Abstract
The pleiotropic effects of active retinoids are transduced by their cognate nuclear receptors, retinoid X receptors (RXRs) and retinoic acid receptors (RARs), which act as transcriptional regulators activated by two stereoisomers of retinoic acid (RA): 9-cis RA (9-cRA) and all-trans RA (a-tRA). Among nuclear receptors, RXR occupies a central position and plays a crucial role in many intracellular signalling pathways as a ubiquitous heterodimerization partner with numerous other members of this superfamily. Whereas RARs bind both isomers, RXRs exclusively bind 9-cRA. The crystal structure of the ligand-binding domain (LBD) of human RXRα bound to 9-cRA reveals the molecular basis of this ligand selectivity and allows a comparison of both apo and holo forms of the same nuclear receptor. In the crystal, the receptor is monomeric and exhibits a canonical agonist conformation without direct contacts between the ligand and the transactivation helix H12. Comparison with the unliganded RXRα LBD structure reveals the molecular mechanisms of ligand-induced conformational changes and allows us to describe at the atomic level how these changes generate the proper protein interface involved in nuclear receptor–coactivator interaction.
Keywords: conformational change/crystal structure/nuclear receptors/retinoic acid/RXR
Introduction
Members of the nuclear receptor (NR) superfamily exhibit a modular architecture that notably includes two highly conserved functional domains: a DNA-binding domain that specifically binds to cognate cis-acting DNA response elements present in target genes, and a ligand-binding domain (LBD) that contains the ligand-dependent transcriptional activation function AF-2 (Gronemeyer and Laudet, 1995). Our initial crystallographic studies on retinoic acid (RA) NRs (Bourguet et al., 1995; Renaud et al., 1995; Wurtz et al., 1996) have suggested that NR LBDs, upon binding of cognate agonistic ligands, could undergo an apo- to holo-LBD conformational transition, the so-called transconformation (Moras and Gronemeyer, 1998), resulting in a conformational change that promotes their interaction with transcriptional coactivators and components of the general transcription machinery and at the same time dissociation from corepressors (Torchia et al., 1998). RAs, the active retinoid derivatives of vitamin A, regulate complex gene networks involved in morphogenesis, organogenesis, growth, cellular differentiation and homeostasis (Mangelsdorf et al., 1994). Two families of NRs, the retinoic acid receptors (RARs) and the retinoid X receptors (RXRs), are implicated in the transduction of RA signals. RARs can be activated by either all-trans (a-tRA) or 9-cis stereoisomers (9-cRA), while RXRs are exclusively activated by their 9-cis isomer. The various RAR (RARα, β and γ) and RXR (RXRα, β and γ) isotypes are encoded by different genes, while their multiple isoforms, which differ in their N-terminal region, are the results of differential promoter usage and alternative splicing (Leid et al., 1992; Chambon, 1996). Studies performed in in vitro cell cultures, and in vivo genetic studies in mouse, have led to the conclusion that RXR–RAR heterodimers are the functional units transducing the retinoid signal (Kastner et al., 1995; Botling et al., 1997; Minucci et al., 1997). Furthermore, it has been shown that RXRs play a unique role among NRs as they are not only able to heterodimerize with RARs, but also with a number of other members of the NR superfamily (Mangelsdorf et al., 1995).
Our comparative study with the two first crystal structures, those of the RXRα apo-LBD (Bourguet et al., 1995) and the RARγ holo-LBD (Renaud et al., 1995), suggested that ligand-induced transconformations of NR LBDs may generate the surface interacting with the transcriptional coactivators (Torchia et al., 1998) that mediate the AF-2 activation function to the chromatin template and/or transcription machinery. We proposed a ‘mouse trap’ mechanism that most notably involves a major repositioning of the ‘transactivation helix’ H12 that corresponds to the core of the AF-2 activation domain (AF-2 AD) (Wurtz et al., 1996; Moras and Gronemeyer, 1998). This model was subsequently validated by the determination of the crystal structures of the holo-LBDs of a number of additional NRs. However, and even though it is widely accepted, this transconformation model still relies on the assumption that the holo-LBD structure of RXRα is similar to those of RARγ holo-LBD and other NR holo-LBDs.
We now report the crystal structure of 9-cRA-liganded RXRα at 2.5 Å resolution, thus allowing the comparison of the apo and holo states of the same receptor LBD. This comparison supports our conformational switch model, and furthermore reveals how RXR discriminates between the a-tRA and 9-cRA stereoisomers.
Results
Overall structure of the RXRα holo-LBD
The crystal structure of the complex between the human RXRα1 (hRXRα) LBD and its natural agonist ligand 9-cRA (holo-LBD) has been determined by molecular replacement (Table I). The asymmetric unit contains two monomers, but none of the packing contacts is reminiscent of the dimer interface observed in the unliganded RXRα LBD (apo-LBD) homodimer (Bourguet et al., 1995). Crystallization conditions could explain the dissociation of the putative holo-LBD dimer seen in solution (solution studies using small angle scattering and analytical ultracentrifugation techniques will be published elsewhere), but in any event the absence of dimer in the crystalline state most probably reflects the weakening of the homodimeric interface upon ligand binding, thus favouring the formation of heterodimers.
