Abstract
Dolichol-phosphate-mannose (DPM) synthase generates mannosyl donors for glycosylphosphatidylinositols, N-glycan and protein O- and C-mannosylation. In Saccharomyces cerevisiae, this enzyme is encoded by DPM1. We reported previously that mammalian DPM synthase contains catalytic DPM1 and regulatory DPM2 subunits, and that DPM1 requires DPM2 for its stable expression in the endoplasmic reticulum. Here we report that human DPM synthase consists of three subunits. The third subunit, DPM3, comprises 92 amino acids associated with DPM1 via its C-terminal domain and with DPM2 via its N-terminal portion. The stability of DPM3 was dependent upon DPM2. However, overexpression of DPM3 in Lec15 cells, a null mutant of DPM2, restored the biosynthesis of DPM with an increase in DPM1, indicating that DPM3 directly stabilized DPM1. Therefore, DPM2 stabilizes DPM3 and DPM3 stabilizes DPM1. DPM synthase activity was 10 times higher in the presence of DPM2, indicating that DPM2 also plays a role in the enzymatic reaction. Schizosaccharomyces pombe has proteins that resemble three human subunits; S.pombe DPM3 restored biosynthesis of DPM in Lec15 cells, indicating its orthologous relationship to human DPM3.
Keywords: dolichol-phosphate-mannose/endoplasmic reticulum/N-glycan/glycosylphosphatidylinositol/O-mannosylation
Introduction
Dolichol-phosphate-mannose (DPM) is a mannosyl donor important for the biosynthesis of various glycoconjugates. DPM donates all three (four in yeast) mannoses in glycosylphosphatidylinositols (GPIs), the last four mannoses in N-glycan precursors, the first mannose in O-mannosylation of many yeast proteins (reviewed in Kornfeld and Kornfeld, 1985; Hirschberg and Snider, 1987; Abeijon and Hirschberg, 1992; Englund, 1993; Herscovics and Orlean, 1993) and a mannose in C-mannosylation (Doucey et al., 1998). In the absence of DPM, GPI-anchored proteins would not be synthesized and protein N-glycosylation would be impaired. O- and C-mannosylation of proteins would also be arrested. A lack of GPI biosynthesis causes embryonic lethality in mice (Nozaki et al., 1999). It was reported recently that partially deficient DPM biosynthesis causes congenital disorder of glycosylation (CDG) (formerly termed carbohydrate-deficient glycoprotein syndrome or CDGS; Imbach et al., 2000; Kim et al., 2000). Patients deficient in DPM synthesis present with developmental delay, seizures, hypotonia and dysmorphic features. These symptoms may be due to a lack of N-glycan and/or a lack of GPI-anchored proteins.
DPM is synthesized from GDP-mannose and dolichol-phosphate by DPM synthase in the endoplasmic reticulum (ER). DPM synthases in various organisms are grouped into two types: one is a single-component enzyme represented by Saccharomyces cerevisiae Dpm1p (Colussi et al., 1997) and the other is a multicomponent enzyme represented by human DPM synthase, which contains a catalytic subunit DPM1 and a regulatory subunit DPM2 (Maeda et al., 1998). The former group includes the enzymes of S.cerevisiae (Orlean et al., 1988), the fungus Ustilago maydis (Zimmerman et al., 1996), and the protozoa Trypanosoma brucei (Mazhari-Tabrizi et al., 1996) and Leishmania mexicana (Ilgoutz et al., 1999). Those DPM synthases can be expressed in Escherichia coli as functional enzymes (Orlean et al., 1988; Schutzbach et al., 1993; Mazhari-Tabrizi et al., 1996; Zimmerman et al., 1996). In contrast, human DPM1 cannot be stably expressed in E.coli (our unpublished observation) and requires the presence of DPM2 for stable expression in the ER (Maeda et al., 1998). Members of the former group share 50–60% amino acid identity and have a transmembrane domain near the C-terminus, whereas human DPM1 has only ∼30% amino acid identity to the former group members and does not have a transmembrane domain (Colussi et al., 1997; Tomita et al., 1998). Schizosaccharomyces pombe and Caenorhabditis elegans have DPM1 homologues that lack a transmembrane domain like human DPM1, and have 65 and 64% amino acid identity to human DPM1, respectively (Colussi et al., 1997). Clearly, the enzymes of the two groups have different structural and functional characteristics.
To determine whether DPM1 and DPM2 are the only subunits of human DPM synthase, we have isolated the enzyme and found that it consists of three subunits and that the third subunit (termed DPM3) is stabilized by DPM2, and that DPM3, in turn, stabilizes DPM1.
Results
Identification and cloning of DPM3
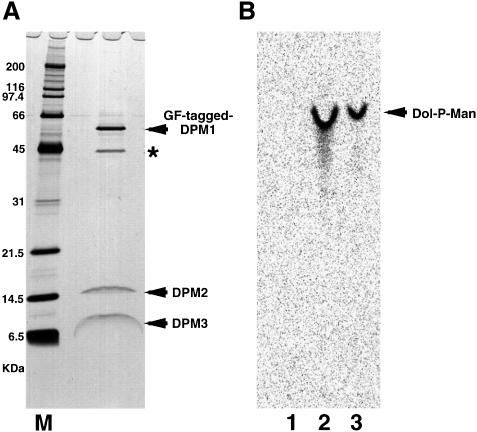
To isolate human DPM synthase, we transfected human B-lymphoblastoid JY25 cells with DPM1 cDNA that was tagged tandemly with GST and FLAG, and isolated the DPM synthase complex by two-step affinity purification using anti-FLAG and glutathione beads. The preparation contained four proteins of 60, 45, 15 and 8 kDa (Figure 1A). The 60 kDa protein was confirmed as tagged DPM1 because it was detected by western blotting with anti-FLAG and anti-GST antibodies (data not shown) and because its mobility on SDS–PAGE was consistent with the calculated molecular size of GST- and FLAG-tagged DPM1. The 45 kDa protein must be a contaminant because it was an abundant protein and its ratio to the three other components was decreased greatly after the second purification step. We determined the N-terminal amino acid sequences of the 15 and 8 kDa proteins. The sequence of the first 30 amino acids of the 15 kDa protein was ATGTDQVVGLGLVAVSLIIFTYYTAXVILL, where X was unknown. This coincided exactly with the sequence from residues 2 to 31 of the predicted human DPM2, when X was assumed to be W, indicating that the first methionine did not remain in the natural DPM2 protein. The sequence of the first 13 amino acids of the 8 kDa protein was MTKLAQWLXGLAI, representing a new protein.
