri (Uchiya et al., 1996) and Providencia stuartii to the alignment does not change the number of conserved residues. The conserved residues are marked with asterisks. Secondary structural elements, assigned according to the Kabsch and Sander criteria using INSIGHT II, are shown above the sequence alignment. Shaded residues belong to a conserved phosphatase sequence motif. The invariant residues in motifs 1 (KX6RP), 2 (PSGH) and 3 (SRX5HX3D) are colored red, green and blue, respectively.
Hexamer structure
An inspection of the crystal structure revealed that the EB-NSAP hexamer is actually a trimer of dimers (Figure 5). The trimer is built up from three dimers through a 3-fold axis, which corresponds to a crystallographic 63 screw axis. On the other hand, the dimers are related by a 2-fold crystallographic symmetry and are stacked along the 3-fold axis. The hexamer looks like a three-wing propeller with the shaft being the 3-fold axis, when viewed down the 3-fold axis (Figure 5A). The dimer interface, involving α 1–2 and 10–12, is fairly extensive and flat without any deep protrusion from one subunit into another. Upon dimer formation, the solvent accessibility of many hydrophobic residues, i.e. Ile25, Leu30, Ile40, Leu43, Leu159, Leu173 and Val199, decreases notably. In addition, two aromatic residues, Trp155 and Tyr177, are almost completely buried. The dimer is also stabilized by hydrogen bonds, many of which are mediated by water molecules. Direct hydrogen bonds are formed between O of Glu23 and NE2 of Gln169, N of Asn26 and OD1 of Asn170, NE1 of Trp155 and OG of Ser202, OE1 of Gln169 and OG1 of Thr207.
Fig. 5. Two orthogonal views of the EB-NSAP hexamer viewed (A) along a 3-fold axis and (B) along a 2-fold axis. Ball-and-stick drawings at the center of (A) represent Ile40 and Leu43, which play important roles in assembling the six subunits. This figure was prepared using MOLSCRIPT (Kraulis, 1993) and RASTER3D (Merritt and Murphy, 1994).
In contrast to the extensive dimer interface, the interactions for the trimer formation made by the 3-fold axis are rather local. The surface areas per subunit buried upon forming the dimer and the trimer, which were calculated using GRASP (Nicholls et al., 1991), are 2091 and 1041 Å2, respectively. However, there are still some hydrophobic and hydrophilic interactions that stabilize the trimeric structure. Residues Val37, Ile40, Leu43, Leu54, Ile121 and Phe126 form hydrophobic interactions. Ile40 and Leu43 seem to be particularly important, because the burial of these residues is accomplished not only by the contacts made by trimer formation but also by dimer formation (Figure 5A). The hydrogen bonds between the guanidine of Arg53 and O of Phe126, OD1 of Asn57 and NZ of Lys142, O of Gly38 and N of Ile40 also seem to be particularly important for trimer formation.
Structure of the enzyme–molybdate complex
Co-crystallization was successful for the complexes of EB-NSAP with vanadate and molybdate, respectively. Since the latter yielded larger crystals, we decided to determine its crystal structure. The molybdate complex crystallizes with three identical subunits in the asymmetric unit. A hexamer is formed by the association of a pair of trimers via a crystallographic 2-fold axis. The three subunits are virtually identical to each other in the three-dimensional structure, as revealed by root mean square deviations (r.m.s.ds) between equivalent Cα positions of 0.43–0.44 Å, given by pairwise superposition calculations. Therefore, the following discussion is based on one of the three subunits.
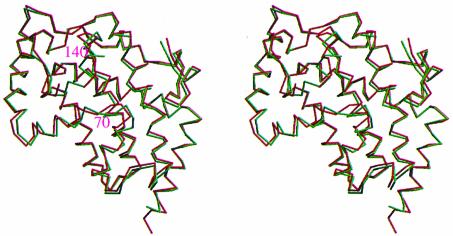
The r.m.s.d. in 220 Cα positions (Gly7–His228, excluding Thr135 and Glu136) is 1.2 Å between the native enzyme and the enzyme–molybdate complex (Figure 6). Substantial structural differences are observed in two regions, Ala68–Gly74 and Thr135–Asn143. The latter is especially surprising, since this region is very flexible and structurally random in the native enzyme, but is less flexible and contains an α-helix ranging from Glu136 to Ser141 in the molybdate complex.
Fig. 6. Stereo view superposition of the native enzyme and the enzyme–molybdate complex. The Cα traces of the native enzyme and the molybdate complex are colored red and green, respectively. Residues 70 and 140, which undergo significant movements upon molybdate binding, are labeled. This figure, which is viewed from the same direction as Figure 2, was prepared using MOLSCRIPT (Kraulis, 1993) and RASTER3D (Merritt and Murphy, 1994).
