Abstract
To investigate the events leading to initiation of DNA replication in mammalian chromosomes, the time when hamster origin recognition complexes (ORCs) became functional was related to the time when Orc1, Orc2 and Mcm3 proteins became stably bound to hamster chromatin. Functional ORCs, defined as those able to initiate DNA replication, were absent during mitosis and early G1 phase, and reappeared as cells progressed through G1 phase. Immunoblotting analysis revealed that hamster Orc1 and Orc2 proteins were present in nuclei at equivalent concentrations throughout the cell cycle, but only Orc2 was stably bound to chromatin. Orc1 and Mcm3 were easily eluted from chromatin during mitosis and early G1 phase, but became stably bound during mid-G1 phase, concomitant with the appearance of a functional pre-replication complex at a hamster replication origin. Since hamster Orc proteins are closely related to their human and mouse homologs, the unexpected behavior of hamster Orc1 provides a novel mechanism in mammals for delaying assembly of pre-replication complexes until mitosis is complete and a nuclear structure has formed.
Keywords: DNA replication/initiation/origin recognition complex
Introduction
How do mammalian cells initiate DNA replication at specific sites along their chromosomes? During the past few years, it has become clear that most, if not all, of the proteins used to initiate DNA replication in the budding yeast, Saccharomyces cerevisiae, are also used to initiate DNA replication in many, perhaps all, eukaryotes. Homologs for many of these proteins have been identified in yeast, fungi, plants, nematodes, frogs, flies and mammals (a current list of the organisms encoding each protein and their literature citations can be found by searching the protein database at www.ncbi.nlm.nih.gov). They include the six Orc proteins that comprise the origin recognition complex (ORC), cell division cycle (Cdc) proteins 6 and 45, six mini-chromosome maintenance (Mcm) proteins and the Cdc7 protein kinase and its cofactor Dbf4. Several of these homologs are required for DNA replication. Orc2 and Dbf4 proteins are required in Drosophila (Landis et al., 1997; Landis and Tower, 1999). Orc, Cdc6, Mcm, Cdk2 and Cdk7/Dbf4 proteins are required in Xenopus (Carpenter et al., 1996; Coleman et al., 1996; Romanowski et al., 1996; Rowles et al., 1996; Walter and Newport, 1997; Hua and Newport, 1998). Mcm, Cdc6, Cdk2 and Cdc7/Dbf4 proteins are required in mammals (Todorov et al., 1994; Krude et al., 1997; Yan et al., 1998; Jiang et al., 1999a; Kumagai et al., 1999). Thus, the mechanism for DNA replication is highly conserved among eukaryotes.
As with yeast, initiation of DNA replication in the differentiated cells of frogs, flies and mammals also begins predominantly at specific replication origins (DePamphilis, 1999; Phi-van and Stratling, 1999). Initiation of bidirectional DNA replication at these sites depends on both proximal (Handeli et al., 1989; Aladjem et al., 1998; Malott and Leffak, 1999) and distal (Aladjem et al., 1995; Kalejta et al., 1998) cis-acting DNA sequences. In addition, the activity of individual replication origins can depend on nuclear structure, chromatin structure, the ratio of initiation factors to DNA, and DNA methylation [reviewed in DePamphilis (1999) and Rein et al. (1999)]. The combination of genetic and epigenetic parameters can account for the changes that can occur in the number and locations of initiation sites as rapidly cleaving embryos develop into differentiated organisms (Hyrien et al., 1995; Sasaki et al., 1999).
One difference that may exist between yeast and metazoan cells is the way in which the ORC interacts with chromatin, a step that is critical to understanding how initiation sites are selected. In yeast, both DNA footprinting (Diffley et al., 1994; Fujita et al., 1998) and immunoprecipitation (Liang and Stillman, 1997) analyses reveal that a complete ORC binds to yeast replication origins immediately after initiation of replication occurs and remains there throughout the cell division cycle. In metazoa, however, the situation is less clear. Some evidence suggests that the ORC dissociates from chromatin during mitosis. G1 phase nuclei from mammalian cells can initiate DNA replication when incubated in a Xenopus egg extract that has been depleted of Xenopus laevis Orc (XlOrc) proteins (Romanowski et al., 1996; Yu et al., 1998), whereas metaphase chromatin from mammalian cells replicates poorly under these conditions (Yu et al., 1998). These data suggest that mammalian Orc proteins are absent from chromatin during metaphase and then rebind at some time before G1 phase begins. Consistent with this conclusion, Orc proteins in activated Xenopus eggs bind to sperm chromatin, whereas Orc proteins in mitotic Xenopus eggs do not (Coleman et al., 1996; Hua and Newport, 1998; Findeisen et al., 1999; Rowles et al., 1999), and both Orc1 and Orc2 proteins are present on chromatin in cultured Xenopus cells during interphase but not during metaphase (Romanowski et al., 1996). However, in Drosophila, Orc2 is present in both interphase and metaphase (Pak et al., 1997), and human Orc2 is present on cells throughout the cell cycle, although metaphase cells per se were not examined (Ritzi et al., 1998). These data appear to contradict the work in Xenopus, suggesting that the behavior of ORC may vary even among metazoa. Therefore, it is difficult to relate the presence of Orc activity to the presence of Orc proteins in metazoa, because different assays for ORC function or Orc proteins have been carried out in different organisms. Moreover, the hypothesis that functional, chromatin-bound ORCs exist on mammalian chromatin throughout G1 phase is difficult to reconcile with the presence of an ‘origin decision point’ (ODP) (Wu and Gilbert, 1996) in mammalian cells; initiation of DNA replication can be induced by a Xenopus egg extract in all G1 nuclei, but initiation events in late G1 nuclei occur at the same replication origins used in vivo whereas initiation events in early G1 nuclei are randomly distributed along the genome (Wu and Gilbert, 1996; Yu et al., 1998; Li et al., 2000).
The work presented here provides a solution to this paradox by showing that the mammalian ORC disassembles, at least in part, during mitosis and then reassembles completely in early G1 nuclei. In Chinese hamster ovary (CHO) cells, CgOrc2 (where Cg indicates Cricetulus griseus, the Chinese hamster) remained stably bound to chromatin throughout the cell cycle, consistent with studies of Drosophila and human cells, but CgOrc1 became unstably bound to chromatin during mitosis, consistent with studies in Xenopus cells. Moreover, the ability to initiate DNA synthesis in hamster cells and the ability to activate specific mammalian replication origins selectively by incubation in Xenopus egg extract were directly related to the stability of CgOrc1 binding to chromatin. These results support the general concept that metazoan ORCs, in contrast to yeast ORCs, dissociate from chromatin during mitosis. In addition, they reveal that mammals selectively destabilize Orc1 protein during the mitotic to G1 transition, thus providing a novel mechanism in mammalian cells for delaying assembly of pre-replication complexes (pre-RCs), consisting of ORC, Cdc6 and one or more copies of Mcm proteins 2–7, until mitosis is complete and a nuclear membrane has reformed. Finally, they strongly suggest that mammalian ORCs are assembled at specific chromosomal sites.
Results
Functional hamster Orc proteins are absent during mitosis and early G1 phase
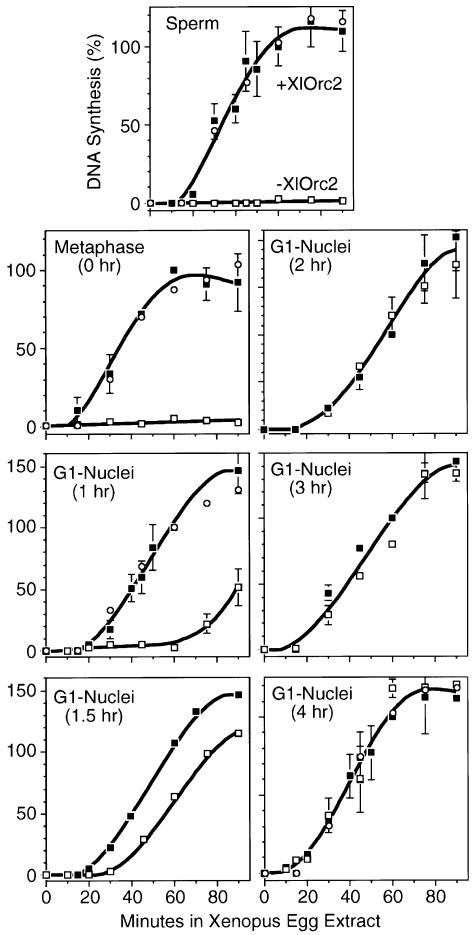
The presence of functional ORCs on mammalian chromatin was assayed by the ability of an Orc-depleted Xenopus egg extract to initiate DNA replication in hamster nuclei or metaphase chromatin. Previous studies have shown that antibodies against XlOrc1 or XlOrc2 co-precipitate other XlOrc proteins (Romanowski et al., 1996; Carpenter and Dunphy, 1998; Tugal et al., 1998), thus depleting egg extract of most, if not all, of their Orc proteins. In our hands, agarose beads coated with anti-XlOrc2 serum removed ≥98% of the XlOrc2 protein from the depleted extract, while <1% of the XlOrc2 protein was lost from the mock-depleted extract (Li et al., 2000). Depletion of XlOrc1 or XlOrc2 proteins from Xenopus egg extract prevents initiation of DNA replication in sperm chromatin or plasmid DNA substrates (Carpenter et al., 1996; Coleman et al., 1996; Romanowski et al., 1996; Rowles et al., 1996, 1999; Yu et al., 1998), demonstrating that initiation of DNA replication in Xenopus egg extract is dependent on a functional interaction between ORC and chromatin. Similarly, the XlOrc-depleted extracts used here did not support DNA replication in Xenopus sperm chromatin (Figure 1), confirming that XlOrc proteins were functionally, as well as physically, absent.