Fig. 1. Purification of DPM synthase complex. GST- and FLAG-tagged hDPM1 was isolated by two-step affinity purification using anti-FLAG and glutathione beads from the digitonin lysates of 3 × 109 JY25 transfectant cells. A part of the purified protein was analysed by SDS–PAGE and silver staining (A) and the rest was used for DPM synthase assay as shown in (B). (A) M, molecular weight markers. The protein indicated by the asterisk is a contaminant. (B) Liposomes only (lane 1), DPM synthase complexes reconstituted in liposomes (lane 2) and membranes of JY25 cells (lane 3) were assayed for DPM synthase activity. The lipids were separated by TLC with a solvent system of chloroform/methanol/H2O (10:10:3).
The isolated protein complexes had DPM synthase activity when they were incorporated into liposome membranes (Figure 1B, lane 2). The Km for GDP-mannose was 240 nM (data not shown), being comparable to that of rat liver DPM synthase, which was reported to be 690 nM (Jensen and Schutzbach, 1985). We therefore concluded that we had isolated a functional DPM synthase.
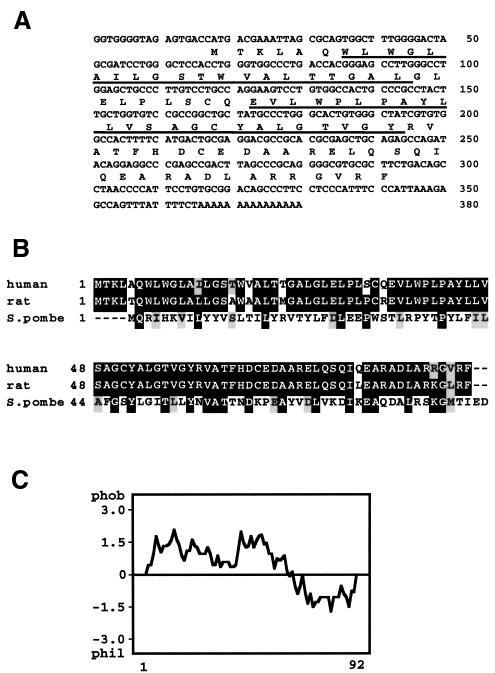
We found several expressed sequence tags (ESTs) (Adams et al., 1991) that correspond to the amino acid sequence of the 8 kDa protein and obtained a 365 bp cDNA that encodes a predicted protein of 92 amino acids (Figure 2A). We termed this gene DPM3 (see below for the functions of DPM3). Some of the DPM3 EST sequences contained a sequence-tagged site (STS), which was mapped to chromosome 1q12–21 (The National Center for Biotechnology Information).
Fig. 2. (A) Nucleotide and protein sequences of human DPM3. The protein sequence is in single-letter code below the nucleotide sequence. The putative transmembrane regions are underlined. Nucleotide numbers are on the right. The accession number of human DPM3 cDNA is AB028128. (B) Alignment of human, rat and S.pombe DPM3 amino acid sequences. Black and grey boxes indicate identical and similar amino acids, respectively. Amino acid numbers are on the left. (C) Hydropathy profile of human DPM3 drawn according to the Kyte and Doolittle program (Kyte and Doolittle, 1982).
We found rat, C.elegans and S.pombe DPM3 homologues in the databases. Rat DPM3 (GenBank accession No. AI111977) had 92 amino acids with 97% identity to human DPM3 (Figure 2B). The C.elegans DPM3 (GenBank accession No. Z70684) consisted of 95 amino acids with 25% identity to human DPM3. The sequence of the C-terminal 36 amino acids of C.elegans DPM3 was 50% identical to that of human DPM3 (not shown). The S.pombe DPM3 (GenBank accession No. AL035581.1, gene ID SPBC1677.02) consisted of 90 amino acids with 31% identity to human DPM3 (Figure 2B). We found no obvious homologue of DPM3 in the genome of S.cerevisiae.
DPM3 is a membrane protein in the ER
The predicted human DPM3 protein had two putative transmembrane domains in the N-terminal portion and a hydrophilic C-terminal portion (Figure 2C). There was no known ER retention signal or N-glycosylation consensus sequence.
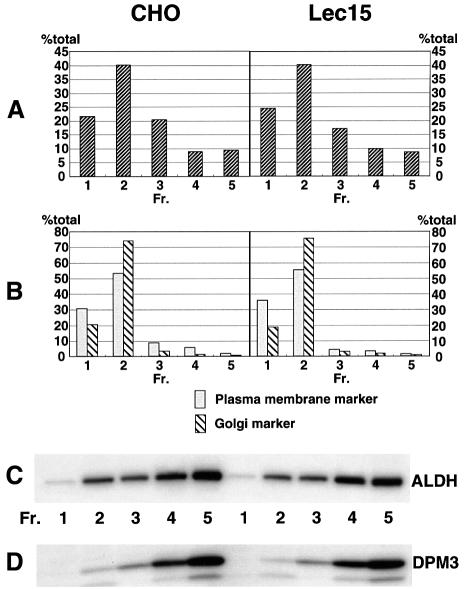
To determine the intracellular expression site of DPM3, we transiently transfected CHO cells with cDNAs of human DPM3 and FLAG-tagged aldehyde dehydrogenase (ALDH), as a marker of the ER (Masaki et al., 1994). We also transfected Lec15 cells, a null mutant of DPM2 (Maeda et al., 1998), with the same cDNAs to see whether a lack of DPM2 affects localization of DPM3. We disrupted the transfectant cells and fractionated nuclei-free samples into the ER, Golgi and plasma membrane by sucrose density gradient centrifugation. DPM3 was found by immunoprecipitation and western blotting with anti-human DPM3 antibodies mainly in fractions 4 and 5, corresponding to the ER (Figure 3). Only a trace amount of DPM3 was found in fractions 1 and 2 containing the Golgi and the plasma membranes, indicating that DPM3 is an ER protein. The ER localization of DPM3 was independent of DPM2 as shown by analysis with the transfected Lec15 cells (Figure 3), although there is no known ER localization signal sequence in DPM3.
Fig. 3. ER localization of DPM3. CHO cells were transfected with pME-Py-hDPM3 and pME-Py-FLAG-ALDH, as an ER marker. Two days later, the post-nuclear supernatants were fractionated by sucrose density gradient centrifugation. Fractions were characterized by (A) protein content, (B) organelle-specific marker enzymes (alkaline phosphodiesterase I for the plasma membrane and α-mannosidase II for the Golgi apparatus), (C) FLAG-tagged ALDH to locate the ER and (D) hDPM3. The protein contents and enzyme activities in fractions are shown as percentages of the total.