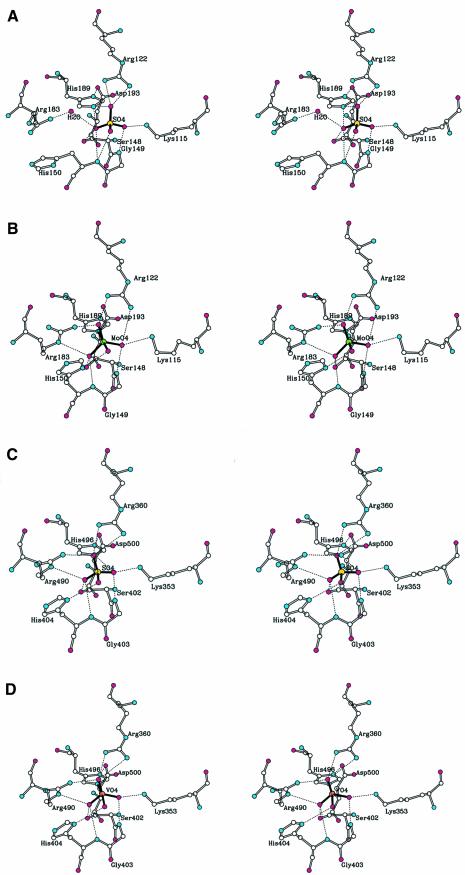
Figure 7A and B shows the active site structures of the native enzyme and the molybdate complex, respectively. Interestingly, His150 and Arg183 adopt different conformations. Both residues form direct hydrogen bonds with the bound oxyanion only in the molybdate complex structure.
Fig. 7. Stereo views of the active site structure. Hydrogen bonds are shown as dashed lines. Sulfur, oxygen, nitrogen, molybdate and vanadium are colored yellow, magenta, cyan, green and orange, respectively. (A) Native EB-NSAP; (B) EB-NSAP complexed with molybdate; (C) apo-CPO; (D) holo-CPO.
Discussion
Structural comparison with CPO from the fungus Curvularia inaequalis
It has been reported that the class A NSAPs share the signature sequence motif KX6RP-PSGH-SRX5HX3D with CPO (Hemrika et al., 1997; Neuwald, 1997; Stukey and Carman, 1997). This enzyme consists of 609 amino acid residues, requires the co-factor vanadium and catalyzes the oxidation of halides (Cl–, Br–, I–) in the presence of hydrogen peroxide. Then, the activated halonium cation is transferred to a halogen acceptor molecule, producing hypohalous acids or organohalogens (Itoh et al., 1987). The tertiary structure of CPO, determined by X-ray crystallography, is mainly helical, with a small β-moiety (Messerschmidt and Wever, 1996). In spite of the lack of overall sequence identity, there is remarkable similarity in the active site structures of EB-NSAP and CPO. The binding site of the sulfate in apo-CPO is comparable to that of the sulfate in EB-NSAP (Figure 7A and C), and so is the binding site of the vanadate in holo-CPO to that of the molybdate in the EB-NSAP–molybdate complex (Figure 7B and D). Therefore, it is not very surprising that apo-CPO shows phosphatase activity (Hemrika et al., 1997). In contrast to the similarity in the active site structure, a significant difference is observed in the accessibility to the active site from the bulk solution; EB-NSAP has a much larger opening near the active site, which is quite consistent with the lack of specificity of the enzyme.
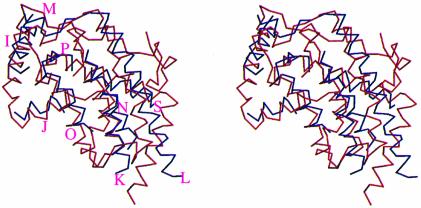
As for the overall tertiary structure, the topology of the core of the EB-NSAP structure shows reasonable similarity to the fold of the active site containing part of the CPO structure. The best superposition, which was calculated for 137 Cα atoms, between the core of EB-NSAP and the relevant portion of CPO yielded an r.m.s.d. of 4.2 Å (Figure 8), although there is no significant sequence homology other than KX6RP-PSGH-SRX5HX3D. Therefore, not only the similarity of the active site structures but also the similarity of the overall three-dimensional structures suggests that the class A NSAPs and CPO share the same ancestor.