Table I. Data collection and statistics of crystallographic data and refinement.
| Data sets | BM14 | ID14-EH2 | ID14-EH4 |
|---|---|---|---|
| Wavelength (Å) | 0.96 | 0.93 | 0.93 |
| Resolution (last shell) (Å) | 15–3.2 (3.3–3.2) | 40–2.8 (2.9–2.8) | 15–2.5 (2.6–2.5) |
| Total reflections | 46 568 | 71 272 | 87 788 |
| Unique reflections | 8871 | 14 441 | 22 453 |
| Redundancy | 5.2 | 4.9 | 3.9 |
| Completeness (last shell) (%) | 99.6 (99.2) | 96.8 (99.5) | 98.9 (95.5) |
| Rsym (last shell) (%) | 5.3 (25.4) | 7.4 (24.9) | 4.5 (19.4) |
|
I/σ (last shell) |
30.6 (6.5) |
18.2 (8.8) |
22.7 (5.3) |
| Refinement statistics | |||
| Reflections in working set (95% at 2σ) | 19 261 | ||
| Reflections in test set (5% at 2σ) | 981 | ||
| Rcrysta (%) | 22.5 | ||
| Rfreeb (%) | 25.7 | ||
| R.m.s.d. bonds (Å) | 0.011 | ||
| R.m.s.d. angles (°) | 1.50 | ||
| R.m.s.d. between monomers 1 and 2 (Å) | 0.046 | ||
| Monomers 1 and 2 | |||
| non-hydrogen protein atoms | 1766 and 1707 | ||
| non-hydrogen ligand atoms | 22 and 22 | ||
| solvent molecules | 127 and 78 | ||
| Average B factor (Å2) | |||
| non-hydrogen protein atoms | 47.2 and 57.9 | ||
| non-hydrogen ligand atoms | 41.7 and 48.2 | ||
| solvent molecules | 58.3 and 62.8 | ||
R.m.s.d is the root mean square deviation from ideal geometry.
aRsym = Σhkl Σi |Ihkl,i – <Ihkl,i>|/Σhkl Σi |Ihkl,i| where <Ihkl,i> is the average intensity of the multiple hkl,i observations for symmetry-related reflections.
bRcryst = Σ|Fobs – Fcalc|/Σ|Fobs|, where Fobs and Fcalc are observed and calculated structure factors. Rfree is calculated from a randomly chosen 5% of reflections (2σ) and Rcryst is calculated for the remaining 95% of reflections (2σ).
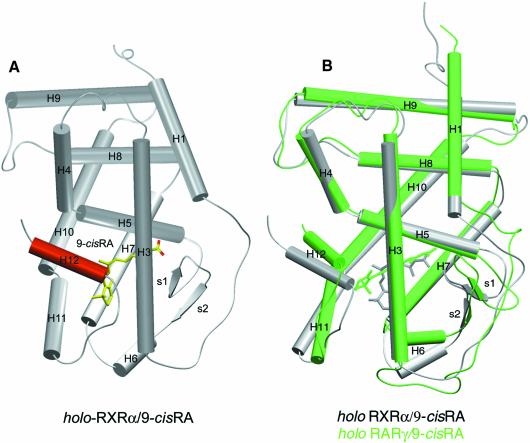
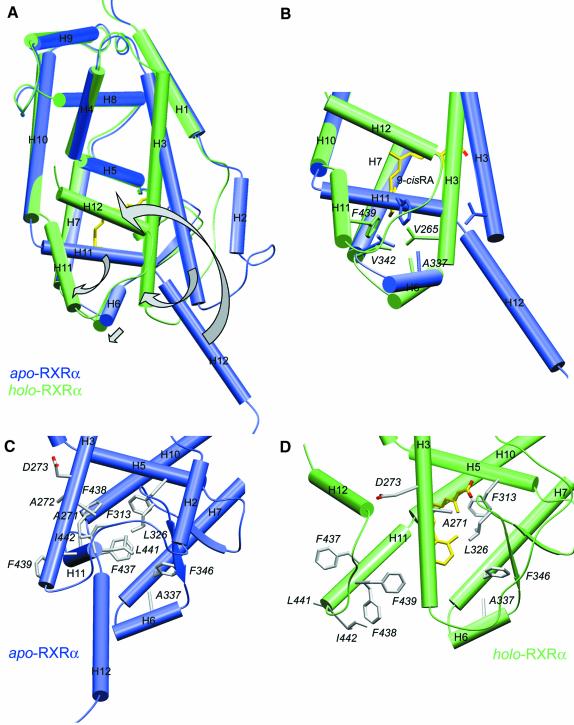
The more compact structure of the RXRα holo-LBD exhibits an ‘agonist conformation’ similar to that observed in RARγ and other holo-LBD structures (Figure 1A). When compared with the RXRα apo structure (Figure 4A), two major differences are observed: (i) three helices, H3, H11 and the transactivation helix H12, undergo large conformational changes; and (ii) helix H2 of hRXRα apo-LBD is replaced by a disordered connecting peptide in an extended conformation that may be necessary to allow the N-terminal part of helix H3 to reach its new position.
Fig. 1. Overall structure of human RXRα LBD. (A) General view of RXRα holo-LBD. α-helices are represented as cylinders and β-sheets as arrows. The whole structure is coloured in grey except for the transactivation helix H12 containing the AF2 function, which is highlighted in red. The 9-cRA ligand molecule is depicted in yellow and red for carbon and oxygen atoms, respectively. (B) Superimposition of holo-LBD structures of 9-cRA-liganded RXRα and RARγ. Protein and its ligand are depicted in grey and green for RXRα and RARγ, respectively.
Fig. 4. The conformational changes induced by ligand binding in hRXRα. (A) Superimposition of unliganded (in blue) and liganded (in green) hRXRα monomers showing the main conformational differences affecting helices H3, H6, H11, H12 and the connecting helix H2. The arrows show the main structural changes affecting helices H3, H6, H11 and H12 upon ligand binding. The ligand is depicted in yellow and red for carbon and oxygen atoms, respectively. (B) Superimposition of helices H3, H6, H7, H11 and H12 regions between the apo (in blue) and holo (in green) forms of hRXRα. For the sake of clarity other regions have been omitted. The arrows in (A) emphasize the conformational changes occurring upon ligand binding, showing the binding site closure at the level of the β-ionone ring of the ligand molecule. Residues Val265 (H3), Ala337 (H6), Val342 (H7) and Phe439 (H11) involved in binding site closure are labelled. The ligand is depicted in yellow and red for carbon and oxygen atoms, respectively. (C and D) Comparison of the holo (in green) versus apo (in blue) hRXRα H3, H6, H11 and H12 regions. The same orientation is used in both views. In the apo form residues from helix H11 occupy the binding pocket. The ligand is depicted in yellow and red for carbon and oxygen atoms, respectively.