DPM3 associates directly with DPM1 and DPM2
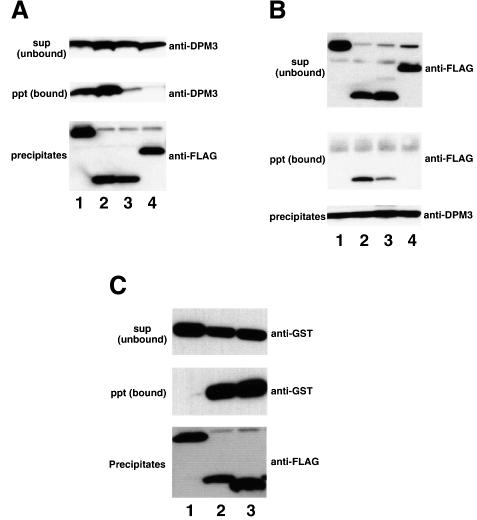
We analysed the physical association of DPM3 with DPM1 and DPM2. For this, we used Lec15 cells. A cDNA of human DPM3 was co-transfected with one of the cDNAs of FLAG-tagged human DPM1, rat DPM2, rat DPM2FY/LS (a mutant of rat DPM2 in which Phe21 and Tyr23 were changed to Leu21 and Ser23) and human SL15 into Lec15 cells. From their detergent extracts, FLAG-tagged proteins were immunoprecipitated and co-preci pitation of DPM3 was analysed by western blotting (Figure 4A). Alternatively, DPM3 was immunoprecipitated first and co-precipitation of FLAG-tagged proteins was analysed by western blotting (Figure 4B). As shown in Figure 4A, DPM3 was co-precipitated with FLAG-tagged DPM1 in the absence of DPM2 (lane 1), indicating a direct association of DPM3 and DPM1. DPM3 was also co-precipitated with FLAG-tagged DPM2 (lane 2) as expected, because, in this case, all three components were present. Only a small amount of DPM3 was co-precipitated with the mutant DPM2FY/LS (lane 3), indicating that Phe21 and/or Tyr23 in DPM2 are important in the association with DPM3. co-precipitation of DPM3 with FLAG-tagged SL15 was negligible (lane 4), indicating that DPM3 is not associated with SL15, a protein involved in usage of DPM (Camp et al., 1993; Ware and Lehrman, 1996).
Fig. 4. Association of DPM1 and DPM2 with DPM3. (A and B) Human DPM3 was co-expressed in Lec15 cells with FLAG-tagged hDPM1 (lane 1), rDPM2 (lane 2), rDPM2FY/LS (lane 3) and hSL15 (lane 4), and their associations were analysed by immunoprecipitation. The lysates were immunoprecipitated with anti-FLAG (A) or anti-DPM3 (B). In (A), immunoprecipitates (middle and lower panels) and the supernatant (upper panel) were analysed by western blotting against anti-hDPM3 (upper and middle panels) and anti-FLAG (lower panel) antibodies. In (B), immunoprecipitates (middle and lower panels) and the supernatant (upper panel) were analysed by western blotting against anti-FLAG (upper and middle panels) and anti-hDPM3 (lower panel) antibodies. (C) GST-tagged human DPM2 was co-expressed in Lec15 cells with FLAG-tagged hSL15 (lane 1), hDPM3 (lane 2) and hDPM3dC (lane 3). Their lysates were immunoprecipitated with anti-FLAG beads. Precipitates (middle and lower panels) and the supernatant (upper panel) were analysed by western blotting against anti-GST (upper and middle panels) and anti-FLAG (lower panel) antibodies.
When DPM3 was first immunoprecipitated (Figure 4B), FLAG-tagged DPM2 but not SL15 was co-precipitated (lanes 2 and 4), and FLAG-tagged DPM2FY/LS was co-precipitated inefficiently (lane 3) as expected from the above result. However, FLAG-tagged DPM1 was not co-precipitated (lane 1), contrary to the above result. Since anti-DPM3 antibodies were made against the C-terminal portion, this lack of co-precipitation may be due to steric hindrance of the C-terminal portion of DPM3 by associated DPM1 that prevented binding of the antibody.
If the C-terminal portion of DPM3 is involved in association with DPM1, the N-terminal portion of DPM3 may be involved in association with DPM2. To test this possibility, we generated a truncated DPM3, DPM3dC, in which 27 C-terminal amino acids were deleted, and assessed its association with DPM2. We transfected Lec15 cells with a cDNA of GST-tagged DPM2 together with one of the cDNAs of FLAG-tagged DPM3dC, DPM3 and SL15, immunoprecipitated FLAG-tagged proteins from their detergent extracts and analysed co-precipitation of GST-tagged DPM2 (Figure 4C). GST-tagged DPM2 was co-precipitated with FLAG-tagged DPM3dC (lane 3) as well as with FLAG-tagged DPM3 (lane 2), but not with FLAG-tagged SL15 (lane 1), indicating that the N-terminal 65 amino acid portion of DPM3 is involved in association with DPM2.
DPM3 stabilizes DPM1 in the absence of DPM2 while DPM2 is necessary for stable expression of DPM3
We reported that the expression level of DPM1 was low in the absence of DPM2 due to instability of DPM1 (Maeda et al., 1998). Because DPM3 associated directly with DPM1, we next tested whether transient overexpression of DPM3 in the absence of DPM2 results in an increase in the expression of DPM1. We transfected Lec15 cells with a cDNA of GST-tagged DPM1 together with an empty vector, DPM3 cDNA or DPM2 cDNA, as a positive control, and compared expression levels of GST-tagged DPM1 in these cells (Figure 5A). cDNAs of GST-tagged ALDH and free GST were also co-transfected to assess transfection efficiency. Transient transfection of DPM3 cDNA in fact increased the level of GST-tagged DPM1 10-fold (lane 1 versus lane 3). This increase was much higher than that caused by transfection of DPM2 cDNA (lane 2). Transfection of DPM3dC did not increase DPM1 expression (data not shown), further supporting the idea that DPM3 associates with DPM1 via the C-terminal domain.