Fig. 8. A least-square superposition of the relevant portion of CPO (blue) with EB-NSAP (red). Residues 254–268 (αI), 280–288 (αJ), 300–314 (αK), 325–366 (αL to αM), 401–419 (αN), 479–501 (αO to αP) and 552–565 (αS) of CPO are superimposed with residues 38–52 (α2), 56–64 (α3), 70–84 (α4 to α5), 85–126 (α6 to α9), 145–163 (α10) and 170–206 (α11 to α12) of EB-NSAP. The α-helices of CPO are labeled. This figure, which is viewed from the same direction as Figure 2, was prepared using MOLSCRIPT (Kraulis, 1993) and RASTER3D (Merritt and Murphy, 1994).
Active site and mechanism
As shown in Figure 7A and B, the active site of EB-NSAP consists of Lys115, Arg122, Ser148, Gly149, His150, Arg183, His189 and Asp193. All of the residues are found in the conserved phosphatase sequence motif KX6RP-(X12–54)-PSGH-(X31–54)-SRX5HX3D (Hemrika et al., 1997; Neuwald, 1997; Stukey and Carman, 1997).
His189, located at the bottom of the phosphate binding pocket, is obviously equivalent to the conserved histidine residue in motif 3 (SRX5HX3D) of the conserved sequence motif, which has been predicted to attack the substrate’s phosphoryl group to produce a phosphoenzyme catalytic intermediate (Hemrika et al., 1997; Neuwald, 1997; Stukey and Carman, 1997). This histidine is presumably essential for catalytic activity, since EB-NSAP is irreversibly inhibited by K2PtCl4, which was used as a heavy atom derivative and gave a single binding site near NE2 of His189. In addition, the crystal structure of the enzyme–molybdate complex reveals that NE2 of His189 and the molybdenum form a covalent bond (Figure 7B). A cluster of positively charged residues around His189 must attract the negatively charged phosphate group and probably prevents protonation of the histidine residue, even at a low pH, by reducing the pKa value of the histidine residue.
The phosphate transfer reaction must be terminated by protonation of the substrate leaving group. A candidate for this proton donor is the conserved histidine residue in motif 2 (PSGH), as suggested by Stukey and Carman (1997), Neuwald (1997) and Hemrika et al. (1997). This speculation is supported by a mutational study of glucose-6-phosphatase; the replacement of His119 (equivalent to the histidine residue in motif 2) with alanine resulted in a loss of enzymatic activity (Lei et al., 1995). The side-chain of the corresponding histidine residue of EB-NSAP, His150, is not proximal to the bound sulfate in the crystal structure of the native enzyme, and hydrogen-bonds only to water molecules (Figure 7A). However, His150 changes the χ1 conformation from trans to gauche(+) upon formation of the phosphohistidine intermediate so that it can interact with the phosphate, as revealed by the structure of the enzyme–molybdate complex (Figure 7B).
Asp193 at the exit of motif 3 stabilizes the side-chain conformation of His189 through a direct hydrogen bond with a distance of 3.06 Å. Neuwald (1997) proposed that the aspartate residue and the histidine residue of motif 3, corresponding to Asp193 and His189 of EB-NSAP, establish a charge-relay system to facilitate the formation of the phosphoenzyme catalytic intermediate. In the last step of the catalytic reaction, the intermediate is attacked by a water molecule, so that the covalent bond between His189 and the phosphate is cleaved and inorganic phosphate is released from the enzyme.
Lys115 and Arg122 in motif 1 (KX6RP) play crucial roles in holding the phosphate group of a substrate close to His189 so that the nucleophilic attack by His189 readily takes place. Also anchoring the phosphate to the proper place are the side-chain of Ser148 and the amide nitrogen atoms of Gly149 and His150 in motif 2. The first residue of motif 2, Pro147, has a cis-peptide bond, and thus is not easily replaced by any other amino acid. The side-chain of Ser182 at the entrance of motif 3 forms hydrogen bonds with both the main-chain carbonyl oxygen of Glu178 and the guanidine group of Arg60; therefore, this residue seems to be important to construct the protein structure. The side-chain of Arg183 moves 2.0 Å toward the molybdate to form two hydrogen bonds with two of the three equatorial oxygen atoms of the molybdate in the structure of the molybdate complex. Arg183 apparently helps to stabilize the phosphoenzyme intermediate.