The 9-cRA binding pocket of RXRα
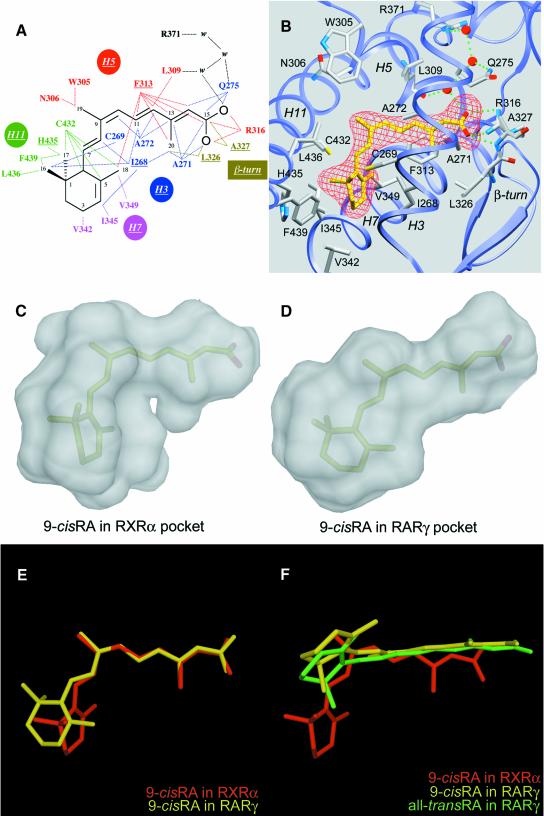
9-cRA is buried in an essentially hydrophobic pocket formed by residues located on helices H3, H5, H7 and H11, and the β-turn (Figure 2A and B). These residues are conserved in all three RXR (α, β and γ) isotypes, thus leaving little hope of finding α, β or γ isotype-specific ligands. The elongated pocket is sealed by Arg316 of helix H5 on one side and by the transactivation helix H12 on the other side. However, in contrast to the situation in RARγ LBD bound to either 9-cRA or a-tRA, the ligand does not interact with H12 and the H11–H12 loop. A similar observation was made in the case of oestrogen receptor α (ERα) bound to oestradiol (Brzozowski et al., 1997). The carboxylate group of 9-cRA is tightly sandwiched between the side chains of residues Phe313, Ala271 and Ala272, which form a hydrophobic tunnel. Phe313 plays a key role by interacting with isoprenic carbons (C11–C14), carbon C15 of the carboxylate group and methyl groups at positions 18 and 20. Ala271 and Ala272 are in van der Waals contact with C20 and the C11 and C12 isoprenic carbon atoms, respectively. The ionic interaction between the carboxylate group and Arg316 is similar to that seen in the a-tRA and 9-cRA RARγ holo-LBD structures. The carboxylate group participates in a water-mediated hydrogen bond network involving the backbone carbonyl group of Leu309 and the side chains of Gln275 and Arg371. Furthermore, it forms a hydrogen bond with the backbone amide group of the β-turn residue Ala327. This latter interaction is also observed in the RARγ holo-LBD structure. The β-sheet establishes van der Waals bonds through Leu326 and Ala327 that contact the C20 methyl group and C15, respectively. At the other side of the pocket, residue Cys432 makes contacts with the C6 β-ionone ring carbon and the isoprenic C7 carbon, and residue His435 interacts with the methyl group at position 17. The β-ionone ring carbons C3 and C4 interact with residues Val342 and Ile345, respectively, and the methyl group at position 16 interacts with residue Cys269. Val349 makes a weak contact with the C18 methyl group. Near the kink of the ligand, Ile268 contacts both methyl groups 16 and 18 as well as the β-ionone carbons C3 and C4. The closest neighbours of methyl group 19 are residues Trp305 and Asn306.
Fig. 2. The bound 9-cRA in RXRα and the comparison of the 9-cis retinoic binding modes in RXRα and RARγ. (A) Schematic drawing showing the interactions between the protein and the ligand molecule. Only contacts closer than 4.2 Å are indicated as dotted lines. The H3, H5, H7, H11 and β-turn residues are indicated. (B) The ligand molecule shown in an Fo – Fc electron density omit map contoured at 2.0 SD. Water molecules are displayed as red spheres. Only residues closer than 4.2 Å are displayed. Direct and water-mediated hydrogen bond implicated residues are shown with green dotted lines. The same orientation is shown in both pictures. (C and D) The probe-occupied ligand cavities in RXRα (C) and RARγ (D) calculated by VOIDOO (Kleywegt and Jones, 1994) and MSMS with a probe radius of 1.4 Å are displayed in white using DINO (Philippsen, 1999). The occupation of both cavities by the 9-cRA ligand molecule is shown by transparency. Ligand atoms of 9-cRA are displayed in pale green and pink for carbon and oxygen atoms, respectively. (E) Superimposition of 9-cRA molecules from holo hRXRα (yellow) and hRARγ (red). (F) Relative orientations of 9-cRA and a-tRA molecules after superimposition of liganded hRXRα and hRARγ proteins. 9-cRA from liganded hRXRα is coloured in red, 9-cRA from liganded hRARγ is coloured in yellow and a-tRA molecule from liganded hRARγ is coloured in green.