Fig. 1. Orc proteins were required to initiate DNA synthesis in hamster chromatin. Xenopus sperm chromatin (1.6 × 105) or nuclei prepared by digitonin lysis (25 000/µl) were tested for their ability to replicate when incubated in complete Xenopus egg extract (+XlOrc2, filled squares), XlOrc2-depleted extract (–XlOrc2, open squares) or mock-depleted extract (open circles) supplemented with [α-32P]dATP and [α-32P]dCTP. The amount of acid-precipitable 32P-labeled DNA was expressed as % DNA synthesis relative to the incorporation observed after 1 h of incubation in complete extract. The mean ± SEM is given for three or more independent experiments in complete or depleted extract. One experiment was performed with mock-depleted extract. The amount of DNA synthesized by 1 h (0.4 pmol dAMP × 10–6/nucleus) was equivalent to 10–15% of the genome replicated, comparable to previous studies (Gilbert et al., 1995; Wu et al., 1997).
Hamster cells were synchronized in mitosis and released into G1 phase; metaphase chromatin (0 h) or G1 phase nuclei (1–6 h) were then isolated by permeabilizing the cells with digitonin. When these substrates were incubated in either complete or mock-depleted Xenopus egg extract, the time courses for DNA synthesis (Figure 1) were typical of those reported previously (Gilbert et al., 1995). However, when the same substrates were incubated in XlOrc-depleted extract, a clear transition was observed from dependence to independence of DNA replication on XlOrc proteins (Figure 1). Hamster metaphase chromatin had the same dependence on XlOrc proteins as Xenopus sperm chromatin, and DNA synthesis was not detected during the first hour of incubation in nuclei isolated 1 h after metaphase. Hamster nuclei isolated 1.5 h after metaphase were able to function partially in the absence of Xenopus Orc proteins, while nuclei isolated ≥2 h after metaphase no longer required the presence of XlOrc proteins to initiate DNA synthesis. Therefore, hamster Orc proteins are not functional until 1–2 h after metaphase.
Acquisition of site-specific DNA replication in hamster nuclei coincides with independence from XlOrc proteins
Previous studies using Xenopus egg extract to initiate DNA replication in hamster G1 phase nuclei revealed that site-specific initiation did not appear until mid-G1 phase (the ODP). Before that point, DNA synthesis appeared ‘randomly’ distributed throughout the dihydrofolate reductase (DHFR) gene region. After that point, DNA synthesis was largely confined to the 55 kb intergenic region between the DHFR gene and the 2BE2121 gene as a broad peak of DNA synthesis centered at the ori-β/β′ locus. Therefore, we considered the possibility that the ODP coincided with the functional association of hamster Orc proteins with hamster chromatin.
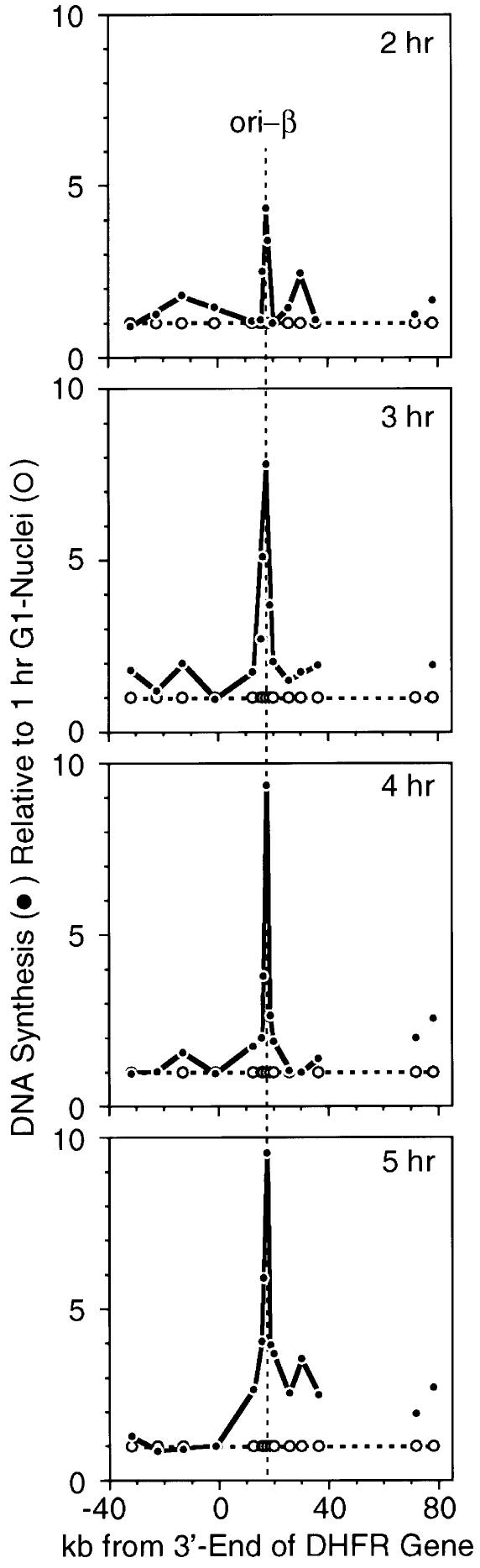
In an effort to confirm and refine the ODP, and to relate it to the results reported here, the ODP was mapped using a simplified experimental protocol that allowed Xenopus egg extract to initiate DNA replication selectively at the same primary initiation sites used by CHO cells in vivo, resulting in much sharper peaks of origin activity than previously reported (Li et al., 2000). Hamster G1 phase nuclei were incubated in Xenopus egg extract for 45 min to label the initial burst of newly synthesized DNA with [α-32P]dATP and [α-32P]dCTP, most of which consisted of RNA-primed DNA chains from 0.5 to ∼2 kb in length. Under these conditions, site-specific initiation events were not observed in 1 h G1 nuclei when DNA synthesis was dependent on the presence of XlOrc proteins (Figure 2). However, ori-β was selectively activated in G1 nuclei isolated ≥2 h after release from mitosis, and this activation, like total DNA synthesis (Figure 1), was independent of Xenopus Orc proteins (Li et al., 2000; data not shown). Therefore, the appearance of ori-β activity resulted from activation of hamster pre-RCs by Xenopus egg extract.
Fig. 2. The ability of Xenopus egg extract to activate ori-β selectively in hamster nuclei increases as hamster cells progress through G1 phase. Nuclei were isolated 1, 2, 3, 4 or 5 h after release of CHOC 400 cells from mitosis and incubated for 45 min in Xenopus egg extract supplemented with [α-32P]dATP and [α-32P]dCTP. The resulting 32P-labeled DNA was hybridized to specific DNA probes, and the results from each time point were then divided by the results from 1 h G1 nuclei. Thus, the data for 1 h G1 nuclei are represented by the horizontal dashed line (open circles), and the increase above this level of DNA synthesis at each subsequent time point by the solid line (filled circles).
To determine the time course for the appearance of hamster pre-RCs at ori-β, nuclei were isolated at various times after metaphase and incubated in Xenopus egg extract. The newly synthesized 32P-labeled DNA present after 45 min was then hybridized to specific DNA sequences, and the counts/base pair of 32P-labeled DNA that hybridized to each probe was divided by the corresponding value obtained with 1 h G1 nuclei (Figure 2). This ratio provided a quantitative assessment of the extent to which ori-β could be selectively activated as cells progressed through G1 phase. Pre-RCs were first detected at ori-β 1.5–2 h after metaphase, and their assembly within the cell population was complete 4–5 h after metaphase, or ∼2 h before the onset of S phase (Figure 3). This result was similar to that of Wu and Gilbert (1996) except that the ODP determined here occurred ∼1 h earlier.