Conformational change upon molybdate binding
The formation of a covalent bond between the molybdate and His189 causes the His150 side-chain to move toward the metal oxyanion. The movement of the side-chain leaves a hole within the protein structure, which is filled by the protein itself. The side-chain of Leu70, which protrudes into the solvent in the native enzyme, follows the movement of the His150 side-chain. Consequently, the side-chain of Leu70 moves 6.7 Å toward the protein interior. This movement induces a structural change of the region ranging from Ala68 to Gly74 (Figure 6). The position and orientation of the His150 side-chain in the enzyme–molybdate complex are stabilized by two hydrogen bonds (NE2 of His150 to O of Ala68 and OE1 of Glu136). Thr135 and Glu136 are disordered in the native enzyme, but become ordered in the molybdate complex. Consequently, the loop ranging from Asn133 to Asn143, which is highly flexible in the native enzyme, forms a less flexible structure that contains an α-helix ranging from Glu136 to Ser141. This newly formed α-helix extends over the active site and covers the bound molybdate. Accompanying the α-helix formation, the side-chain of Leu140 moves 6.2 Å toward the molybdate and is situated near the metal oxyanion; the distance between CD1 of Leu140 and one of the equatorial oxygen atoms of the molybdate is 3.3 Å. The existence of the hydrophobic side-chain near the covalent phosphohistidine is thought to make the approach of a water molecule less frequent. When the phosphoenzyme intermediate is formed, the α-helix probably stabilizes the intermediate by protecting the covalent phosphohistidine from an attack by a water molecule. The phosphoenzyme intermediate would be more susceptible to hydrolysis if the conformational change did not occur.
The conformational change described above is supposed to make the hydrolysis of the phosphoenzyme intermediate more rate-limiting compared with the formation of the intermediate. If alcohol attacks the phosphoenzyme intermediate instead of water, the phosphate is transferred to the alcohol to produce phosphoryl alcohol. Although the newly formed α-helix stabilizes the phosphoenzyme intermediate by preventing its decomposition by hydrolysis, the intermediate is not necessarily resistant to an attack by alcohol, which is considered to approach the active site from a different direction and to bind to a depression surrounded by Leu16, Ser71, Ser72 and Glu104. Therefore, it is not surprising that EB-NSAP has a significant PPi-glucose phosphotransferase activity. If glucose-6-phosphatase, which belongs to the same superfamily as EB-NSAP, undergoes a similar conformational change, then this may account for the view that the PPi-glucose phosphotransferase and ATP-glucose phosphotransferase activities of glucose-6-phosphatase are potentially the most potent glucose phosphorylating systems that have been characterized for the liver (Nordlie, 1971).
Materials and methods
Strains, plasmids and culture conditions
Escherichia blattae JCM1650 was used as the DNA donor. Escherichia coli JM109 (Vierira and Messing, 1982) was used as the host strain for DNA manipulation and expression. Plasmids pUC18, pUC19 and pUC118 (Vierira and Messing, 1982) were used as vectors for E.coli. LB medium (Sambrook et al., 1989) was used for the culture of E.blattae and E.coli. These microorganisms were grown aerobically at 37°C. For the selection of E.coli transformants, 50 µg/ml ampicillin was added to the medium.
Cloning and sequencing of the EB-NSAP gene
All basic recombinant DNA procedures, such as isolation and purification of DNA, restriction enzyme digestion, ligation of DNA and bacteria transformation, were done as described by Sambrook et al. (1989). The E.blattae chromosomal DNA library was constructed by inserting partial Sau3A1 digested fragments of 4–8 kb into the BamHI site of pUC118. Escherichia coli JM109 transformants were grown on LB plates containing 50 µg/ml ampicillin and 1 mM IPTG for 16 h. About 1 ml of 0.1 M MES [2-(N-morpholino)-ethanesulfonic acid]–NaOH buffer (pH 6.0) containing 4 mM pNPP was poured onto the surface of the plates to visualize the colonies of the transformants with the acid phosphatase activity. Four of 8000 transformants were phosphatase positive. The shortest plasmid rescued from one candidate transformant, designated as pEAP300, was used for further study. The subcloning results showed that the phosphatase activity was retained on a 1.2 kb PstI–HincII fragment. The 1.2 kb fragment was subcloned into pUC19 and the resultant plasmid was designated as pEAP320.
The DNA was sequenced by the dideoxynucleotide chain termination method with the Dye Terminator Cycle sequencing kit (Perkin-Elmer, Norwalk, CT) and a DNA sequencer (model 373A, Perkin-Elmer). To generate shorter clones suitable for sequencing, exonuclease III deletions were prepared using a kilo-sequencing kit (Takara Shuzo, Kyoto, Japan).
The sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession No. AB020481.