The accessible volume of the cavity in the RXRα LBD is 489 Å3. Since 9-cRA has a volume of 291 Å3, the ligand occupies only 59% of the cavity, which can be compared to values of 66, 63, 67 and 56% calculated for a-tRA, oestradiol (Brzozowski et al., 1997), progesterone (Williams and Sigler, 1998) and vitamin D3 (Rochel et al., 2000) in their respective LBDs. Unoccupied volume is visible at two places: near the C19 methyl group close to Trp305 and Asn306, and in the vicinity of the C18 methyl group close to Val349 (Figure 2C). These pockets could easily accommodate substituent side groups for synthetic analogues of 9-cRA such as SR11237, thus offering interesting possibilities for the design of new, more specific ligands (Vivat et al., 1997; Peet et al., 1998). The size of such substituents is critical, however; for example, some synthetic RXR antagonists display bulky groups at the level of the C19 methyl group of 9-cRA. Their antagonistic effect may result from the displacement of residue Trp305 and the subsequent disruption of the hydrophobic interactions that stabilize the agonist position of helix H12.
Discussion
Ligand-binding specificity of RARs
RXR exclusively binds 9-cRA, whereas RAR binds both 9-cRA and a-tRA stereoisomers (Heyman et al., 1992; Levin et al., 1992). The comparison of the 9-cRA molecules as seen in the structures of RXRα holo-LBD and RARγ holo-LBD reveals two major differences. (i) A more pronounced bending angle in RXRα (70 versus 60° in RARγ) (Figure 2E). The intrinsic geometry of the RXRα ligand-binding pocket (LBP) selects the 9-cis isomer because the sharper bend imposed by the induced fit cannot be reached by the all-trans isomer due to its flexure limit (Klaholz et al., 1998). The slightly lower affinity of RXRα for 9-cRA (KdRXR9c = 1.5 nM) compared with RARγ (KdRAR9c = 0.8 nM and KdRARat = 0.2 nM) (Allegretto et al., 1993; Allenby et al., 1993) could be explained by a smaller number of hydrophobic contacts (67 for RXRα–9-cRA versus 83 and 94 for RARγ–9-cRA and RARγ–a-tRA, respectively, with a 4.2 Å distance cut-off), which reflects the lower occupancy of the cavity in RXRα compared with that of RARγ (Figure 2C and D). (ii) A different relative orientation of the β-ionone ring with a rotation of ∼90° around the C9–C10 bond in RXRα (Figure 2F). This is the consequence of the different orientation and location of the β-ionone binding sites, which are 9 Å apart in the proteins. In RARγ holo-LBD, the β-ionone ring points towards helix H12 and makes hydrophobic contacts with it, whereas in RXRα it points to the bottom of the LBD, away from helix H12 (Figure 1A).
The superimposition of the two complexes (Figure 1B) reveals an additional difference in the relative position of the bound ligands. While the carboxylate group of all bound retinoids is anchored by an interaction with the conserved arginine residue of helix H5, the isoprenic chain being similarly oriented, a 2.7 Å shift of the ligand towards the centre of the cavity is observed in RXRα when compared with the 9-cRA–RARγ complex (Figure 2F). The natural flexibility of the arginine side chain allows such movement as would permit the binding of a longer ligand by sliding along the tunnel (Figure 2B).
The effects of point mutations on hRXRα residues Phe313 and Leu436 revealed that their substitutions generate two classes of RXR proteins with altered ligand specificities and responsiveness: a first class exhibiting decreased activation by 9-cRA and at the same time an increased activation by synthetic ligands, and a second class still responsive to 9-cRA, but insensitive to synthetic ligands (Peet et al., 1998). Mutation of the corresponding conserved leucine residue (Leu525) in ERα also strongly affects the transcriptional potency of synthetic ligands (Ekena et al., 1997). The present structure accounts for their roles in RXRα ligand specificity and activation by 9-cRA: Phe313 at one entry of the binding pocket is in very close contact with the carboxylate end of the ligand, whereas Leu436 is part of the cavity’s lid and participates to stabilize helix H12 in the agonist position.
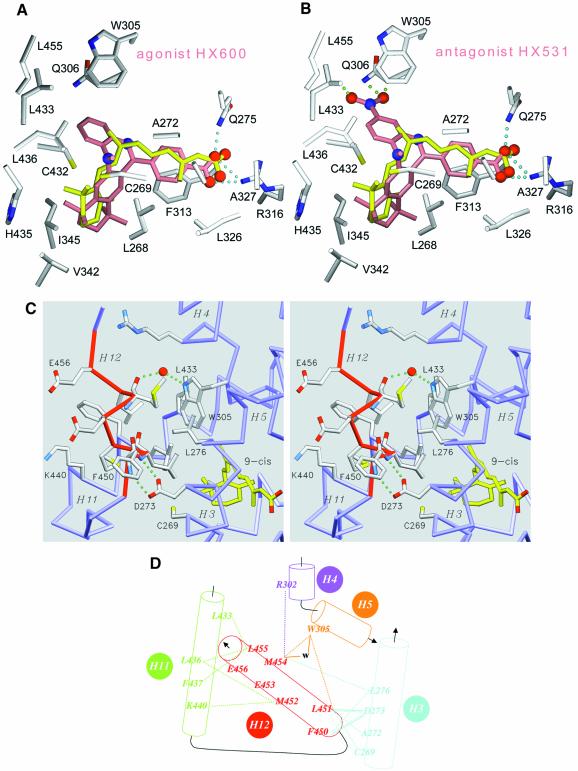
Modelling studies were performed considering synthetic compounds with known agonistic or antagonistic activity towards RXR. Agonist HX600 and antagonist HX531 (Umemiya et al., 1997a, b) were docked in the RXRα LBP (Figure 3A and B). All these compounds carry a carboxylate moiety that establishes a similar set of anchoring ionic and hydrogen interactions to those observed in the RXRα LBD bound to 9-cRA (Figure 3A). Agonist HX600 occupies the LBP and maximizes the cavity occupancy ratio at the level of residues Leu433, Trp305 and Gln306. In contrast, for the antagonist HX531 the additional bulky NO2 group causes steric hindrance with the side chains of residues Gln306, Trp305 and Leu433 (Figure 3B); the latter two residues being involved in the stabilization of helix H12 agonist conformation (see below). Docking of HX531 in the model of RXR under its antagonist conformation as observed in the RXR–RAR heterodimer (Bourguet et al., 2000) also suggests the necessity of additional conformational adaptation of either the ligand or the protein.