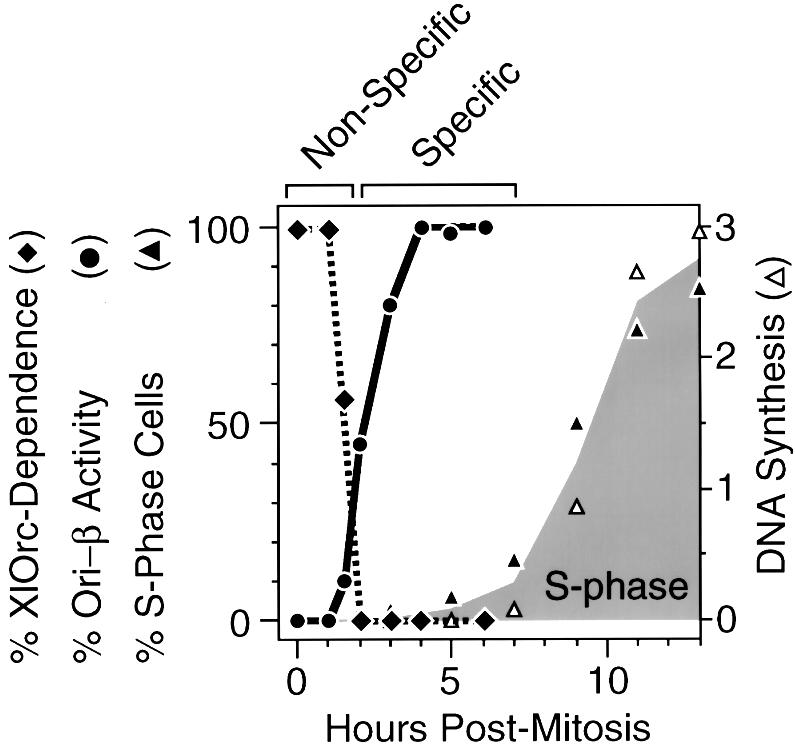
Fig. 3. Non-specific initiation of DNA replication in hamster G1 nuclei was dependent on Xenopus Orc proteins, but site-specific initiation (e.g. ori-β) was not. % XlOrc dependence (filled diamonds) is the % DNA synthesis observed after CHOC 400 nuclei had been incubated for 1 h in Xenopus egg extract (Figure 1). % Ori-β activity (filled circles) is the height of the peak at ori-β observed at the indicated time relative to the height observed 4 h after metaphase (Figure 2). % S-phase cells (filled triangles) is the percentage of cells labeled with BrdU in vivo. DNA synthesis (open triangles) is the amount of [3H]thymidine-labeled cells incorporated in vivo (104 c.p.m./105 cells).
The transition from XlOrc dependence to XlOrc independence of DNA replication in hamster chromatin coincided with the transition from non-specific (‘random’) to site-specific initiation of DNA replication in hamster chromatin (Figure 3). Before the ODP, DNA replication in hamster chromatin depended on the presence of XlOrc proteins and was initiated ‘randomly’ throughout the chromatin. After the ODP, DNA replication in hamster chromatin was independent of XlOrc proteins and occurred preferentially at specific sites such as ori-β. Therefore, the ability of Xenopus egg extract to activate selectively specific replication origins in hamster nuclei in vitro required prior assembly of hamster pre-RCs in vivo.
CgOrc1 and CgOrc2 levels are constant during mitosis and G1 phase
The results summarized in Figure 3 suggested that CgOrc proteins were not functionally associated with hamster chromatin until mid-G1 phase. To monitor the presence of these proteins, the CgORC1 and CgORC2 genes were cloned and expressed, and antibodies were produced against the expressed proteins. CgORC1 encodes an 850 amino acid polypeptide (CgOrc1) with a predicted mol. wt of 96 kDa. CgORC2 encodes a 576 amino acid polypeptide (CgOrc2) with a predicted mol. wt of 66 kDa. Hamster amino acid sequences are closely related to their human and mouse homologs (77% similar for Orc1; 88% similar for Orc2), particularly in their C-terminal ends [93% for Orc1 (CgOrc1 amino acids 484–850); 97% for Orc2 (CgOrc2 amino acids 297–576); Figure 4]. The antisera readily detected proteins of the expected molecular weights in hamster cells that co-migrated during gel electrophoresis with samples of the expressed proteins (Figure 5A).
Fig. 4. Comparison of the predicted amino acid sequences of mammalian Orc1 (A) and Orc2 (B) proteins. DDBJ/EMBL/GenBank identification numbers for Orc1 are: Chinese hamster (Cg), AF254572; mouse (Mm), 4034785; and human (Hs), 4758850; those for Orc2 are: Chinese hamster, AF254573; mouse, 2498710; and human, 5453830. Identical amino acids are in black. Similar amino acids are in gray.
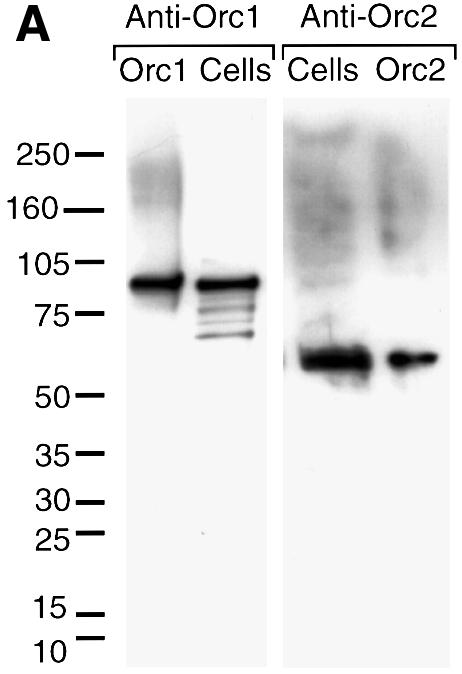
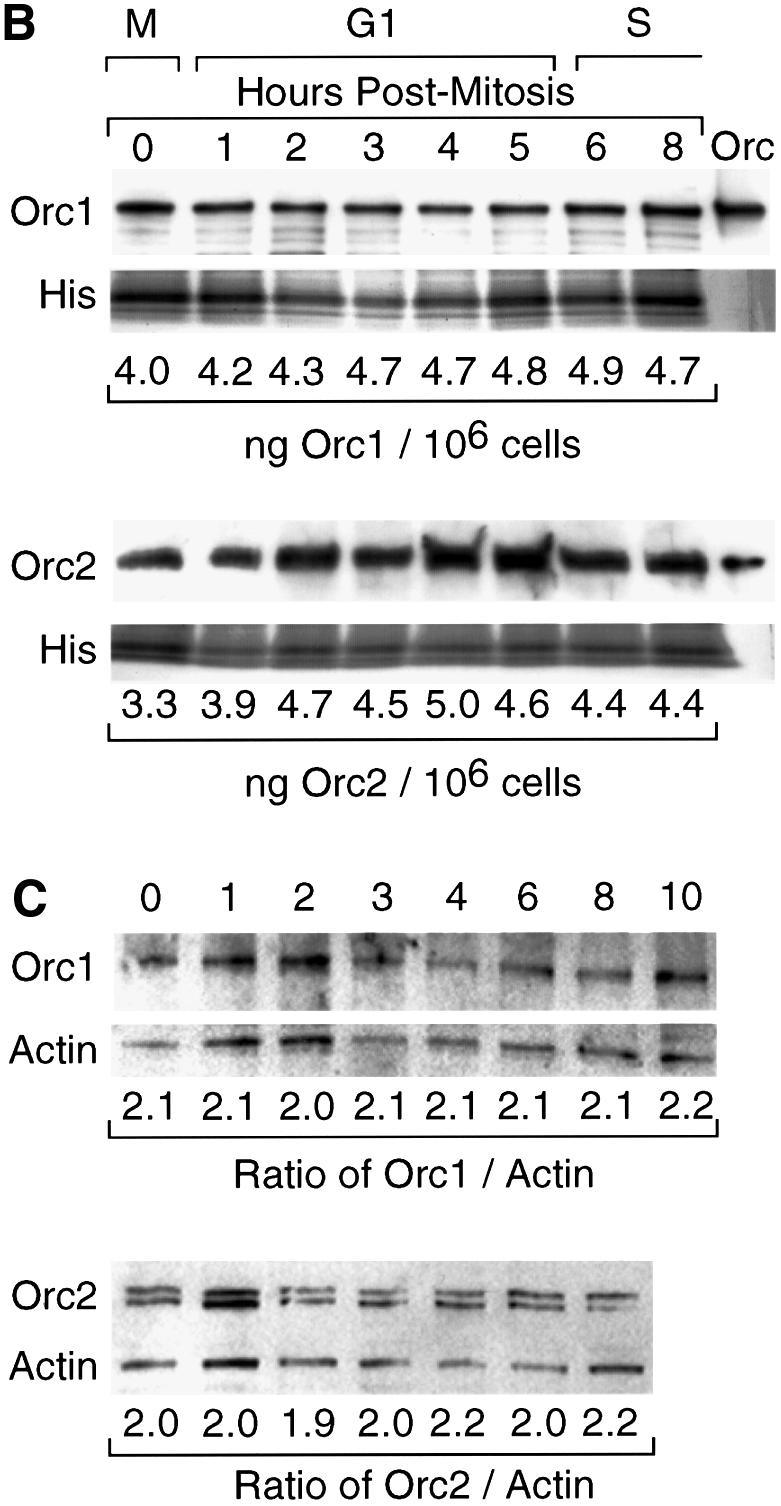
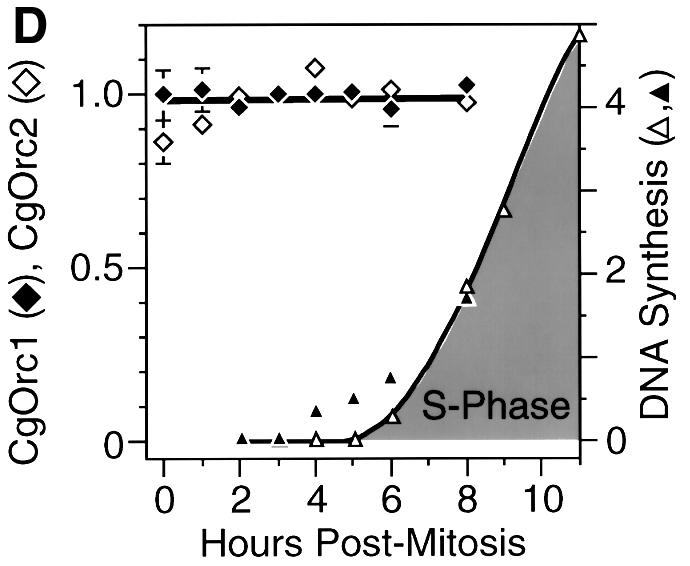
Fig. 5. Total CgOrc1 and CgOrc2 protein levels remained constant during mitosis and G1 phase. CHOC 400 cells were arrested in mitosis and then released into G1 phase. At the indicated time, cells were lysed with SDS. One aliquot was fractionated by electrophoresis in a 4–14% polyacrylamide gradient gel and assayed for Orc1 while another aliquot was fractionated and assayed for Orc2. (A) Antibody specificity was confirmed by immunoblotting samples of G1 phase cell extracts run in parallel with purified CgOrc1 or CgOrc2 protein and molecular weight markers. (B) Amounts of Orc1 and Orc2 at each time point were determined by subjecting the top portion of each gel to immunoblotting while the bottom portion of the gel was stained to detect histones. Individual bands were quantified by densitometry. Relative amounts of Orc protein were determined from the Orc:histone ratio in each lane. Total amounts of Orc protein were estimated from the intensities of samples of purified Orc proteins run in parallel. (C) At the indicated time, cells were permeabilized with digitonin and processed as above except that samples were fractionated in a 12% polyacrylamide gel and the entire gel was subjected to immunoblotting simultaneously with the indicated anti-CgOrc IgG and anti-actin IgM. (D) The mean ± SEM at each time point was determined for six experiments. DNA synthesis is the amount of [3H]thymidine-labeled cells incorporated in vivo (DNA synthesis, 103 c.p.m./105 cells, open triangles) and the fraction of cells labeled with BrdU in vivo (filled triangles, plotted on the left y-axis).