Assay of phosphatase activity
The phosphatase activity was assayed by monitoring the rate of hydrolysis of pNPP. The reaction mixture contained 100 mM MES–NaOH buffer (pH 6.0), 10 mM pNPP, and the enzyme solution in a total volume of 1 ml. The reaction was incubated for 10 min at 30°C and then was stopped by adding 0.2 ml of 2 N KOH. The quantity of p-nitrophenol released was measured at 410 nm, and was calculated using a molar extinction coefficient of 18 300 M–1cm–1. One unit of phosphatase activity was defined as the amount of enzyme that produced 1 µmol of p-nitrophenol per minute. When other phosphate esters were used as substrates, the quantity of inorganic phosphate released from various phosphate esters was measured by the method of Drewes (1972).
The PPi-glucose phosphotransferase activity was assayed in a standard reaction mixture containing 100 µmol of sodium acetate buffer (pH 5.0), 40 µmol of glucose, 100 µmol of Na4P2O7·10H2O, and the enzyme solution in a total volume of 1 ml. The reaction was incubated for 10 min at 30°C and then the reaction was stopped by adding 0.2 ml of 1 N NaOH. The quantity of glucose-6-phosphate formed was measured enzymatically with glucose-6-phosphate dehydrogenase by a modification of the method of Noltman et al. (1961). One unit of phosphotransferase activity was defined as the amount of enzyme that produces 1 µmol of glucose-6-phosphate per minute under the assay conditions.
Enzyme purification
All operations were carried out at 4°C, and potassium phosphate buffer was used. A recombinant E.coli JM109 strain harboring pEAP320 was cultured and harvested by centrifugation. Washed cells (28 g from 8 l of culture broth) were suspended in 0.1 M potassium phosphate buffer (pH 7.0) and were disrupted with a Dyno-mill (WA Bachofen, Switzerland). Cell debris was removed by centrifugation. The crude extract was fractionated by salting out with ammonium sulfate to 30–60% saturation, followed by dialysis against 0.1 M potassium phosphate buffer (pH 6.0). The dialysate was purified by SP-Toyopearl column chromatography with a linear gradient of KCl (0–0.3 M) in 0.1 M potassium phosphate buffer (pH 6.0). The active fraction was further purified by Butyl-Toyopearl column chromatography with a linear gradient of ammonium sulfate (35–15%) in 0.1 M potassium phosphate buffer (pH 7.0). The active fraction was precipitated by adding 60% ammonium sulfate and was stored at 4°C.
To assess the purity of the purified EB-NSAP, SDS–PAGE was performed by the method of Laemmli (1970) and the proteins in the gels were stained with Coomassie Brilliant Blue R-250. The purified enzyme appeared as a single band on an SDS–PAGE gel.
The protein concentration was assayed by the method of Bradford (1976) using a dye reagent concentrate (Bio-Rad laboratories) with bovine serum albumin as the standard.
To sequence the N-terminal amino acids, the purified enzyme was covalently bound to Sequelon-AA and DITC membranes, and was then analyzed with an automatic protein sequencer 6625 Prosequencer (Milligen/Biosearch) using phenylthiohydantoin derivatives.
Crystallization and data collection
Crystallization was performed using the vapor diffusion method in hanging drops at 20°C. Crystals were grown from drops containing protein (10 mg/ml, 20 mM HEPES pH 8.0) and reservoir (43–45% PEG 400, 0.1 M Tris–HCl pH 8.0) in a 1:1 ratio. Hexagonal prisms appeared after a few days and grew to a maximum size of 0.3 × 0.3 × 1.2 mm within 10 days. The crystals suffered from severe radiation damage at room temperature, so they were flash-cooled at –173°C after being transferred to a cryoprotectant solution in which the PEG400 concentration was increased by 5%. The crystals belong to space group P6322 with unit cell dimensions a = b = 124.7 Å, c = 97.7 Å, and with one monomer in the asymmetric unit, giving a calculated value of 72% for the solvent content. The best native data set was collected from two crystals aligned differently at beamline 6B of the Synchrotron Radiation Source at the National Laboratory for High Energy Physics (Photon Factory), Tsukuba. Measured intensities were integrated using DENZO (Otwinowski and Minor, 1997) and were scaled with SCALA (CCP4, 1994) and AGROVATA (CCP4, 1994). Data collection statistics are summarized in Table I.