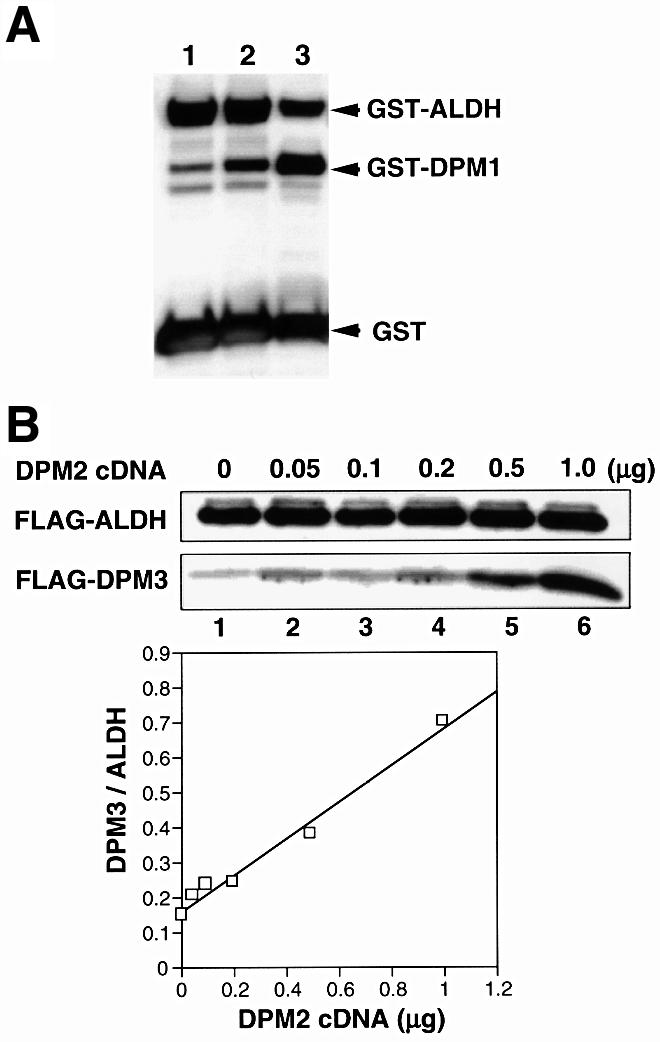
Fig. 5. (A) Stabilization of DPM1 by DPM3 in the absence of DPM2. A mixture of cDNAs of GST-tagged DPM1, GST-tagged ALDH and free GST was co-transfected into Lec15 cells with an empty vector (lane 1), DPM2 cDNA (lane 2) or DPM3 cDNA (lane 3). Two days later, expression of GST and GST-tagged proteins was assessed by western blotting. (B) Stabilization of DPM3 by DPM2. A mixture of 3 µg of cDNA of FLAG-tagged DPM3 and 6 µg of cDNA of FLAG-tagged ALDH was co-transfected with increasing amounts of DPM2 cDNA (0–1 µg) into Lec15 cells. The total amount of cDNA was kept at 10 µg by adding an empty vector. Two days later, expression of FLAG-tagged DPM3 and ALDH was assessed by western blotting. The ratio of FLAG-tagged DPM3 to ALDH was plotted as a function of the amount of DPM2 cDNA.
Because the above result raised the possibility that DPM2-mediated stabilization of DPM1 could be an indirect effect via stabilization of DPM3, we next asked whether DPM2 stabilizes DPM3. We transfected Lec15 cells with a limited amount of cDNA of FLAG-tagged DPM3 (3 µg for 5 × 106 cells) and increasing amounts of DPM2 cDNA (0–1 µg) to see whether expression of FLAG-tagged DPM3 depends upon the dose of DPM2 cDNA (Figure 5B). A constant amount of cDNA of FLAG-tagged ALDH (6 µg) was also co-transfected to normalize transfection efficiency. The expression of FLAG-DPM3 increased with an increase in transfected DPM2 cDNA, whereas the expression of FLAG-ALDH remained at a similar level. The level of FLAG-DPM3 increased ∼5-fold with the highest dose of DPM2 cDNA. Therefore, DPM2 stabilizes DPM3, and DPM3, in turn, stabilizes DPM1.
DPM2 is important for high specific activity of DPM synthase
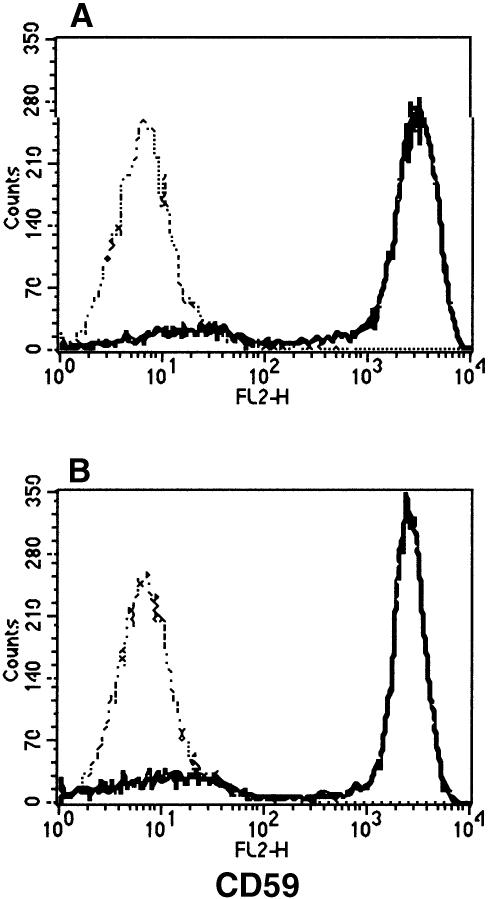
We previously found that DPM1 has DPM synthase activity in the absence of DPM2 if it is stably expressed (Maeda et al., 1998). We next tested whether DPM1 stabilized by DPM3 has DPM synthase activity without DPM2. We first confirmed by RT–PCR analysis that Lec15 cells do not express DPM2 mRNA but express DPM3 mRNA at a wild-type level (data not shown). It would appear that an endogenous level of DPM3 protein generated by this endogenous DPM3 mRNA in Lec15 cells is not sufficient for DPM1 to express a detectable level of DPM synthase activity. We transiently transfected Lec15 cells with DPM3 cDNA driven by an SRα promoter to force a high level of expression. For this, we used Lec15.B5 cells in which a cDNA of human CD59, a reporter GPI-anchored protein, was stably transfected. CD59 was not expressed on Lec15.B5 cells due to a lack of GPI caused by defective synthesis of DPM. If DPM is synthesized, GPI would be synthesized and, in turn, CD59 would be GPI anchored and expressed on the cell surface. Transfection of DPM3 cDNA into Lec15.B5 cells induced the surface expression of CD59 at a level similar to that induced by transfection of DPM2 cDNA, indicating restoration of DPM synthase activity by DPM3 in the absence of DPM2 (Figure 6).