To quantify the relative concentrations of CgOrc1 and CgOrc2 during mitosis and G1 phase, hamster cells were isolated at different times after their release from metaphase and lysed with sodium dodecyl sulfate (SDS). Separate aliquots of the lysate were then fractionated by gel electrophoresis and subjected to immunoblotting with CgOrc1 or CgOrc2 antiserum. The bottom portion of each gel was stained separately to detect histones and the amount of Orc protein in each lane was normalized to the amount of histone present. Samples of purified CgOrc1 or CgOrc2 protein of known concentrations were included during the gel fractionation step in order to identify the correct protein and to generate a standard curve for quantification.
CgOrc1 and CgOrc2 were present in similar amounts (∼30 000 and ∼43 000 molecules/cell, respectively) throughout mitosis and G1 phase of the CHOC 400 cell cycle (Figure 5B) (CHOC 400 is a CHO cell line that contains ∼1000 tandemly integrated copies of the 273 kb DHFR gene amplicon). Measurements at other time points indicated that the levels of these proteins remained constant throughout the cell cycle (data not shown). To determine whether or not these proteins were present in the substrates that were used in the previous DNA replication assays (Figures 1 and 2), aliquots of nuclei prepared by digitonin lysis were assayed for CgOrc1 or CgOrc2 as described above (Figure 5C). The validity of using histones as an internal standard was confirmed by simultaneously immunoblotting α-actin. The higher percentage gel used in these assays revealed the presence of two CgOrc2 proteins, similar to what has been observed with Drosophila Orc2 (Huang et al., 1998). The results confirmed that the two hamster Orc proteins were present in similar amounts in nuclei prepared using digitonin, and in whole cells (Figure 5D). Taken together with the DNA replication assays, these data reveal that hamster Orc proteins are not functional during mitosis and early G1 phase.
CgOrc1 and CgOrc2 differ in their association with chromatin as cells transit from mitosis to G1 phase
To investigate whether the inability of Orc proteins to function during the M–G1 transition could result from an inability to bind stably to hamster chromatin, the amount of CgOrc1 and CgOrc2 that remained associated with nuclear pellets was determined by immunoblotting as described above except that cells were lysed with the non-ionic detergent Triton X-100, and then the nuclei were washed in a Triton X-100 buffered salts solution containing ATP. This procedure permeabilizes nuclei to large molecules and allows weakly bound proteins to be eluted from chromatin while stabilizing pre-RCs. Two different protocols, A and B, were tested (see Materials and methods) in case subtle differences affected the outcome. These protocols measure the amount of Orc bound to chromatin, because human Orc2 has been shown to be tightly bound to oligonucleosomes (Ritzi et al., 1998), and both CgOrc1 and CgOrc2 can be released from the pellet fraction by digestion with micrococcal nuclease (data not shown). CgOrc1 and CgOrc2 were quantified routinely in separate aliquots of the same lysate.
The amount of CgOrc2 stably bound to chromatin during mitosis and G1 phase was constant at ∼2.6 ng/106 cells (∼24 000 molecules/cell) (Figure 6), or about half of the total CgOrc2 present in these cells. In contrast, the amount of CgOrc1 in the same extracts changed dramatically from an average ∼0.3 ng CgOrc1/106 cells during metaphase to an average of ∼4.1 ng CgOrc1/106 cells (∼26 000 molecules/cell) by 4 h into G1 phase (Figure 7). The amount of CgOrc1 stably bound to chromatin in metaphase cells varied among experiments from <0.1 to 1.3 ng/106 cells, while the amount of Orc1 in late G1 nuclei was comparatively stable. CgOrc1 that was not bound tightly to chromatin appeared to be unstable under these extraction conditions, since it was largely absent from the supernatant fractions, and was presumably degraded during sample preparation (data not shown). The same results were obtained using protocols A and B, and with cells that had been synchronized either with or without nocodazole present (Figure 7A).
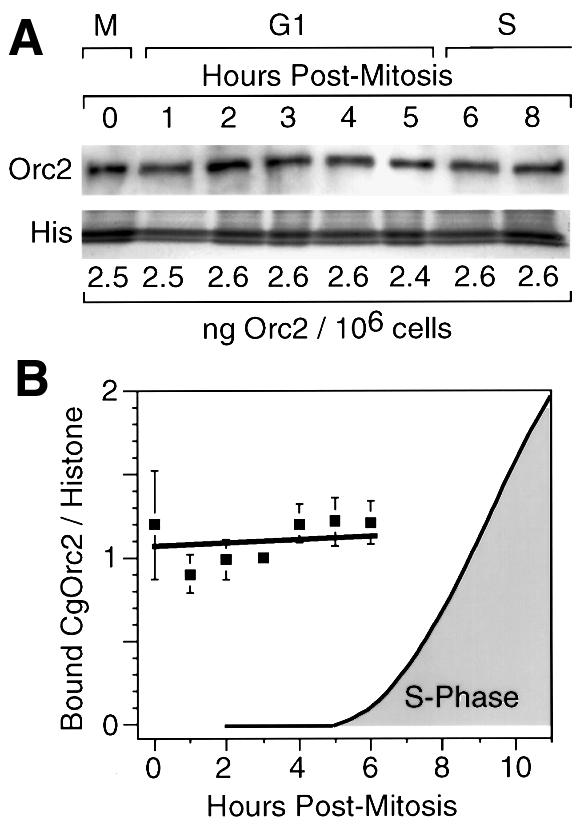
Fig. 6. CgOrc2 remained tightly bound to chromatin during mitosis and G1 phase. Nuclei were prepared in the presence of Triton X-100, NaCl and ATP using protocol A or B (see Materials and methods). One aliquot was used to quantify the amount of CgOrc2 that remained bound, as described in Figure 5. Another aliquot was used to quantify the amount of CgOrc1 (see Figure 7). (A) The amount of CgOrc2 using protocol B was quantified as in Figure 5. (B) The CgOrc2:histone ratio in each gel lane was determined in four separate experiments (two by protocol A and two by protocol B). Each set of results was then normalized to a ratio of 1 for the 3 h time point, and then combined to give a mean ± SEM (filled squares). S phase was determined as in Figure 5D.