Table I. Data collection and phasing statistics.
| Data set | Native | Pt1 | Pt2a | Hg | MoO4 |
|---|---|---|---|---|---|
| Data collection | |||||
| device | PF BL-6B | R-AXIS IIC | PF BL-6B | R-AXIS IIC | PF BL-6B |
| no. of crystals | 2 | 1 | 1 | 1 | 2 |
| resolution | 1.9 | 2.7 | 2.5 | 2.7 | 2.4 |
| no. of unique reflections | 34 749 | 9993 | 14 622 | 11 604 | 32 057 |
| redundancy | 4.0 | 4.4 | 3.3 | 4.5 | 5.1 |
| completeness (%) | 92.7 | 80.1 | 91.4 | 92.7 | 89.7 |
| Rmergeb | 7.4 | 8.6 | 8.3 | 7.4 | 8.5 |
| MIR phasing (100–2.8 Å) | |||||
| heavy atom concentration | 1 mM K2PtCl4 | 1 mM K2PtCl4 | 5 mM K2HgI410 mM KI | ||
| soak time (d) | 4.5 | 4 | 5 | ||
| Risoc | 0.199 | 0.204 | 0.186 | ||
| no. of heavy atom sites | 1 | 1 | 5 | ||
| Rcullis (acentric/centric)d | 0.52/0.54 | 0.51/0.55 | 0.85/0.81 | ||
| phasing power (acentric/centric)e | 2.62/1.66 | 2.67/1.67 | 0.99/0.75 | ||
| mean figure of merit | 0.623 |
aAnomalous dispersion data included in the refinement.
bRmerge = Σ[Σi|Ii – <I>|/|Σ|<I>|] where Ii and <I> are the observed and the mean intensity of related reflections, respectively.
cRiso = Σ|FPH – FP|/Σ|FP| where FPH and FP are the structure factor amplitudes for derivative and native crystals, respectively.
dRcullis = Σ||FPH ± FP| – FH||/Σ|FPH – FP| where FH is the calculated structure factor amplitude due to the scattering by the heavy atoms.
ePhasing power = FH/E, the r.m.s. heavy atom structure factor amplitudes divided by the lack of closure error.
Phasing
The crystal structure was solved by the MIR method with anomalous scattering using two derivatives, K2PtCl4 and KHgI4-KI (Table I). Two derivative data sets were collected using a Rigaku R-AXIS IIc imaging-plate detector with a Rigaku rotating-anode generator. A second K2PtCl4 data set was collected at the Photon Factory to measure the anomalous differences more precisely, and was then processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997). The coordinates of the only platinum site from K2PtCl4 were readily determined from the difference Patterson map using RSPS (CCP4, 1994). The heavy atom parameters were refined and the phases were calculated by MLPHARE (CCP4, 1994). In the difference Fourier map produced using these phases, the five mercury sites from KHgI4-KI were located. After the heavy atom parameters of the two derivatives were refined together, the calculated MIR phases were improved by solvent flattening combined with histogram matching using DM (CCP4, 1994). The solvent-flattened map was good enough to allow most of the residues to be fitted into the density unambiguously.
Refinement and quality of the model
The initial model was built into a 2.8 Å map using QUANTA97, and was then refined with X_PLOR 3.851 (Brünger et al., 1987) using data to 1.9 Å. The electron densities for the first six residues at the N-terminus, residues 135–136 and the C-terminal residue remained unclear, presumably due to disorder. The final model consists of 222 residues out of a total of 231 residues, 236 water molecules and one sulfate ion, which we believe comes from the ammonium sulfate used to precipitate the protein for storage. The refinement statistics are summarized in Table II. The coordinates and structure factors have been deposited with the Protein Data Bank (ID code 1D2T).
Table II. Refinement statistics and quality of the model.
| Native | MoO4 complex | |
|---|---|---|
| Resolution range (Å) | 8–1.9 | 10–2.4 |
| Number of reflections | 32 601 | 30 944 |
| Number of atoms | 1939 | 5225 |
| R-factor (%) | 21.5 | 21.8 |
| Rfree (%)a | 25.7 | 28.0 |
| Average B | ||
| protein atoms (Å2) | 26.0 | 38.4 |
| water molecules (Å2) | 45.3 | 50.0 |
| Root mean square deviations | ||
| bond lengths (Å) | 0.017 | 0.012 |
| bond angles (°) | 2.988 | 1.585 |
| Ramachandran plot | ||
| most favored regionsb (%) | 92.7 | 84.6 |
| additionally allowed regionsb (%) | 7.3 | 14.9 |
| generously allowed regionsb (%) | 0.0 | 0.5 |
| disallowed regionsb (%) | 0.0 | 0.0 |
aR-factor based on 10% of the data excluded from refinement.
bResidue regions as defined by PROCHECK (Laskowski et al., 1993).