Fig. 6. Restoration of the surface expression of GPI-anchored proteins on Lec15 cells by DPM3. Lec15.B5 cells transfected with DPM3 cDNA (solid line in A), DPM2 cDNA (solid line in B) and an empty vector (broken lines) were stained for CD59.
To correlate the amounts of DPM1 protein and DPM synthase activity in the presence and absence of DPM2, we transfected Lec15 cells with a cDNA of GST-tagged DPM1 in combination with DPM2 or DPM3 cDNA, and assessed expression levels of GST-tagged DPM1 and DPM synthase activity. Expression of GST-tagged DPM1 was normalized by expression of GST-tagged ALDH in the same cells (Figure 7A). DPM synthase activity was normalized by dolichol-phosphate-glucose (Dol-P-Glc) synthase activity of the same cells (Figure 7B). The amount of DPM1 protein in DPM2 transfectants was about one-third of that in DPM3 transfectants (Figure 7A), whereas DPM synthase activity was three times higher in DPM2 transfectants than in DPM3 transfectants (Figure 7B). Therefore, the activity of DPM synthase per amount of GST-tagged DPM1 was ∼10 times higher in the presence than in the absence of DPM2 (Figure 7C), indicating that DPM2 plays a role in the enzymatic reaction. The difference in the enzyme activity in the presence and absence of DPM2 was similar even at 6 µM GDP-mannose, ∼10 times the Km, indicating that the affinity for GDP-mannose is not responsible for the increase in enzyme activity (data not shown).
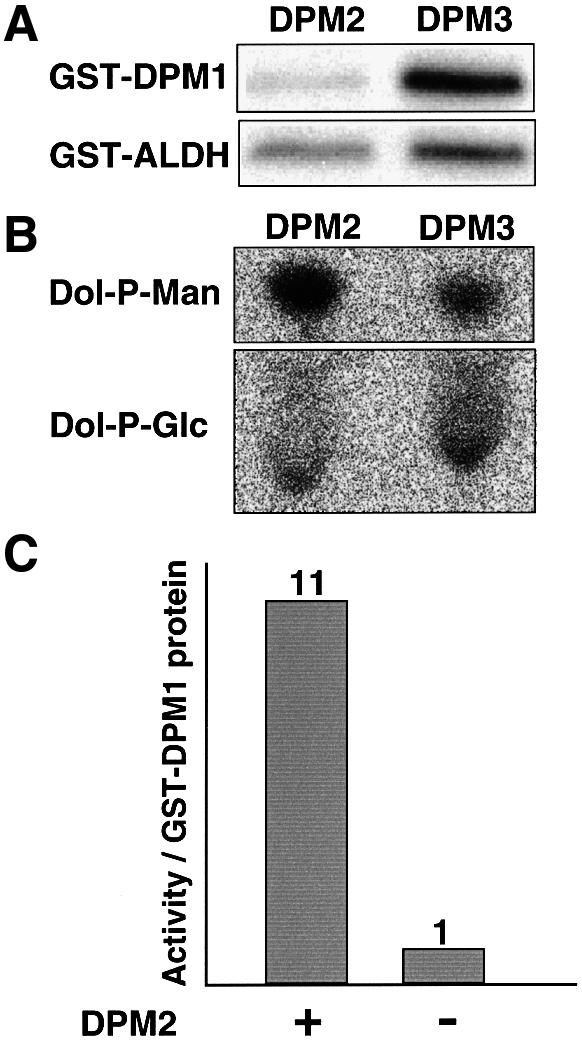
Fig. 7. Enzyme with DPM2 had a higher specific activity. A mixture of cDNAs of GST-tagged DPM1 and GST-tagged ALDH was co-transfected into Lec15 cells with DPM2 cDNA and DPM3 cDNA. The microsomal membranes were prepared 2 days after transfection and used for measuring the amount of GST-tagged proteins (A) and DPM and Dol-P-Glc synthase activities (B). The intensity of the DPM spot divided by that of the Dol-P-Glc spot was taken as DPM synthase activity. The intensity of the band of GST–DPM1 divided by that of the band of GST–ALDH was taken as the protein level of DPM1. The specific activity of DPM synthase was determined by dividing DPM synthase activity by the protein level of DPM1 (C).
Schizosaccharomyces pombe has a DPM synthase similar to the human counterpart
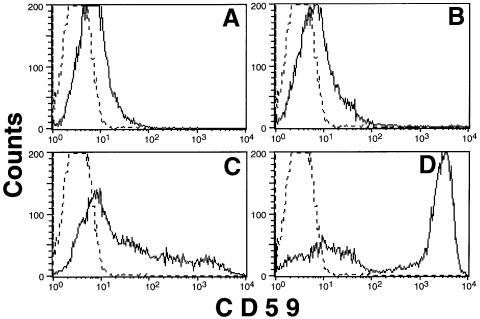
Schizosaccharomyces pombe has three proteins, SpDPM1 (GenBank accession No. AF007873), SpDPM2 (GenBank accession No. AL121794, gene ID SPBC21B10.11) and SpDPM3 (Figure 2B), that resemble human DPM1, DPM2 and DPM3, respectively. It was reported that SpDPM1 is a component of DPM synthase (Colussi et al., 1997). To test whether SpDPM3 is a functional homologue of human DPM3, we cloned it into a mammalian expression vector and transfected it into Lec15.B5 cells (Figure 8). Transfection of SpDPM3 partially restored the surface expression of GPI-anchored protein CD59 (Figure 8C), whereas transfection of an empty vector had no effect (Figure 8A), indicating that SpDPM3 had a weak activity in stabilizing hamster DPM1 in the absence of DPM2. Transfection of SpDPM1 into Lec15 cells did not cause significant surface expression of CD59 (Figure 8B), but co-transfection of SpDPM1 and SpDPM3 caused full surface expression of CD59 (Figure 8D). Therefore, SpDPM3 must have stabilized SpDPM1 in Lec15 cells. Transfection of SpDPM2 had no effect (data not shown), suggesting that SpDPM2 is not compatible with the mammalian system.
Fig. 8. Schizosaccharomyces pombe DPM3 is functional in mammalian cells. Lec15.B5 cells were transiently transfected with an empty vector (A), SpDPM1 (B), SpDPM3 (C) and SpDPM1 plus SpDPM3 (D). Two days later, the cells were stained with anti-CD59 antibody (solid lines) or the second reagent only (broken lines).