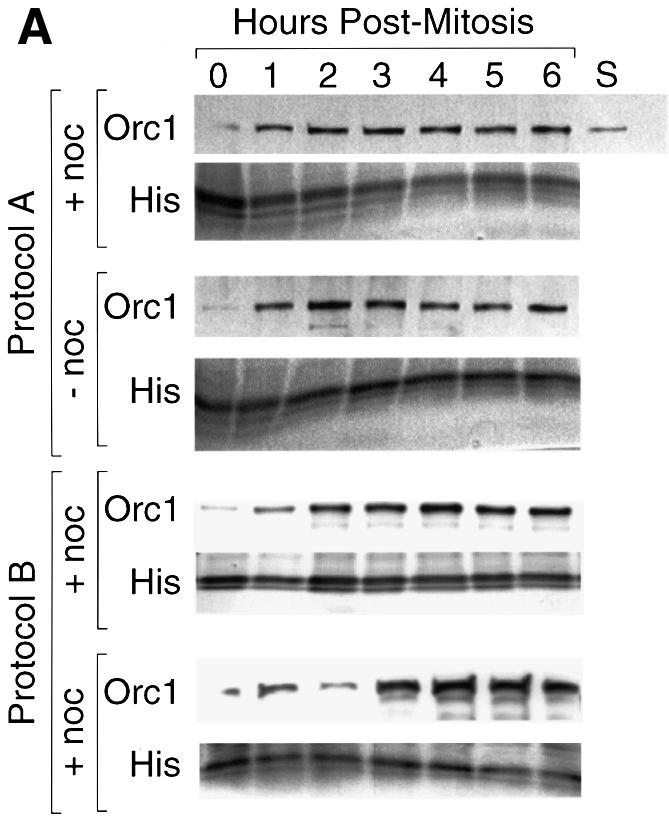
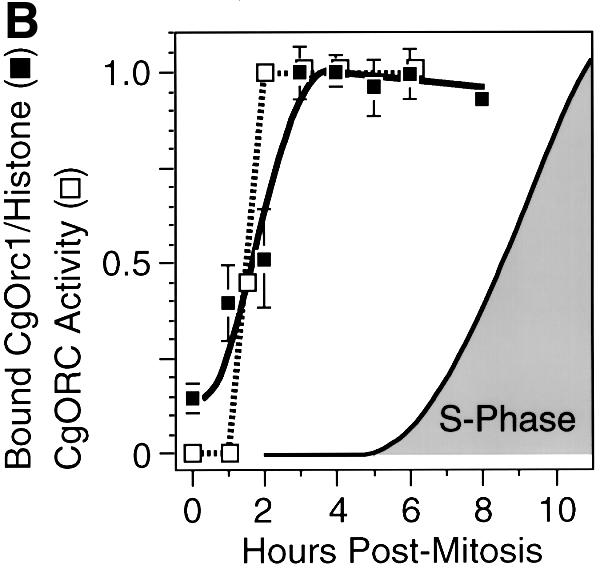
Fig. 7. CgOrc1 binds weakly to hamster chromatin during mitosis but strongly during early G1 phase. (A) CgOrc1 was quantified in aliquots of the same Triton X-100, NaCl, ATP prepared nuclei described in Figure 6. Cells were synchronized at metaphase in the presence (+ noc) or absence (– noc) of nocodazole. (B) The CgOrc1:histone ratio in each gel lane was determined, and then normalized to the maximum value in that experiment. The mean ± SEM for six experiments was determined (filled squares) and compared with the % CgORC activity (open squares), the reciprocal of the % XlOrc-dependence in Figure 4.
These results revealed that individual hamster Orc proteins can behave differently during the transition from metaphase to G1 phase, because CgOrc2 binds tightly to chromatin throughout mitosis and G1 phase, whereas CgOrc1 binds weakly to chromatin during mitosis and early G1 phase, and then strongly during late G1 phase. This transition from weak to strong Orc1 binding coincided with the appearance of a functional hamster ORC in G1 nuclei (Figure 7B). Since measuring ORC activity required that the nuclei had to be incubated for 1 h in an XlOrc-depleted extract (Figure 1), ORC activity may appear more quickly than CgOrc1 binding to chromatin, because CgOrc1 may continue binding to chromatin in vitro. These results suggest that assembly of pre-RCs at specific chromosomal sites in mammalian nuclei begins with the stable binding of Orc1.
Binding and activity of Mcm3 protein mimics Orc1
Previous studies in yeast and in Xenopus egg extracts have shown that Mcm proteins do not bind to chromatin before ORC binding (Romanowski et al., 1996 and references therein). To confirm that pre-RC assembly in hamster nuclei is delayed until G phase, Xenopus egg extract was depleted of XlMcm3 protein and then tested for its ability to initiate DNA replication in hamster G1 nuclei. Since XlMcm3 in Xenopus egg extracts exists as part of an Mcm2–7 complex, depletion of XlMcm3 depletes the other XlMcm proteins as well (Thommes et al., 1997). As previously observed with XlOrc-depleted extracts, XlMcm-depleted extracts were not able to initiate replication in Xenopus sperm chromatin, hamster metaphase chromatin or hamster early G1 nuclei, but were able to initiate replication in hamster late G1 nuclei (Figure 8B). The extent to which hamster nuclei were dependent on XlMcm proteins was inversely related to the amount of CgMcm3 protein stably bound to hamster chromatin (Figure 8). Chromatin-bound CgMcm3, which sometimes appeared as a doublet [probably due to differences in its phosphorylation (Todorov et al., 1995)], increased at least 9-fold as cells transited from mitosis to late G1 phase. Taken together, these results are consistent with the delayed binding of CgOrc1 to hamster chromatin.
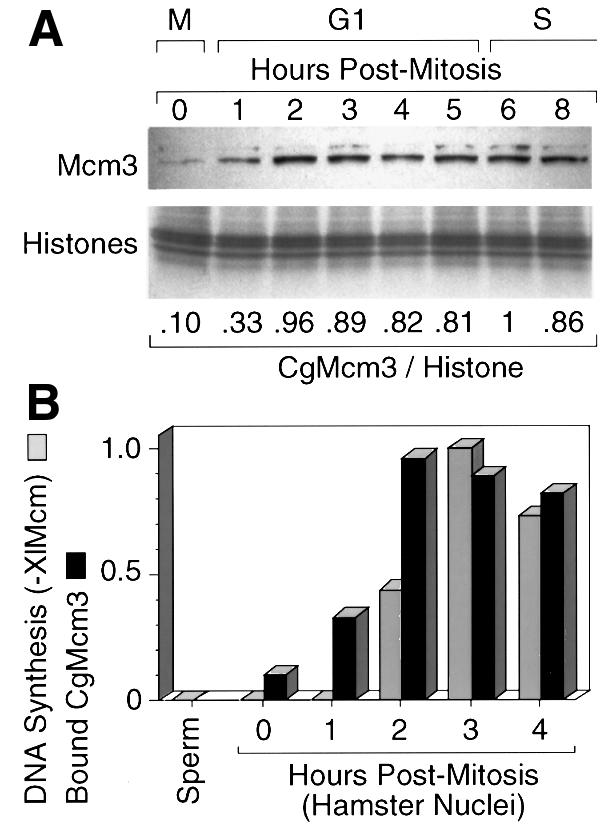
Fig. 8. CgMcm3 bound weakly to hamster chromatin during mitosis but strongly during early G1 phase. (A) CgMcm3 was quantified in nuclei prepared with Triton X-100, NaCl and ATP using protocol A. The Mcm3:histone ratio in each gel lane was normalized to the maximum value (6 h after metaphase). (B) The relative amounts of bound CgMcm3 were compared with the relative amounts of DNA synthesis observed when CHOC 400 digitonin-prepared nuclei were incubated in a Xenopus egg extract depleted of XlMcm3 protein. The amount of 32P-labeled DNA synthesized after a 1 h incubation in XlMcm3-depleted extract was compared with the amount synthesized in a mock-depleted extract, and the –XlMcm3:+XlMcm3 ratio is reported. Xenopus sperm chromatin was included as a control.
Discussion
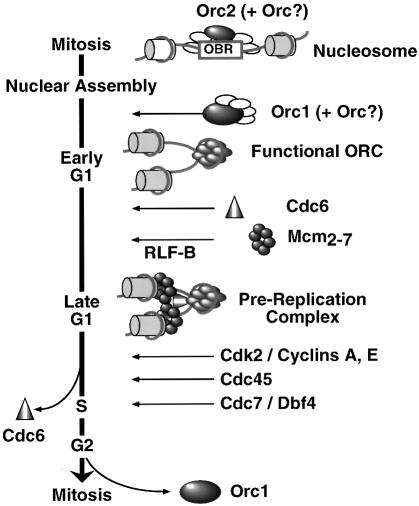
The results presented here and elsewhere suggest that the sequence of events leading to the assembly of pre-RCs in hamster cells (Figure 9) differs from that described for yeast in that the mammalian ORC is not functional during the transition from mitosis to G1 phase, because Orc1 is not stably bound to chromatin during this period. The fact that hamster Orc1 and Orc2 are closely related to their mouse and human homologs strongly suggests that the events outlined in Figure 9 apply to all mammals. The results presented here are consistent with the cell cycle-dependent changes observed in an ORC-like in vivo footprint at the human lamin B2 origin (Abdurashidova et al., 1998). These changes suggested the presence of a pre-RC during G1 phase that is modified to a post-replication complex during S phase (ORC only) and finally the absence of an ORC at this origin during mitosis.
Fig. 9. Assembly of pre-replication complexes in mammalian cells. See the text for a description.
Immunoblotting analyses revealed that both Orc1 and Orc2 were present at equivalent concentrations throughout the CHO cell cycle, but that CgOrc1 could be eluted from mitotic and early G1 chromatin under conditions where CgOrc2 remained bound (Figures 5–7). Incubation of hamster metaphase chromatin or G1 nuclei in Orc-depleted Xenopus egg extract revealed that loss of CgOrc1 binding to chromatin was accompanied by loss of hamster ORC function (Figures 1 and 3). Therefore, at least one of the hamster Orc proteins disassembles from ORC during the M–G1 transition. CgOrc1 then rebound stably to chromatin during early G1 phase, concomitant with the appearance of functional hamster ORCs (Figures 1, 3 and 7), and functional pre-RCs at specific mammalian replication origins (Figures 2, 3 and 7). The behavior of Orc1 was mimicked by Mcm3 protein, consistent with the requirement for a complete chromatin-bound ORC before binding of Mcm proteins (Figure 8). Therefore, the appearance of ORC activity is limited by the behavior of Orc1 and possibly other Orc proteins as well. Before the establishment of hamster pre-RCs at specific chromosomal sites, Xenopus Orc proteins were capable of initiating DNA replication randomly throughout the hamster genome (Figure 3). Whether or not changes in chromosomal architecture during G1 phase also contribute to the random initiation observed under these conditions (Yu et al., 1998) remains to be determined.