Fig. 3. The agonist conformation of transactivation helix H12 in the holo form. Docking of retinoid agonist (HX630) and antagonist (HX531) in hRXRα LBP. (A and B) Agonist (HX600) and antagonist (HX531) compounds docked in the LBP of hRXRα. Protein atoms are coloured in grey for carbon, blue for nitrogen, red for oxygen and yellow for sulfur. The oxygen and nitrogen atoms of docked compounds are depicted as red and blue spheres, respectively. 9-cRA is coloured in yellow, whereas docked ligands are colored in salmon. Cyan dotted lines represent the structurally conserved hydrogen interaction between the carboxylic moiety of ligands and residues Arg316 and Gln275 of the protein. Green dotted lines underline steric clashes through close interatomic contacts between ligand and protein atoms (the distance between consecutive dots is 0.5 Å). (C) Detailed stereoview of helix H12 contacts showing the exposed glutamic residues Glu453 and Glu456 involved in transactivation and the interactions stabilizing helix H12 in its agonist position. Helix H12 is depicted in red. 9-cRA ligand atoms are coloured in yellow for carbon and red for oxygen, respectively. Protein atoms are coloured in grey for carbon, blue for nitrogen, red for oxygen and yellow for sulfur. The protein backbone is coloured in blue. A water molecule is drawn as a red sphere and hydrogen bonds are depicted as green dotted lines. For the sake of clarity only a few side chains are labelled. (D) Schematic drawing of interactions stabilizing H12 in its agonist conformation. van der Waals interactions and hydrogen bonds are represented as dotted and continuous lines, respectively. A water molecule is referred to as w.
The transactivation helix H12 adopts a canonical agonist conformation upon 9-cRA binding
In RXRα, holo-LBD helix H12 leans over the ligand-binding cavity and seals it. It is packed against helices H3, H4, H5 and H11, and held in place mainly by a set of tight hydrophobic interactions involving residues Leu451, Met424, Met454 and Leu455 of helix H12 and several residues of the core LBD such as Cys269, Ala272 and Leu276 (H3), Arg302 (H4), Trp305 (H5), Leu433, Leu436, Phe437 and Lys440 (H11) (Figure 3C and D). Two additional interactions exist: (i) a water-mediated hydrogen bond between the indole NH atom of Trp305 of helix H5 and the backbone carbonyl group of Met454 of helix H12; and (ii) two hydrogen bonds between the carboxylate of Asp273 of helix H3 and the backbone amide groups of Phe450 and Leu451 of helix H12. This last stabilizing interaction involving a conserved acidic amino acid on helix H3 and backbone amide groups of helix H12 is also present in both ERα and progesterone receptor (PR) but not in RARγ holo-LBDs. Unlike RARγ and vitamin D3 receptor (VDR) (Rochel et al., 2000) holo-LBDs, but like in thyroid hormone receptor α (TRα) holo-LBD (Wagner et al., 1995), the potential salt bridge between Arg302 of helix H4 and Glu453 of the conserved motif Glu453MetLeuGlu456 that constitutes the core of the AF-2 AD is not formed. In RXRα, Arg302 points towards the solvent and is involved in a crystal packing contact. Note that residues Cys269 and Ala272 of helix H3 and Trp305 of helix H5, which participate in the correct positioning and stabilization of helix H12, are also involved in ligand binding, suggesting an indirect influence of the ligand on the fine positioning of H12. Globally, in RARγ, ERα and PR, but also in TRα and peroxisome proliferator-activated receptor γ (PPARγ) LBDs, H12 makes similar contacts with the homologous residues that form the H12 interaction groove (data not shown), in keeping with a high structural homology of the agonist conformation of all NRs. In this position, H12 residues contribute, as in TRβ (Darimont et al., 1998), ERα (Shiau et al., 1998) and PPARγ (Nolte et al., 1998), to the formation of the hydrophobic cleft that constitutes the coactivator binding site. Note that the amino acid residues that constitute the H12 interaction groove are much less conserved than those forming the coactivator binding site; with the exception of Leu276 none of them belongs to the highly conserved NR signature motif (Wurtz et al., 1996).
Ligand-induced conformational changes: comparison between RXRα apo- and holo-LBD structures
The elucidation of the structure of the RXRα holo-LBD allows us to investigate the effects of ligand binding on receptor conformation. It supports the concept of a canonical LBD ‘agonist structure’ valid for all NRs (Figure 4A). The volumes of apo and holo monomers, 39 110 and 35 870 Å3, respectively, show the effective compaction resulting from ligand binding. The volume of 35 870 Å3 is close to those of 37 410 and 39 370 Å3 found for its homologues RARγ and ERα holo-LBDs (32 and 31% amino acid identity, 51 and 48% amino acid similarity). The main conformational changes affect the N-terminal part of helix H3 and helices H11 and H12. Helix H6 and the two-stranded β-sheet undergo small structural adaptations, while all other helices remain essentially unaffected.
The apo form exhibits an additional helix (H2) in the region connecting helix H1 and helix H3. In the holo form this segment unfolds and the flexible loop region sticks to the protein. This different conformation of loop H1–H3 probably illustrates the dynamics of this region, which may act as a molecular spring accompanying the movement of helix H3, which undergoes a very large conformational change from residues Asp263 to Thr278. This N-terminal part of H3 rotates by ∼90° around its helical axis, while kinking at the level of residue Thr278 to pack against the LBP. The resulting global shift of the N-terminal end of H3 is ∼13 Å. This H3 movement, which is permitted by the displacement of helix H11 from its apo position, accompanies the binding of the ligand in an induced fit mechanism. It has three effects: (i) it brings residues Ile268, Cys269, Ala271, Ala272 and Gln275 of H3 into contact with the ligand; (ii) it seals the binding pocket at the level of the β-ionone ring region by bringing four residues into close van der Waals contact: Val265, Ala337, Val342 and Phe439 from helices H3, H6, H7 and H11, respectively (Figure 4B); and (iii) it positions residues Asp273, Cys269, Ala272 and Leu276 to establish the proper set of helix H12-stabilizing van der Waals interactions.