Crystal structure determination of the enzyme–molybdate complex
Crystallization was performed using the vapor diffusion method in sitting drops at 20°C. Crystals were grown from drops containing protein (10 mg/ml, 20 mM HEPES pH 8.0) and reservoir (40% PEG 400, 1 mM Na2MoO4, 0.1 M Tris–HCl pH 8.0) in a 1:1 ratio. Rhombic prisms appeared after a few days and grew to a maximum size of 0.3 × 0.3 × 0.3 mm within 1–2 weeks. The crystals belong to space group P3121 with unit cell dimensions a = b = 86.6 Å, c = 205.3Å, and with three monomers in the asymmetric unit, giving a calculated value of 59% for the solvent content. The best data set was collected at 15°C using the macromolecular-oriented Weissenberg camera (Sakabe, 1991) installed at beamline 6B of the Synchrotron Radiation Source at the National Laboratory for High Energy Physics (Photon Factory), Tsukuba. Two crystals whose c-axes were aligned either parallel or perpendicular to the rotation axis were used. Measured intensities were integrated using WEIS (Higashi, 1989), and were scaled with SCALA (CCP4, 1994) and AGROVATA (CCP4, 1994). Data collection statistics are summarized in Table I. The structure of the molybdate complex was solved by molecular replacement using the structure of the native enzyme as a search model. The model consisted of three monomers, which were correlated by a 3-fold axis to each other. The rotation and translation functions, which were solved with AMoRe (Navaza, 1994), gave a clear solution and confirmed that the space group was P3121 (translation functions in P3221 did not yield a better solution). Rigid-body refinement in the resolution range of 10–3 Å resulted in an R-factor of 0.424. Subsequent model building was carried out using QUANTA97 and all refinement was done in X_PLOR 3.851 (Brünger et al., 1987) using data to 2.4 Å. The final model for one of the three monomers in the asymmetric unit consists of 222 residues (Gly7–His228) and one MoO4 group. The asymmetric unit contains 125 water molecules. The electron density is continuous across the whole sequence, but no interpretable density was present for the first six residues at the N-terminus and the last three residues at the C-terminus of all monomers. Refinement statistics are summarized in Table II. The coordinates and structure factors have been deposited with the Protein Data Bank (ID code 1EOI).
Acknowledgments
Acknowledgements
We thank Tom James (UCSF) for reading the manuscript and Hisashi Kawasaki (Ajinomoto) for helpful discussions. This study was partially supported by the Sakabe project of the Tsukuba Advanced Research Alliance.
References
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Kuriyan,J. and Karplus,M. (1987) Crystallographic R-factor refinement by molecular dynamics. Science, 235, 458–460. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Drewes P.A. (1972) Direct colorimetric determination of phosphorus in serum and urine. Clin. Chim. Acta, 39, 81–88. [DOI] [PubMed] [Google Scholar]
- Guddat L.W., McAlpine,A.S., Hume,D., Hamilton,S., de Jersey,J. and Martin,J.L. (1999) Crystal structure of mammalian purple acid phosphatase. Structure, 7, 757–767. [DOI] [PubMed] [Google Scholar]
- Hemrika W. and Wever,R. (1997) A new model for the membrane topology of glucose-6-phosphatase: the enzyme involved in von Gierke disease. FEBS Lett., 409, 317–319. [DOI] [PubMed] [Google Scholar]
- Hemrika W., Renirie,R., Dekker,H.L., Barnett,P. and Wever,R. (1997) From phosphatases to vanadium peroxidases: a similar architecture of the active site. Proc. Natl Acad. Sci. USA, 94, 2145–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T. (1989) The processing of diffraction data taken on a screenless Weissenberg camera for macromolecular crystallography. J. Appl. Crystallogr., 22, 9–18. [Google Scholar]
- Itoh N., Izumi,Y. and Yamada,H. (1987) Haloperoxidase-catalyzed halogenation of nitrogen-containing aromatic heterocycles represented by nucleic bases. Biochemistry, 26, 282–289. [Google Scholar]
- Kasahara M., Nakata,A. and Shinagawa,H. (1991) Molecular analysis of the Salmonella typhimurium phoN gene, which encodes nonspecific acid phosphatase. J. Bacteriol., 173, 6760–6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde T., Sträter,N., Fröhlich,R., Witzel,H. and Krebs,B. (1996) Mechanism of Fe(III)–Zn(II) purple acid phosphatases based on crystal structures. J. Mol. Biol., 259, 737–748. [DOI] [PubMed] [Google Scholar]
- Kostrewa D., Grüninger-Leitch,F., D’Arcy,A., Broger,C., Mitchell,D. and van Loon,A.P.G.M. (1997) Crystal structure of phytase from Aspergillus ficcum at 2.5 Å resolution. Nature Struct. Biol., 4, 185–190. [DOI] [PubMed] [Google Scholar]