A previous study on the ability of Orc-depleted Xenopus egg extract to initiate DNA replication in hamster nuclei concluded that hamster ORC activity was absent from metaphase chromatin but present in both early and late G1 phase nuclei (Yu et al., 1998). However, in this study, only metaphase cells, 1.5 h G1 nuclei and 5 h G1 nuclei were assayed, and DNA synthesis did not begin until 30 min of incubation had elapsed. In our experiments, 1 and 1.5 h G1 nuclei did not initiate DNA synthesis in the absence of XlOrc proteins until the equivalent of 2 h had elapsed after metaphase (Figure 1), the time elapsed in vivo before nuclei were isolated plus the time elapsed in vitro before DNA synthesis began. Thus, the 1.5 h data from Yu et al. (1998) are equivalent to the 2 h data presented here. Yu et al. (1998) did not examine hamster Orc proteins, but later data from the same laboratory showed that CgMcm2 does not associate with hamster chromatin until 40–60 min after metaphase, and then continues to bind during G1 phase with kinetics similar to those observed here (Figure 8) for Mcm3 (Dimitrova et al., 1999). This result is consistent with a delay in the functional association of CgOrc1 with chromatin as reported here (Figure 7B), and with the appearance of pre-RCs at ori-β during mid-G1 phase (Figure 3; Wu and Gilbert, 1996).
ORC and regulation of the metazoan cell cycle
In yeast, where ORC remains stably bound to chromatin throughout the cell cycle (see Introduction), conversion of ORCs into pre-RCs is delayed until mitosis is completed, because Cdk1/cyclin B simultaneously promotes mitosis and inhibits binding of Cdc6 to ORC (Dahmann et al., 1995; Piatti et al., 1996), apparently by phosphorylation of Cdc6 protein (Jallepalli et al., 1997). Thus, the first step in regulating pre-RC assembly in yeast occurs at the level of Cdc6 binding to chromatin. In mammals, assuming that Cdc6 cannot bind to chromatin until a functional ORC has been assembled, the ability of mammalian Orc1 to dissociate and reassociate with chromatin during the M–G1 transition would be the primary mechanism for delaying assembly of pre-RCs until mitosis is complete and a nuclear membrane has formed. In addition, the fact that human (Hs) Orc1 binds HsCdc6 (Saha et al., 1998) suggests that release of Orc1 during mitosis may facilitate formation of an Orc1–Cdc6 complex that could expedite reassembly of pre-RCs during G1 phase.
The significant diversity (as low as 27% similarity) between Orc proteins from yeast, flies, frogs and mammals could account for the differences observed in the stability of their ORCs during the cell cycle and whether or not they remain bound to chromatin. Saccharomyces cerevisiae ORC is stably bound to chromatin throughout the cell cycle (Diffley et al., 1994; Liang and Stillman, 1997; Fujita et al., 1998), while Xenopus ORC appears to be stable but released from chromatin during metaphase, because both Orc1 and Orc2 dissociate from chromatin during mitosis (Coleman et al., 1996; Romanowski et al., 1996; Hua and Newport, 1998; Findeisen et al., 1999; Rowles et al., 1999). Mammalian ORC (and possibly fly ORC) partially dissociates during the M–G1 transition. In Drosophila, DmOrc2 remains bound to chromosomes throughout the cell cycle (Pak et al., 1997), whereas the amount of nuclear bound Drosophila (Dm) Orc1 is greatest during late G1 and S phases (Asano and Wharton, 1999), suggesting a cell cycle-dependent, differential association of DmOrc proteins with chromatin. Thus, in metazoa, ORCs appear to dissociate partially or completely from chromatin during mitosis and then reassemble during G1 phase.
The affinity of Orc proteins for chromatin may depend on their phosphorylated state. Both XlOrc1 and XlOrc2 are hyperphosphorylated in metaphase-arrested eggs relative to activated eggs (Carpenter and Dunphy, 1998; Tugal et al., 1998), and ORC can be selectively released from chromatin by incubating chromatin in a metaphase extract (Rowles et al., 1999) or with Cdc2/cyclin A (Hua and Newport, 1998; Findeisen et al., 1999). Thus, the same cyclin-dependent protein kinase responsible for driving cells into mitosis may concomitantly disassemble pre-RCs at the end of each cell cycle. Subsequent degradation of cyclins would then allow ORC to assemble on chromatin. Similarly, Cdc6 is phosphorylated by Cdk2/cyclin A at the beginning of S phase and translocated from nuclei to cytoplasm where it remains phosphorylated until the completion of mitosis (Jiang et al., 1999b; Petersen et al., 1999). Therefore, both the assembly of a functional ORC on chromatin and the subsequent binding of Cdc6 to the ORC–chromatin complex may be regulated in metazoa by phosphorylation of specific Orc proteins as well as Cdc6.
ORC and site-specific initiation of DNA replication
The destabilization of metazoan ORCs during each cell cycle provides the cell with a mechanism for changing the number and locations of initiation sites during animal development. Before the onset of zygotic gene expression in the rapidly cleaving embryos of frogs and flies, pre-RCs are assembled at many sites throughout the genome with no indication of site specificity, whereas after zygotic gene expression has occurred, DNA replication begins at specific genomic sites (Hyrien et al., 1995; Sasaki et al., 1999). This transition is accompanied by a large decrease in the ratio of Orc proteins to DNA. Xenopus eggs contain enough XlOrc1 to saturate sperm chromatin with about one ORC every 10 kb (∼20 ng XlOrc1/µg DNA) (Rowles et al., 1999). This is at least 30 times more than the total amount of CgOrc1 in hamster cells (0.7 ng CgOrc1/µg DNA) (based on there being 4.7 ng CgOrc1/106 cells and 6.6 pg DNA/cell). In contrast, based on the amounts of CgOrc1 and CgOrc2 bound to hamster chromatin (∼25 000 molecules/cell), the hamster genome contains about one ORC per 240 kb (based on there being 9.1 × 105 kb/pg DNA, 6.6 pg DNA/cell and 2.5 × 104 ORC bound in each cell), consistent with the average size of replicons in hamster and other mammalian cells [200–300 kb (Ockey and Saffhill, 1976; Yurov and Liapunova, 1977)]. The fact that a single Xenopus egg contains ∼105 more ORCs than a single Xenopus somatic cell (Tugal et al., 1998) undoubtedly contributes to their ability to initiate replication at a much greater range of DNA sequences than observed in cultured somatic cells.
Several observations suggest that pre-RCs are assembled at specific chromosomal loci in differentiated mammalian cells. First, Orc-depleted Xenopus egg extract can selectively activate ori-β and ori-β′ in hamster G1 phase nuclei, two origins of bidirectional replication that are activated at the beginning of the hamster S phase (Figure 3; Li et al., 2000). Therefore, pre-RCs must have been assembled at these initiation sites during G1 phase in vivo. Secondly, the appearance of these pre-RCs coincided with the strong binding of CgOrc1 and CgMcm3 to chromatin (Figures 7B and 8B). Thirdly, since CgOrc2, and possibly other Orc proteins, remained bound to the chromatin during mitosis and G1 phase (Figure 6), at least some Orc2 must have been bound at specific sites such as ori-β in order for Orc1 to form a functional ORC. In fact, Drosophila ORC has recently been shown to bind specifically to ACE3, an element controlling the origin of DNA replication (Austin et al., 1999). Thus, the release and reassociation of Orc1 offer a novel mechanism to select which initiation sites will be used during the subsequent round of DNA replication. While this role for Orc1 may have little significance in cultured cells where the same origins are used each cell cycle, it could provide a mechanism for reprogramming the numbers and locations of pre-RCs during animal development. As the cellular ratio of Orc proteins to DNA changes during animal development, simply limiting the amount of Orc1 present would reduce the number of ORCs that could form. ORCs will be assembled at those origins that are most accessible and that bind Orc proteins most tightly. Incomplete ORCs that remain may be displaced from chromatin by replication forks or transcription passing through the origin. Such a mechanism may account for the observations that the amount of DmOrc1 per embryo decreases dramatically during Drosophila development, and that expression of ectopic DmOrc1 promotes rather than inhibits DNA synthesis, suggesting that the level of Orc1 protein governs the activity of replication origins (Asano and Wharton, 1999).