Discussion
In this study, we have demonstrated that human DPM synthase consists of three subunits, namely DPM1, DPM2 and DPM3. Mutations of DPM1 and DPM2 are responsible for defective biosynthesis of DPM in two mutant cell lines, Thy-1– class E cells and CHO Lec15 cells, respectively (Maeda et al., 1998; Tomita et al., 1998). There is no genetic evidence that DPM3 is necessary for DPM biosynthesis due to a lack of mutant cells defective in DPM3; however, we have shown that DPM3 stabilizes DPM1 and that DPM3 is itself stabilized by DPM2. A complex of DPM1, DPM2 and DPM3, when incorporated into liposomes, showed DPM synthase activity with a Km for GDP-mannose comparable to that of DPM synthase purified from rat liver. We concluded, therefore, that DPM1, DPM2 and DPM3 are necessary for the generation of human DPM synthase.
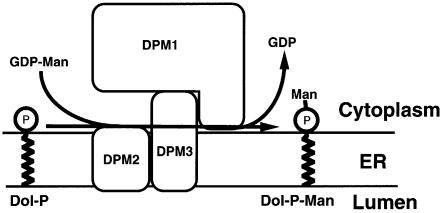
Figure 9 shows a schematic representation of a structural model of human DPM synthase. DPM2, a very hydrophobic protein of 84 amino acids, associates with the N-terminal hydrophobic portion (∼60 amino acids) of DPM3. DPM3, via its C-terminal hydrophilic portion (∼30 amino acids), associates with DPM1 (260 amino acids). The level of DPM3 was five times higher in the presence than in the absence of DPM2. The level of DPM1 was enhanced 10-fold by overexpression of DPM3. These results suggest that the DPM2-dependent stabilization of DPM1 reported previously by us (Maeda et al., 1998) was an indirect effect mediated by DPM3. When DPM2 was overexpressed in Lec15 cells, the level of DPM1 was enhanced only 3- to 4-fold (Figure 5A). It is likely that the endogenous level of DPM3 was limiting.
Fig. 9. Schematic representation of human DPM synthase. Hydrophobic DPM2 associates with the N-terminal hydrophobic portion of DPM3. The hydrophilic C-terminal portion of DPM3 associates with catalytic DPM1, which is hydrophilic.
We reported that the C-terminal 24 amino acids of DPM1 are necessary for association with DPM2 and that Phe21 and Tyr23 in DPM2 are important for association with DPM1 (Maeda et al., 1998). Because the present results suggest that DPM2 associates with DPM1 indirectly through DPM3, these previous observations should be interpreted as indicating that the C-terminal 24 amino acids of DPM1 are necessary for association with DPM3 and that DPM2 associates with DPM3 using its first transmembrane domain, which includes Phe21 and Tyr23.
DPM1 stabilized with DPM3 in the absence of DPM2 had DPM synthase activity, indicating that DPM2 is not essential for the enzymatic reaction. However, the specific enzymatic activity was 10 times higher when DPM2 was present (Figure 7). Therefore, DPM2 plays a role in the enzymatic reaction. Since DPM2 is very hydrophobic, it seems possible that DPM2 facilitates usage of dolichol-phosphate. Alternatively, association of DPM2 might cause a steric change in DPM3 that leads to enhanced enzyme activity.
The fission yeast S.pombe has three genes whose products resemble human DPM1, DPM2 and DPM3. SpDPM3 was compatible with mammalian DPM1, causing partial restoration of DPM synthesis in Lec15 cells. co-expression of SpDPM1 and SpDPM3, but not expression of SpDPM1 alone, in Lec15 cells restored the full expression of GPI-anchored proteins, indicating that SpDPM3 stabilized SpDPM1. SpDPM2 was incompatible with the mammalian system, so we have no evidence that SpDPM2 is in fact functional as a subunit of DPM synthase.
In contrast to human and S.pombe, S.cerevisiae has a single-component DPM synthase. It is not clear why a basic enzyme such as DPM synthase has diverged into two groups. It is particularly interesting that two yeasts, S.cerevisiae and S.pombe, have different types of the enzyme. The multicomponent system should be suitable for regulation. Schizosaccharomyces pombe and/or its ancestral yeasts might have required or might require regulation of DPM synthesis under certain conditions.
Deficiency of DPM synthase causes CDG type Ie (Imbach et al., 2000; Kim et al., 2000). Four patients with mutations in DPM1 have been reported. DPM1 resides on chromosome 20q13 (Kinoshita et al., 1997). All four lost wild-type DPM1 alleles but had one or two copies of a common mutant DPM1 allele with a point mutation that causes an amino acid substitution. Owing to this mutant allele, some DPM synthase activity remained (Imbach et al., 2000; Kim et al., 2000). Perhaps complete deficiency of DPM would cause embryonic lethality because systemic deficiency of GPI anchor causes lethality during embryogenesis in mice (Nozaki et al., 1999). Because both DPM2 and DPM3 are involved in the stability of DPM1, a defect in DPM2 or DPM3 would also cause reduced expression of DPM1, resulting in DPM synthase deficiency and CDG. DPM2 and DPM3 reside on chromosomes 9q33 (our unpublished data) and 1q12–21, respectively. Their mutations would, therefore, be inherited in an autosomal recessive manner.
Materials and methods
Cells and antibodies
Wild-type CHO-K1 and Lec15 (Lec15.2) mutant cells (gifts from Dr M.A.Lehrman, Texas Southwestern Medical Center, Dallas, TX) (Camp et al., 1993) were cultured in Ham’s F-12 medium supplemented with 10% fetal calf serum (FCS). Lec15.B5 cells, and Lec15 stably transfected with cDNAs of human DAF and CD59 (Maeda et al., 1998) were maintained in 600 µg/ml G-418. Human B-lymphoblastoid JY25 cells (a gift from Dr M.L.Shin, University of Maryland, Baltimore, MD) (Hollander et al., 1988) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS. Anti-CD59 monoclonal antibody 5H8 was a gift from Dr M.Tomita (Showa University, Tokyo) (Sugita et al., 1994).