The present structure confirms the crucial role of helix H11 (Bourguet et al., 1995; Wurtz et al., 1996; Vivat et al., 1997). In the absence of the ligand, it stabilizes the apo form by filling the pocket with hydrophobic residues. In the holo form, it is displaced by helix H3, moves away and rotates by 180° around its own axis, generating a proper binding site and helping the repositioning of helix H12 (Figure 4C and D). In both structures, helix H11 remains unchanged to position His435. While in the RXRα apo-LBD, helix H11 is kinked at position Leu436 and then extends to position Asp444, thereby filling the unoccupied binding pocket, in the RXRα holo-LBD, helix H11 adopts a regular α-helical conformation in the continuity of H10 until position Ile442. In RXRα apo-LBD, the LBP cavity is filled with several side chains from helix H11 (Leu441, Phe437 and Phe438) that occupy the β-ionone ring position, while two hydrophobic side chains from the same helix (Leu436 and Phe439) are exposed to the solvent. In the RXRα holo-LBD, 9-cRA occupies the binding pocket, and the side chains of Leu441, Phe437 and Phe438 are outside, exposed to the solvent, while the side chains of Leu436 and Phe439 are internalized and form the lower part of the binding pocket. In conclusion, H11 exposes one face towards the solvent in the apo state and the other face in the holo state. The movements of helix H11 emphasize the role of solvation versus desolvation processes in the structural transition. These entropic effects are expected to be general, as suggested by structural and mutational studies on the ER.
The most striking conformational change affects helix H12, which is completely repositioned upon ligand binding. In the apo form, H12 protrudes from the protein core and is exposed to the solvent, whereas in the holo form it rotates and folds back towards the LBP, thus inducing the compaction of the LBD. Comparison with the RXRα apo-LBD structure reveals that upon ligand binding some key residues of the AF-2 AD core, such as Phe450, Glu453 and Glu456 involved in coactivator interaction and transactivation are exposed to the solvent. In the RXRα apo structure, residue Glu453 is hydrogen bonded to the backbone carbonyl group of residue Pro461 and the side chain of residue Asn262 on loop H2–H3, whereas residue Phe450 is close to residue Pro264 on helix H3. Thus, ligand binding, acting first through the rearrangement of helix H3, induces the repositioning of helix H12 by expelling helix H11. It must be emphasized that the ligand plays an indirect role in the stabilization of H12 through a set of hydrophobic interactions that stabilize the compact core of the LBD (Figures 3A, B and 4B).
Conclusion
We have shown here that RXR, a unique ubiquitous heterodimeric partner for several NR signalling pathways, exhibits a canonical holo-LBD conformation. Like its heterodimerization partners, e.g. RAR, VDR, PPAR and TR, the liganded RXRα LBD crystallizes as a monomer. The present structure unequivocally supports the existence of a structural switch with a ligand-induced transition that triggers the repositioning of helix H12 in an ‘agonist conformation’ and generates an ‘activated’ LBD able to interact subsequently with coactivators. This agonistic conformation of the 9-cRA RXR complex raises the question of RXR subordination, that is why are agonist-liganded RXRs transcriptionally inactive in RXR–RAR heterodimers, unless RAR is itself liganded (Vivat et al., 1997; Dilworth et al., 1999). Further crystallographic studies may reveal the underlying mechanism. Our present results also show how ligand binding promotes protein rearrangements mainly through helix rotations and translations, and suggest that the correlated desolvation processes could be important for the establishment of proper protein–protein interactions.
The specificity of RXR for the 9-cRA stereoisomer results from a different shape and size of its LBP, as compared with that of RAR. The RXR LBP exhibits a more pronounced kink, which prevents binding to a-tRA, the binding site of the β-ionone moiety being rotated by ∼90° towards the bottom core of the protein. As a consequence, and similar to some steroid receptors (see above), the ligand does not contact helix H12. The structure shows that the size of the pocket could accommodate slightly bulkier ligands at the level of the C18 and C19 methyl groups of 9-cRA.
The comparison of RXRα apo- and holo-LBD structures suggests that the molecular mechanism that leads to the holo structure relies upon a dynamic equilibrium of the LBD between an apo form where the ligand binding pocket is partly filled by the hydrophobic end of H11 and a holo form where H11 extends its helical structure leaving the LBP unoccupied and accessible for ligand binding. The bending of H3 may favour the dynamics of the equilibrium. Upon ligand binding, through a path similar to that described for RARγ (Blondel et al., 1999), helix H11 would assume its stable straight conformation, while helix H3 clamps the ligand and locks it in. The subsequent reorganization of H12, which seals the LBP in the agonist conformation, is most likely to be entropy driven. Indeed, solvent exclusion seems to play an important role in the various steps. The essentially hydrophobic ligand pocket results from the concerted and cooperative motions of helices H3 and H11, which clamp the ligand. H3 rotates and bends back towards the domain core, bringing together residues essential for ligand binding and H12 positioning. H11 rotates by half a turn around its axis and is tilted away to open the cavity for ligand binding. This movement brings hydrophobic residues previously exposed to the solvent inside the pocket towards the ligand, and concomitantly exposes others that could be subsequently implied in contacts with coactivators.