Materials and methods
Cloning hamster ORC1 and ORC2 genes
The following PCR-based strategy was used to clone CgORC1 and CgORC2 from the Stratagene Uni-ZAP XR cDNA library. Sequence information about CgORC sequences was first obtained using total CHO K1 RNA as a template for random-primed reverse transcription (Perkin-Elmer kit). Specific primers based on conserved sequences (between human and Xenopus ORC1 or human and mouse ORC2) were then used to amplify the putative hamster ORC sequences. The ORC1 primers (AGGCCAAAAAGCAAAACCATGCGG and CCGTCGTGCATC TCCAGACAGTGCTGCTACC) were expected to yield an ∼400 bp DNA product covering the last third of the open reading frame. The ORC2 primers (ATTTAAAGAAGATTCTTCTTTAGAACTC and TTGTGGTCCCTAAATTCAGTTAACTGGG) were expected to yield an ∼500 bp product covering the last third of the open reading frame. TaKaRa Ex-Taq was used under conditions described by the manufacturer. ORC1 PCR was performed for 45 cycles (94°C for 45 s; 47°C for 45 s; 72°C for 45 s). ORC2 PCR was performed for four cycles (94°C for 45 s; 45°C for 45 s; 72°C for 45 s) followed by 41 cycles (94°C for 45 s; 52°C for 45 s; 72°C for 45 s). DNA amplification products of the appropriate size were purified from low-melting-point agarose gels following digestion of the gel with β-agarase (New England Biolabs), and sequenced directly. These native CgORC sequences allowed us to take advantage of the cDNA library vector sequences in order to use PCR to clone the genes.
CgORC1 and CgORC2 were cloned in three steps (described below) using the high-fidelity Pfu polymerase (Stratagene) for sequence amplification. After addition of a 3′-terminal A, the reaction products were inserted into the pCR2.1 T/A cloning vector (Invitrogen). The sequences were determined by automated dye-terminator sequencing (ABI) of both strands of three independent clones for each piece.
The 5′-ends were obtained by placing one primer within the fragment of known ORC sequence and the second primer in the cloning vector. In this way, the 5′-end was progressively extended until an open reading frame was identified. The 5′-portion of ORC1 was obtained using the M13 reverse primer and the internal primer GGCACGAGGTGGGTGCGCA, and a 45 cycle amplification (94°C for 30 s; 70°C for 30 s; 72°C for 60 s). The middle portion of ORC1 was obtained using the internal primers ATCTGCAGGCGTGCCACCGAGA and GAGTAGGCCAATCAA AGAGGTTGTACATGAC, and a 45 cycle amplification (94°C for 45 s; 62°C for 45 s; 68°C for 7.5 min). The 3′-end was obtained using the internal primer GTCATGTACAACCTCTTTGATTGGCCTACTC and the M13 forward primer, and a 45 cycle amplification (94°C for 60 s; 72°C for 45 s; 72°C for 7.5 min). The 5′-end of CgORC2 was obtained using the M13 reverse primer and the internal primer GGTCAC TCTTCTCCAATTCACATTCTGGC, and a 45 cycle amplification (94°C for 30 s; 67°C for 30 s; 72°C for 1.5 min). The middle portion of ORC2 was obtained using the internal primers TTTGTGGGA GATGATgATGTTC (the lower case ‘g’ is a mismatch to the hamster sequence) and GTAAATCTCTCTTGGACCCCAAACCATAGAGC, and a 45 cycle amplification (94°C for 45 s; 60°C for 45 s; 68°C for 5 min). The 3′-end of ORC2 was obtained using the internal primer GACATCTGATAGAACCCTGCAGAAGC and the M13 forward primer, and a 45 cycle amplification (94°C for 60 s; 68°C for 45 s; 68°C for 7.5 min). The complete cDNAs were ligated together after digestion with appropriate restriction enzymes, and inserted into pBluescript II SK+ (Stratagene).
An N-terminal FLAG epitope was attached to facilitate purification. A primer was designed to incorporate a KpnI site at the 5′-end of the open reading frame, such that the coding region would be in-frame with a FLAG-coding baculovirus vector. The primer used for this purpose was GGTTGGCCGGTACCAGCACACTGCAGTTAAAGG. The resulting sequence after ligation would replace the native start codon with the FLAG epitope (MDYKDDDK), followed by GT and the rest of the ORC2 sequence.
Antibodies
His6-tagged XlOrc2 protein was produced from a baculovirus expression vector (Carpenter et al., 1996), extracted from Sf9 cells and purified over a Ni-NTA column (Qiagen) using batch elution with 250 mM imidazole. CgOrc1 protein was produced in Escherichia coli by David Gilbert (SUNY Health Science Center, Syracuse, NY) using the cDNA described above. N-terminally FLAG-tagged CgOrc2 protein was expressed in Sf9 cells using a baculovirus expression vector and purified over an anti-FLAG affinity column (Kodak) using batch elution with 0.1 M glycine–HCl pH 2.5. Eluates were immediately neutralized by the addition of 1 M NaH2PO4 pH 8.0. The protein was further purified to >95% purity (as judged by Coomassie Blue staining) by elution from polyacrylamide gels (Bio-Rad). Antisera were produced in rabbits by Covance Laboratories, and the IgG fractions were purified by chromatography over Staphylococcus protein A beads (Pierce). Anti-actin monoclonal mouse IgM was purchased from Calbiochem.
Cells and nuclei
Where indicated, cells were synchronized in mitosis using nocodazole and then released into G1, and their progress was monitored by incorporation of either bromodeoxyuridine (BrdU) or [3H]thymidine into parallel cultures (Gilbert et al., 1995). Consistent with previous reports (Wu and Gilbert, 1996), all of the cells contained nuclei by 1 h after metaphase. For DNA replication studies, nuclei were prepared by lysing cells with 40 µg/ml digitonin in 0.5× transport buffer (10 mM HEPES pH 7.3, 55 mM KOAc, 2.5 mM NaOAc, 1 mM Mg(OAc)2, 0.5 mM EGTA) as previously described (Gilbert et al., 1995). To wash nuclei free of proteins that are not tightly bound to chromatin, nuclei were prepared as described by Fujita et al. (1997) (protocol A) or by Todorov et al. (1995) (protocol B). In general, cells were washed twice with phosphate-buffered saline, then once with CSK buffer (see below), then lysed with CSK buffer containing Triton X-100 and the nuclei washed once with CSK buffer containing Triton X-100. In protocol A, CSK buffer consisted of 10 mM HEPES pH 7.5, 150 mM NaCl, 300 mM sucrose, 1 mM MgCl2, 1 mM Mg2+-ATP, 1 mM EGTA, 1 mM dithiothreitol and 10 µg/ml each of aprotinin, leupeptin and pepstatin, and the cells were lysed with 0.1% Triton X-100. In protocol B, CSK buffer consisted of 10 mM HEPES pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM Mg2+-ATP and 10 µg/ml each of aprotinin, leupeptin and pepstatin, and the cells were lysed with 0.5% Triton X-100.
DNA replication in Xenopus egg extract
Extracts were prepared from Xenopus eggs and, where indicated, depleted of Xenopus Orc and Mcm proteins as described by Chong et al. (1997) with the modifications reported by Li et al. (2000). DNA replication analyses are described in Li et al. (2000).
Western blot analysis
Orc and Mcm proteins were assayed by immunoblotting protocols as described in Harlow and Lane (1999). Protein samples were fractionated by electrophoresis under denaturing conditions using a precast mini-gel (Bio-Rad) at 100 V for 1.5 h. Standard amounts of purified Orc1 or Orc2 were included, as were molecular weight markers (Amersham, RPN 800). Proteins from the top portion of each gel were transferred to a membrane by electroblotting. Proteins were first reacted with specific rabbit IgG antibodies (1 in 2000 dilution) and then detected with a 1 in 50 000 dilution of goat anti-rabbit IgG conjugated with peroxidase (Pierce SuperSignal Substrate western blotting kit). The bottom portion of each gel was stained separately with GelCode Blue (Pierce) to reveal histones. Standard curves were generated using purified CgOrc1 or CgOrc2 in parallel. The relative amounts of proteins in each gel lane were determined by densitometry using NIH Image.
Acknowledgments
Acknowledgements
We thank Y.Nakatani (NICHD) for the FLAG coding baculovirus vector, K.Kaneko for CHO RNA, T.Rein for sequencing PCR products, R.Knippers for anti-HsMcm3 antiserum (Ritzi et al., 1998), J.Blow for anti-XlMcm3 antiserum (Thommes et al., 1997), P.Carpenter for baculovirus expressing XlOrc2 protein (Carpenter et al., 1996) and D.Gilbert (SUNY Health Science Center, Syracuse, NY) for expressing CgOrc1 protein in E.coli using the CgORC1 gene described here.