Plasmids
pME-Py, pME-Py-rDPM2, pME-Py-hDPM1, pME-Py-GST-hDPM1, pME-Py-GST-ALDH, pGL3-FLAG-rDPM2 and pGL3-FLAG-rDPM2FY/LS were described previously (Maeda et al., 1998). Human SL15 cDNA was found in the EST database by searching with the hamster SL15 sequence, and sequences of clones with GenBank accession Nos H27246 and H77811 were determined. The DDBJ/EMBL/GenBank accession No. of human SL15 cDNA is D86918. pME-Py-hSL15-FLAG was generated by inserting a sequence encoding FLAG amino acids RDYKDDDDK before the sequence of the last seven amino acids of human SL15. This tagged SL15 cDNA restored usage of DPM in CHO Lec35 mutant cells (Camp et al., 1993), indicating that it expresses functional SL15. pME-Py-hDPM3 was constructed by subcloning human DPM3 cDNA from an EST clone (GenBank accession No. R97442) into pME-Pyori18Sf– expression vector (Ohishi et al., 1996). In human DPM3dC, the last 27 amino acids were replaced by the dipeptide Thr–Arg followed by a stop codon. To fuse FLAG at the N-termini of hDPM3 and hDPM3dC, and GST at the N-terminus of hDPM2, we used pME-Py-GST-PIG-A and pME-Py-FLAG-PIG-A, both of which have a SalI site that connects the tags with PIG-A (Maeda et al., 1998). pME-Py-FLAG-hDPM3, pME-Py-FLAG-hDPM3dC and pME-Py-GST-hDPM2 were obtained by replacing PIG-A of pME-Py-FLAG-PIG-A and pME-Py-GST-PIG-A with the coding regions of the respective genes. In these plasmids, FLAG and GST tag sequences were followed by a SalI site (GTCGAC, which is translated into valine and aspartate) and then ACC in the case of DPM3 and DPM3dC, or TCC in the case of DPM2. pMEEB-GST-FLAG-hDPM1 (GFD1) was constructed by replacing PIG-A of pMEEB-GST-PIG-A (Watanabe et al., 1998) with an XhoI–SpeI fragment from pME-Py-FLAG-hDPM1 by subcloning it into SalI–SpeI sites of pMEEB-GST-PIG-A. This resulted in a fusion protein bearing an insertion of six amino acids, Val-Glu-Val-Ser-Thr-Met, between GST and FLAG-hDPM1. pME-Py-GST-rPIG-L was constructed by subcloning the GST-rPIG-L region from pMEEB-GST-rPIG-L (Nakamura et al., 1997) into pME-Py. pSG5-GST-hDPM2 was constructed by subcloning GST-hDPM2 from pME-Py-GST-hDPM2 into pSG5 expression vector (Stratagene). pSG5-GST-hDPM2, which uses an SV40 promoter and induces a lower expression level than pME-Py-GST-hDPM2, was used to obtain expression of hDPM2 comparable with that of other proteins. Coding sequences of SpDPM1, SpDPM2 and SpDPM3 were amplified by PCR from an S.pombe cDNA library (a gift from Dr H.Nojima, Osaka University) using primers designed from their nucleotide sequences obtained from GenBank and cloned into pME-Pyori18Sf–.
Transfection
CHO, Lec15 and Lec15.B5 cells (5 × 106 cells in 0.4 ml of culture medium) were electroporated at 260 V and 960 µF. JY25 cells (107) suspended in 0.8 ml of HEPES-buffered saline with 20 µg of DNA were electroporated at 250 V and 960 µF. Electroporations were carried out in a Gene Pulser (Bio-Rad).
Purification of DPM synthase complex and cloning of DPM3 cDNA
JY25 cells were transfected with pMEEB-GFD1, selected and maintained in 200 µg/ml hygromycin. A sample of 5 × 109 cells was solubilized with 360 ml of lysis buffer A [1% digitonin, 20 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 2 µg/ml leupeptin and 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (pABSF)] at 4°C for 6 h. After removal of insoluble material by centrifugation at 18 800 g for 5 min twice, the soluble fraction was mixed with 0.32 ml of M2 anti-FLAG beads (Sigma) and agitated overnight. The beads were collected and washed with 10 ml of lysis buffer A four times, and the bound proteins were eluted with 0.3 ml of 1 mg/ml FLAG peptide in buffer A four times. These eluates were collected, pre-cleared with 80 µl of Sepharose CL-6B (Pharmacia) and incubated with 50 µl of glutathione beads (Pharmacia) for 3 h. Then the beads were washed with 0.5 ml of buffer A three times and precipitates were eluted with 60 µl of buffer B (1% digitonin, 50 mM Tris–HCl pH 9.5, 150 mM NaCl and 20 mM reduced glutathione). The eluted proteins were subjected to 15–25% gradient SDS–PAGE and transferred to PVDF membrane (ProBlott, Applied Biosystems Inc.). The N-terminal sequences of the 15 and 8 kDa bands were determined with a 494 sequencer (Applied Biosystems Inc.). Based on the protein sequence, we found several sequences of human, rat and C.elegans DPM3 homologues in the EST database using the Basic Local Alignment Search Tool (Altschul et al., 1990) and determined the sequence of a human DPM3 clone (ID 199465) from the I.M.A.G.E. Consortium (GenBank accession No. R97442) (Lennon et al., 1996).
Reconstitution of DPM synthase complex in liposomes
A mixture of 3.9 mg of egg yolk phosphatidylcholine and 9.1 mg of phosphatidylethanolamine (Sigma) mixed in 1 ml of chloroform was dried under an N2 stream, dissolved in 6.5 ml of buffer consisting of 50 mM HEPES–NaOH pH 7.4, 25 mM KCl and 0.5 mM EDTA under sonication in a bath (UT-53N, SHARP, Japan) for 5 min and stored at –80°C. The lipids were thawed, sonicated and used as liposomes. From 3 × 109 JY25 cells transfected with pMEEB-GFD1, DPM synthase complex was purified as described above. Before the elution with buffer B, about half of the glutathione beads were suspended in 1.5 ml of a buffer containing 2 mg/ml liposomes and 20 mM reduced glutathione, and agitated for 3 h at 4°C. The liposomes were separated from glutathione beads using nylon mesh and stored at –80°C. DPM synthase activities were measured as described below using liposomes containing 100 µg of lipid and normal JY25 membranes containing 100 µg of protein. The rest of the glutathione beads were eluted with buffer B and the eluted solution was subjected to 15–25% gradient SDS–PAGE followed by silver staining.
Production of anti-human DPM3 antibodies in rabbits
We immunized rabbits with a recombinant fusion protein consisting of GST and the 35 C-terminal amino acids of human DPM3. A cDNA of the C-terminal portion of human DPM3 was cloned after amplification by PCR using, as 5′ primer, 5′-CGCCGGATCCGGCTATCGTGTGGCCACTTTTC, and as 3′ primer, 5′-GTGGTCTAGAAACTGGCTCTTTAATGGGAAATGG, and fused with the GST sequence in a pGEX4T-3 expression vector (Pharmacia). The fusion protein expressed in E.coli DH5α was purified with glutathione beads and injected into rabbits. After three immunizations, anti-GST-hDPM3 serum was obtained. The anti serum but not the pre-immune serum reacted with FLAG-tagged hDPM3 on western blotting.