- Kostrewa D., Wyss,M., D’Arcy,A. and van Loon,A.P.G.M. (1999) Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2.4 Å resolution. J. Mol. Biol., 288, 965–974. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1993) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 26, 283–290. [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–290. [Google Scholar]
- Lei K., Pan,C., Liu,J., Shelly,L.L. and Chou,J.Y. (1995) Structure–function analysis of human glucose-6-phosphatase, the enzyme deficient in glycogen storage disease type 1a. J. Biol. Chem., 270, 11882–11886. [DOI] [PubMed] [Google Scholar]
- Lindqvist Y., Schneider,G. and Vihko,P. (1993) Three-dimensional structure of rat acid phosphatase in complex with L(+)-tartrate. J. Biol. Chem., 268, 20744–20746. [PubMed] [Google Scholar]
- Lindqvist Y., Schneider,G. and Vihko,P. (1994) Crystal structures of rat acid phosphatase complexed with the transition-state analogs vanadate and molybdate: implications for the reaction mechanism. Eur. J. Biochem., 221, 139–142. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Messerschmidt A. and Wever,R. (1996) X-ray structure of a vanadium-containing enzyme: chloroperoxidase from the fungus Curvularia inaequalis.Proc. Natl Acad. Sci. USA, 93, 392–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt A. and Wever,R. (1998) X-ray structures of apo and tungstate derivatives of vanadium chloroperoxidase from the fungus Curvularia inaequalis.Inorg. Chim. Acta, 273, 160–166. [Google Scholar]
- Navaza J. (1994) AMoRe: an automated procedure for macromolecular replacement. Acta. Crystallogr. A, 50, 157–163. [Google Scholar]
- Neuwald A.F. (1997) An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci., 6, 1764–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B. (1991) Protein folding and association: insight from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Noltmann E.A., Gubbler,C.J. and Kuby,S.A. (1961) Glucose 6-phosphate dehydrogenase (Zwischenferment). J. Biol. Chem., 236, 1225–1230. [PubMed] [Google Scholar]
- Nordlie R.C. (1971) Glucose-6-phosphatase, hydrolytic and synthetic activities. In Boyer,P.D. (ed.), The Enzymes Volume IV Hydrolysis. 3rd edn. Academic Press, New York, NY, pp. 543–601. [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pan C., Lei,K., Annabi,B., Hemrika,W. and Chou,J.Y. (1998) Transmembrane topology of glucose-6-phosphatase. J. Biol. Chem., 273, 6144–6148. [DOI] [PubMed] [Google Scholar]
- Pond J.L., Eddy,C.K., Mackenzie,K.F., Conway,T., Borecky,D.J. and Ingram,L.O. (1989) Cloning, sequencing and characterization of the principal acid phosphatase, the phoC+ product, from Zymomonas mobilis.J. Bacteriol., 171, 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe N. (1991) X-ray diffraction data collection system for modern protein crystallography with a Weissenberg camera and an imaging plate using synchrotron radiation. Nucl. Instrum. Methods Phys. Res. A, 303, 448–463. [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schneider G., Lindqvist,Y. and Vihko,P. (1993) Three-dimensional structure of rat acid phosphatase. EMBO J., 12, 2609–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträter N., Klabunde,T., Tucker,P., Witzel,H. and Krebs,B. (1995) Crystal structure of a purple acid phosphatase containing a dinuclear Fe(III)–Zn(II) active site. Science, 268, 1489–1492. [DOI] [PubMed] [Google Scholar]
- Stukey J. and Carman,G.M. (1997) Identification of a novel phosphatase sequence motif. Protein Sci., 6, 469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaller M.C., Berlutti,F., Schippa,S., Lombardi,G. and Rossolini,G.M. (1994) Characterization and sequence of PhoC, the principal phosphate-irrepressible acid phosphatase of Morganella morganii.Microbiology, 140, 1341–1350. [DOI] [PubMed] [Google Scholar]
- Thaller M.C., Schippa,S. and Rossolini,G.M. (1998) Conserved sequence motifs among bacterial, eukaryotic and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci., 7, 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiya K., Tohsuji,M., Nikai,T., Sugihara,H. and Sasakawa,C. (1996) Identification and characterization of phoN-Sf, a gene on the large plasmid of Shigella flexneri 2a encoding a nonspecific phosphatase. J. Bacteriol., 178, 4548–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierira J. and Messing,J. (1982) The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene, 19, 259–268. [DOI] [PubMed] [Google Scholar]
- Vincent J.B., Crowder M.W. and Averill,B.A. (1992) Hydrolysis of phosphate monoesters: a biological problem with multiple chemical solutions. Trends Biochem. Sci., 17, 105–110. [DOI] [PubMed] [Google Scholar]