References
- Abdurashidova G., Riva,S., Biamonti,G., Giacca,M. and Falaschi,A. (1998) Cell cycle modulation of protein–DNA interactions at a human replication origin. EMBO J., 17, 2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aladjem M.I., Groudine,M., Brody,L.L., Dieken,E.S., Fournier,R.E., Wahl,G.M. and Epner,E.M. (1995) Participation of the human β-globin locus control region in initiation of DNA replication. Science, 270, 815–819. [DOI] [PubMed] [Google Scholar]
- Aladjem M.I., Rodewald,L.W., Kolman,J.L. and Wahl,G.M. (1998) Genetic dissection of a mammalian replicator in the human β-globin locus. Science, 281, 1005–1009. [DOI] [PubMed] [Google Scholar]
- Asano M. and Wharton,R.P. (1999) E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J., 18, 2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin R.J., Orr-Weaver,T.L. and Bell,S.P. (1999) Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev., 13, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter P.B. and Dunphy,W.G. (1998) Identification of a novel 81-kDa component of the Xenopus origin recognition complex. J. Biol. Chem., 273, 24891–24897. [DOI] [PubMed] [Google Scholar]
- Carpenter P.B., Mueller,P.R. and Dunphy,W.G. (1996) Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature, 379, 357–360. [DOI] [PubMed] [Google Scholar]
- Chong J.P., Thommes,P., Rowles,A., Mahbubani,H.M. and Blow,J.J. (1997) Characterization of the Xenopus replication licensing system. Methods Enzymol., 283, 549–564. [DOI] [PubMed] [Google Scholar]
- Coleman T.R., Carpenter,P.B. and Dunphy,W.G. (1996) The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell, 87, 53–63. [DOI] [PubMed] [Google Scholar]
- Dahmann C., Diffley,J.F. and Nasmyth,K.A. (1995) S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr. Biol., 5, 1257–1269. [DOI] [PubMed] [Google Scholar]
- DePamphilis M.L. (1999) Replication origins in metazoan chromosomes: fact or fiction? BioEssays, 21, 5–16. [DOI] [PubMed] [Google Scholar]
- Diffley J.F., Cocker,J.H., Dowell,S.J. and Rowley,A. (1994) Two steps in the assembly of complexes at yeast replication origins in vivo.Cell, 78, 303–316. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S., Todorov,I.T., Melendy,T. and Gilbert,D.M. (1999) Mcm2, but not RPA, is a component of the mammalian early G1 phase prereplication complex. J. Cell Biol., 146, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeisen M., El-Denary,M., Kapitza,T., Graf,R. and Strausfeld,U. (1999) Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6, and MCM in egg extracts of Xenopus laevis. Eur. J. Biochem., 264, 415–426. [DOI] [PubMed] [Google Scholar]
- Fujita M., Kiyono,T., Hayashi,Y. and Ishibashi,M. (1997) In vivo interaction of human MCM heterohexameric complexes with chromatin. Possible involvement of ATP. J. Biol. Chem., 272, 10928–10935. [DOI] [PubMed] [Google Scholar]
- Fujita M., Hori,Y., Shirahige,K., Tsurimoto,T., Yoshikawa,H. and Obuse,C. (1998) Cell cycle dependent topological changes of chromosomal replication origins in Saccharomyces cerevisiae.Genes Cells, 3, 737–749. [DOI] [PubMed] [Google Scholar]
- Gilbert D.M., Miyazawa,H. and DePamphilis,M.L. (1995) Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol. Cell. Biol., 15, 2942–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handeli S., Klar,A., Meuth,M. and Cedar,H. (1989) Mapping replication units in animal cells. Cell, 57, 909–920. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1999) Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hua X.H. and Newport,J. (1998) Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol., 140, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Fanti,L., Pak,D.T., Botchan,M.R., Pimpinelli,S. and Kellum,R. (1998) Distinct cytoplasmic and nuclear fractions of Drosophila heterochromatin protein 1: their phosphorylation levels and associations with origin recognition complex proteins. J. Cell Biol., 142, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O., Maric,C. and Mechali,M. (1995) Transition in specification of embryonic metazoan DNA replication origins. Science, 270, 994–997. [DOI] [PubMed] [Google Scholar]
- Jallepalli P.V., Brown,G.W., Muzi-Falconi,M., Tien,D. and Kelly,T.J. (1997) Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev., 11, 2767–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., McDonald,D., Hope,T.J. and Hunter,T. (1999a) Mammalian Cdc7–Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J., 18, 5703–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Wells,N.J. and Hunter,T. (1999b) Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl Acad. Sci. USA, 96, 6193–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta R.F., Li,X., Mesner,L.D., Dijkwel,P.A., Lin,H.B. and Hamlin,J.L. (1998) Distal sequences, but not ori-β/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell, 2, 797–806. [DOI] [PubMed] [Google Scholar]
- Krude T., Jackman,M., Pines,J. and Laskey,R.A. (1997) Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell, 88, 109–119. [DOI] [PubMed] [Google Scholar]
- Kumagai H., Sato,N., Yamada,M., Mahony,D., Seghezzi,W., Lees,E., Arai,K. and Masai,H. (1999) A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol. Cell. Biol., 19, 5083–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis G. and Tower,J. (1999) The Drosophila chiffon gene is required for chorion gene amplification, and is related to the yeast Dbf4 regulator of DNA replication and cell cycle. Development, 126, 4281–4293. [DOI] [PubMed] [Google Scholar]
- Landis G., Kelley,R., Spradling,A.C. and Tower,J. (1997) The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc. Natl Acad. Sci. USA, 94, 3888–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Bogan,J.A., Natale,D.A. and DePamphilis,M.L. (2000) Selective activation of pre-replication complexes in vitro at specific sites in mammalian nuclei. J. Cell Sci., 113, 887–898. [DOI] [PubMed] [Google Scholar]
- Liang C. and Stillman,B. (1997) Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev., 11, 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malott M. and Leffak,M. (1999) Activity of the c-myc replicator at an ectopic chromosomal location. Mol. Cell. Biol., 19, 5685–5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ockey C.H. and Saffhill,R. (1976) The comparative effects of short-term DNA inhibition on replicon synthesis in mammalian cells. Exp. Cell Res., 103, 361–373. [DOI] [PubMed] [Google Scholar]
- Pak D.T., Pflumm,M., Chesnokov,I., Huang,D.W., Kellum,R., Marr,J., Romanowski,P. and Botchan,M.R. (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell, 91, 311–323. [DOI] [PubMed] [Google Scholar]
- Petersen B.O., Lukas,J., Sorensen,C.S., Bartek,J. and Helin,K. (1999) Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J., 18, 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phi-van L. and Stratling,W.H. (1999) An origin of bidirectional DNA replication is located within a CpG island at the 3′ end of the chicken lysozyme gene. Nucleic Acids Res., 27, 3009–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Bohm,T., Cocker,J.H., Diffley,J.F. and Nasmyth,K. (1996) Activation of S-phase-promoting CDKs in late G1 defines a ‘point of no return’ after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev., 10, 1516–1531. [DOI] [PubMed] [Google Scholar]
- Rein T., Kobayashi,T., Malott,M., Leffak,M. and DePamphilis,M.L. (1999) DNA methylation at mammalian replication origins. J. Biol. Chem., 274, 25792–25800. [DOI] [PubMed] [Google Scholar]
- Ritzi M., Baack,M., Musahl,C., Romanowski,P., Laskey,R.A. and Knippers,R. (1998) Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem., 273, 24543–24549. [DOI] [PubMed] [Google Scholar]
- Romanowski P., Madine,M.A., Rowles,A., Blow,J.J. and Laskey,R.A. (1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol., 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- Rowles A., Chong,J.P., Brown,L., Howell,M., Evan,G.I. and Blow,J.J. (1996) Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell, 87, 287–296. [DOI] [PubMed] [Google Scholar]
- Rowles A., Tada,S. and Blow,J.J. (1999) Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci., 112, 2011–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P., Chen,J., Thome,K.C., Lawlis,S.J., Hou,Z.H., Hendricks,M., Parvin,J.D. and Dutta,A. (1998) Human CDC6/Cdc18 associates with Orc1 and cyclin–cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol., 18, 2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Sawado,T., Yamaguchi,M. and Shinomiya,T. (1999) Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster.Mol. Cell. Biol., 19, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thommes P., Kubota,Y., Takisawa,H. and Blow,J.J. (1997) The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J., 16, 3312–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov I.T., Pepperkok,R., Philipova,R.N., Kearsey,S.E., Ansorge,W. and Werner,D. (1994) A human nuclear protein with sequence homology to a family of early S phase proteins is required for entry into S phase and for cell division. J. Cell Sci., 107, 253–265. [DOI] [PubMed] [Google Scholar]
- Todorov I.T., Attaran,A. and Kearsey,S.E. (1995) BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J. Cell Biol., 129, 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugal T., Zou-Yang,X.H., Gavin,K., Pappin,D., Canas,B., Kobayashi,R., Hunt,T. and Stillman,B. (1998) The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p. J. Biol. Chem., 273, 32421–32429. [DOI] [PubMed] [Google Scholar]
- Walter J. and Newport,J.W. (1997) Regulation of replicon size in Xenopus egg extracts. Science, 275, 993–995. [DOI] [PubMed] [Google Scholar]
- Wu J.R. and Gilbert,D.M. (1996) A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science, 271, 1270–1272. [DOI] [PubMed] [Google Scholar]
- Wu J.R., Yu,G. and Gilbert,D.M. (1997) Origin-specific initiation of mammalian nuclear DNA replication in a Xenopus cell-free system. Methods, 13, 313–324. [DOI] [PubMed] [Google Scholar]
- Yan Z., DeGregori,J., Shohet,R., Leone,G., Stillman,B., Nevins,J.R. and Williams,R.S. (1998) Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wu,J.R. and Gilbert,D.M. (1998) Analysis of mammalian origin specification in ORC-depleted Xenopus egg extracts. Genes Cells, 3, 709–720. [DOI] [PubMed] [Google Scholar]
- Yurov Y.B. and Liapunova,N.A. (1977) The units of DNA replication in the mammalian chromosomes: evidence for a large size of replication units. Chromosoma, 60, 253–267. [DOI] [PubMed] [Google Scholar]