Materials and methods
Protein expression and purification
The human RXRα LBD (residues Thr223–Thr462) was cloned as an N-terminal His6-tagged fusion protein in pET15b expression vector and overproduced in Escherichia coli BL21 (DE3) strain. Cells were grown in 2× LB medium and subsequently induced for 5–7 h with 0.8 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 20°C. To ensure full saturation of receptor, 9-cRA was added during all steps of purification. Because of ligand photosensitivity all further manipulations were carried out in dimmed light. The purification procedure included an affinity chromatography step on a cobalt chelating column followed by gel filtration to remove residual unliganded RXRα species. After tag removal by thrombin digestion, protein was further purified on a Q Sepharose column and finally by gel filtration. Purity and homogeneity of the RXRα–9-cRA complex were assessed by SDS and native PAGE, isoelectrofocusing, denaturant and native electrospray ionization mass spectrometry, dynamic light scattering and small angle X-ray or neutron scattering and analytical ultracentrifugation.
Crystallization
Crystals of the RXRα LBD–9-cRA complex were obtained at 22°C using the vapour diffusion technique in hanging and sitting drops. Crystals of 700 × 70 × 70 mm3 were grown in several successive rounds of micro and macro seeding. The reservoir solution contained 2–4.5 mol/l sodium formate, 50 mM Tris pH 7.5–8.5, 5–10% glycerol and 17–23% propane-1,2-diol. Crystals belong to the orthorhombic space group P212121 with unit cell parameters a = 66.1, b = 86.3, c = 90.6 Å and α = β = γ = 90°, and present two monomers per asymmetric unit and a solvent content of 51%.
Data collection, structure determination and refinement
Crystals were directly flash-frozen in liquid ethane using crystallization liquor as cryoprotectant. Three native data sets were collected on three different crystals on beamlines BM14, ID14-EH2 and ID14-EH4 at ESRF (Grenoble, France). Data were processed using DENZO and SCALEPACK programs (Otwinowski and Minor, 1996). The structure was solved by molecular replacement using as probes two models: the apo wild-type RXRα LBD monomer and the apo F313A mutant RXRα LBD monomer (Bourguet et al., 2000), in which helix H12, loop H1–H3, helix H11 and loop H11–H12 had been omitted. Data set 1, the first available, was used for molecular replacement calculations between 15 and 3.5 Å using the AMoRe program (Navaza, 1994; Navaza and Vernoslova, 1995). Both models gave an unambiguous solution with a correlation coefficient of 62.7% and an R-factor of 42.6% after rigid body refinement. The resulting map allowed the positioning of helix H12 and the ligand. Iterative cycles of refinement using rigid body refinement and torsion angle molecular dynamics at 4000 K in CNS (Brünger, 1998) with restraint NCS interspersed with model building in O6 (Jones et al., 1991) yielded the complete structure. Loop regions were omitted from restraint NCS calculations. Further refinements at 2.8 and 2.5 Å were subsequently carried out using the two other higher resolution data sets 2 and 3. Anisotropic scaling and a bulk solvent correction were used and individual B atomic factors were refined anisotropically. Solvent molecules were added using an Fo – Fc Fourier difference map contoured at 2.5σ before a last refinement step. The final model consists of residues 223–458, the ligand and 127 water molecules for monomer 1, and residues 229–458, the ligand and 78 water molecules for monomer 2. In both monomers the H1–H3 connecting regions encompassing residues 245–262 appeared poorly ordered. No clear sequence assignment could be made and only a discontinuous polyalanine segment could be built for monomer 1. The last four amino acids (459–462) are not visible. According to PROCHECK (Laskowski et al., 1993), 91.2% of all residues in the model are in the most allowed main chain torsion angle Ramachandran regions and 6.9% are in the additionally allowed regions.
Acknowledgments
Acknowledgements
We wish to express our gratitude to Drs H.Gronemeyer, W.Bourguet, J.-P.Renaud and G.Tocchini-Valentini for discussion and careful reading of the manuscript and to J.-M.Wurtz for synthetic compound docking. We are grateful to J.Fagart for precious help with the figures. We thank E.Mitchell, A.Thompson and S.MacSweeney for help with the synchrotron data collection on the ESRF beamlines BM14, ID14-EH2 and EH4. We are also grateful to N.Potier for the mass spectrometry measurements. This work was supported by funds from Bristol-Myers-Squibb, the INSERM, the CNRS and grants from the Ministère de La Recherche et de l’Enseignement Supérieur, the Fondation pour la Recherche Medicale and the Association pour la Recherche sur le Cancer (P.F.E.).
References
- Allegretto E.A. et al. (1993) Transactivation properties of retinoic acid and retinoid X receptor in mammalian cells and yeast. J. Biol. Chem., 268, 26625–26633. [PubMed] [Google Scholar]
- Allenby G. et al. (1993) Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acid. Proc. Natl Acad. Sci. USA, 90, 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel A., Renaud,J.-P., Fischer,S., Moras,D. and Karplus,M. (1999) Retinoic acid receptor: a simulation analysis of retinoic acid binding and the resulting conformational changes. J. Mol. Biol., 291, 101–115. [DOI] [PubMed] [Google Scholar]
- Botling J., Castro,D.S., Öberg,F., Nilsson,K. and Perlmann,T. (1997) Retinoic acid receptor/retinoid X receptor heterodimers can be activated through both subunits providing a basis for synergistic transactivation and cellular differentiation. J. Biol. Chem., 272, 9443–9449. [DOI] [PubMed] [Google Scholar]
- Bourguet W., Ruff,M., Chambon,P., Gronemeyer,H. and Moras,D. (1995) Crystal structure of the ligand binding domain of the human nuclear receptor RXRα. Nature, 375, 377–382. [DOI] [PubMed] [Google Scholar]
- Bourguet W., Vivat,V., Wurtz,J.-M., Chambon,P., Gronemeyer,H. and Moras,D. (2000) Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding-domains. Mol. Cell, 5, 289–298. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1998) Crystallography and NMR system: a new software for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Brzozowski A.M. et al. (1997) Molecular basis of agonism and antagonism in the oestrogen receptor. Nature, 389, 753–758. [DOI] [PubMed] [Google Scholar]
- Chambon P. (1996) A decade of molecular biology of retinoic acid receptors. FASEB J., 10, 940–954. [PubMed] [Google Scholar]
- Darimont B.D., Wagner,R.L., Apriletti,J.W., Stallcup,M.R., Kushner,P.J., Fletterick,R.J. and Yamamoto,K.R. (1998) Structure and specificity of nuclear receptor–coactivator interactions. Genes Dev., 12, 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth D.J., Fromental-Ramain,C., Remboutsika,E., Benecke,A. and Chambon,P. (1999) Ligand dependent activation of transcription in vitro by retinoic acid receptor α/retinoid X receptor α heterodimers that mimics transactivation by retinoids in vivo. Proc. Natl Acad. Sci. USA, 96, 1995–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena K., Weis,K.E., Katzenellenbogen,J.A. and Katzenellenbogen,B.S. (1997) Different residues of the human estrogen receptor are involved in the recognition of structurally diverse estrogens and antiestrogens. J. Biol. Chem., 272, 5069–5075. [DOI] [PubMed] [Google Scholar]
- Gronemeyer H. and Laudet,V. (1995) Introduction. In Sheterline,P. (ed.), Protein Profile: Transcription Factors 3: Nuclear Receptors. Academic Press, Liverpool, UK, pp. 1173–1181. [PubMed] [Google Scholar]
- Heyman R.A., Mangelsdorf,D.J., Dyck,J.A., Stein,R.B., Eichele,G., Evans,R.M. and Thaller,C. (1992) 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell, 68, 397–406. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kastner P., Mark,M. and Chambon,P. (1995) Non-steroid nuclear receptors: What are genetic studies telling us about their role in real life? Cell, 83, 859–869. [DOI] [PubMed] [Google Scholar]
- Klaholz B.P., Renaud,J.-P., Mitschler,A., Zusi,C., Chambon,P., Gronemeyer,H. and Moras,D. (1998) Conformational adaptation of agonists to the human nuclear receptor RARγ. Nature Struct. Biol., 5, 199–202. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D, 50, 178–185. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structure coordinates. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leid M., Kastner,P. and Chambon,P. (1992) Multiplicity generates diversity in the retinoic acid signalling pathways Trends Biochem. Sci., 17, 427–433. [DOI] [PubMed] [Google Scholar]
- Levin A.A. et al. (1992) 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXRα. Nature, 355, 359–361. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D.J., Umesono,K. and Evans,R.M. (1994) The retinoids receptors. In Sporn,M.B., Roberts,A.B. and Goodman,D.S. (eds), The Retinoids: Biology, Chemistry and Medicine. Raven Press, New York, NY, pp. 319–349. [Google Scholar]
- Mangelsdorf D.J. et al. (1995) The nuclear receptor superfamily: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minucci S. et al. (1997) Retinoid X receptor RXR within the RXR retinoic acid receptor heterodimer binds its ligand and enhances retinoid dependent gene expression. Mol. Cell. Biol., 17, 644–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D. and Gronemeyer,H. (1998) The nuclear receptor ligand binding domain: structure and function. Curr. Opin. Cell Biol., 10, 384–391. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Navaza J. and Vernoslova,E. (1995) On the fast translation functions for molecular replacement. Acta Crystallogr. A, 51, 445–449. [Google Scholar]
- Nolte R.T. et al. (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature, 395, 137–143. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1996) Processing X-ray data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Peet D.J., Doyle,D.F., Corey,D.R. and Mangelsdorf,D.J. (1998) Engineering novel specificities for ligand-activated transcription in the nuclear hormone receptor RXR. Chem. Biol., 5, 13–21. [DOI] [PubMed] [Google Scholar]
- Philippsen A. (1999) DINO: visualizing structural biology. http://www.bioz.unibas.ch/~xray/dino
- Renaud J.-P., Rochel,N., Ruff,M., Vivat,V., Chambon,P., Gronemeyer,H. and Moras,D. (1995) Crystal structure of the RARγ ligand binding domain bound to all-trans retinoic acid. Nature, 378, 681–689. [DOI] [PubMed] [Google Scholar]
- Rochel N., Wurtz,J.-M., Mitschler,A., Klaholz,B. and Moras,D. (2000) The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol. Cell, 5, 173–179. [DOI] [PubMed] [Google Scholar]
- Shiau A.K., Barstad,D., Loria,P.M., Cheng,L., Kushner,P.J., Agard,D.A. and Greene,G.L. (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell, 95, 927–937. [DOI] [PubMed] [Google Scholar]
- Torchia J., Glass C. and Rosenfeld M.G. (1998) Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol., 10, 373–383. [DOI] [PubMed] [Google Scholar]
- Umemiya H. et al. (1997a) Action mechanism of retinoid-synergistic dibenzodiazepines. Biochem. Biophys. Res. Commun., 233, 121–125. [DOI] [PubMed] [Google Scholar]
- Umemiya H. et al. (1997b) Regulation of retinoidal actions by diazepinylbenzoic acids. Retinoid synergists which activate the RXR–RAR heterodimers. J. Med. Chem., 40, 4222–4234. [DOI] [PubMed] [Google Scholar]
- Vivat V. et al. (1997) A mutation mimicking ligand-induced conformational change yields a constitutive RXR that senses allosteric effects in heterodimers. EMBO J., 16, 5697–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R.L., Apriletti,J.W., McGrath,M.E., West,B.L., Baxter,J.D. and Fletterick,R.J. (1995) A structural role for hormone in the thyroid hormone receptor. Nature, 378, 690–697. [DOI] [PubMed] [Google Scholar]
- Williams S.P. and Sigler,P.B. (1998) Atomic structure of progesterone complexed with its receptor. Nature, 393, 392–396. [DOI] [PubMed] [Google Scholar]
- Wurtz J.-M., Bourguet,W., Renaud,J.-P., Vivat,V., Chambon,P., Moras,D. and Gronemeyer,H. (1996) A canonical structure for the ligand binding domain of nuclear receptors. Nature Struct. Biol., 3, 87–94. [DOI] [PubMed] [Google Scholar]