Isolation of subcellular fractions
CHO and Lec15 cells (3 × 107) were electroporated with 60 µg of pME-Py-hDPM3 and 15 µg of pME-Py-FLAG-ALDH in three cuvettes. After culture for 2 days, the cells were suspended in 3 ml of buffer containing 0.25 M sucrose, 10 mM HEPES–NaOH pH 7.5, 1 mM dithiothreitol (DTT), 1 mM pABSF and 2 µg/ml leupeptin, disrupted by a Dounce homogenizer (Wheaton, type A) with 60 strokes and treated with 2 U/ml DNase for 20 min at 4°C. After centrifugation at 10 000 g for 15 min at 4°C, the post-nuclear supernatants were fractionated by discontinuous sucrose density gradient centrifugation and marker enzyme activities in fractions were measured as described previously (Maeda et al., 1998). From each fraction, FLAG-tagged ALDH and hDPM3 were immunoprecipitated with anti-FLAG beads and anti-DPM3 antiserum plus protein G beads (Pharmacia). The precipitates were analysed by SDS–PAGE/western blotting as described below.
Analysis of the protein complexes
Lec15 cells were co-transfected with 12 µg each of pME-Py-FLAG-hDPM1, -rDPM2, -rDPM2FY/LS and pME-Py-hSL15-FLAG, and 12 µg of pME-Py-hDPM3, cultured for 2 days and solubilized with lysis buffer A. After removal of insoluble material by centrifugation at 18 000 g for 5 min and at 100 000 g for 1 h, the soluble fraction was mixed with anti-DPM3 antiserum followed by protein G beads or anti-FLAG beads and agitated for 2 h. The mixture was separated into the unbound fraction (supernatant) and immunoprecipitates by centrifugation at 4000 g for 2 min. To collect unbound proteins, the supernatant was then incubated with anti-FLAG beads or anti-DPM3 antiserum plus protein G beads. Both precipitates were washed with the lysis buffer, eluted with a sample buffer and analysed by SDS–PAGE/western blotting against biotinylated M2 anti-FLAG antibody followed by horseradish peroxidase (HRP)-conjugated streptavidin (Amersham Life Science) or anti-DPM3 antiserum followed by HRP-conjugated anti-rabbit IgG antibody (Amersham) plus chemiluminescence reactions (Dupont). In some cases, the membranes were incubated in a buffer (62.5 mM Tris–HCl pH 6.8, 2% SDS and 0.1 M 2-mercaptoethanol) at 50°C for 30 min to remove bound antibodies for reprobing.
In some cases, cDNAs of GST- and FLAG-tagged proteins were co-transfected into Lec15 cells. Their lysates were immunoprecipitated with anti-FLAG beads. The unbound fraction was mixed with glutathione beads and precipitates were collected. Both precipitates were analysed by SDS–PAGE/western blotting against goat anti-GST antibody (Pharmacia) followed by HRP-conjugated anti-goat IgG antibody (Organon Teknika).
Assay of DPM and Dol-P-Glc synthases
Lec15 cells (5 × 106) were transfected with 10 µg each of pME-Py-GST-hDPM1 and pME-Py-GST-ALDH, and 7 µg of pME-Py, pME-Py-rDPM2 or pME-Py-hDPM3, cultured for 2 days and then incubated in a medium containing 5 µg/ml tunicamycin (Sigma) for 2 h. They were harvested, destroyed by incubation in a hypotonic buffer (20 mM HEPES–NaOH pH 7.4, 2 µg/ml leupeptin and 1 mM pABSF) on ice for 20 min and by a Teflon homogenizer, and centrifuged at 100 000 g for 1 h to collect membranes. Half of the pellet was used for assays of DPM synthase and Dol-P-Glc synthase, and the rest for detection of GST–DPM1 by western blotting. Assays of DPM and Dol-P-Glc synthases were performed without exogenous Dol-P and with 5 min incubation at 37°C in 0.2 ml of reaction buffer containing 1 µCi of GDP-[3H]mannose or UDP-[3H]glucose, respectively (American Radiolabeled Chemicals), and 5 µg/ml tunicamycin. Then lipids were extracted twice with 0.4 ml of water-saturated n-butanol. Pooled butanol extracts were backwashed once with 0.4 ml of butanol-saturated water and then evaporated. The dried materials were extracted with 30 µl of chloroform/methanol (2:1) and the extracts were separated by thin-layer chromatography on Kieselgel 60 (Merk) with a solvent system of chloroform/methanol/H2O (10:10:3). The radiolabelled lipids were analysed by Image Analyzer BAS 1500 (Fuji Film Co., Tokyo) after 2–4 days exposure.
Acknowledgments
Acknowledgements
We thank Reika Watanabe, Norimitsu Inoue and Kazuhito Ohishi for discussion, and Keiko Kinoshita for technical assistance. This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan.
References
- Abeijon C. and Hirschberg,C.B. (1992) Topography of glycosylation reactions in the endoplasmic reticulum. Trends Biochem. Sci., 17, 32–36. [DOI] [PubMed] [Google Scholar]
- Adams M.D. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1656. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Camp L.A., Chauhan,P., Farrar,J.D. and Lehrman,M.A. (1993) Defective mannosylation of glycosylphosphatidylinositol in Lec35 Chinese hamster ovary cells. J. Biol. Chem., 268, 6721–6728. [PubMed] [Google Scholar]
- Colussi P.A., Taron,C.H., Mack,J.C. and Orlean,P. (1997) Human and Saccharomyces cerevisiae dolichol phosphate mannose synthases represent two classes of the enzyme, but both function in Schizosaccharomyces pombe.Proc. Natl Acad. Sci. USA, 94, 7873–7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucey M.A., Hess,D., Cacan,R. and Hofsteenge,J. (1998) Protein C-mannosylation is enzyme-catalysed and uses dolichyl-phosphate-mannose as a precursor. Mol. Biol. Cell, 9, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund P.T. (1993) The structure and biosynthesis of glycosyl phosphatidylinositol protein anchors. Annu. Rev. Biochem., 62, 121–138. [DOI] [PubMed] [Google Scholar]
- Herscovics A. and Orlean,P. (1993) Glycoprotein biosynthesis in yeast. FASEB J., 7, 540–550. [DOI] [PubMed] [Google Scholar]
- Hirschberg C.B. and Snider,M.D. (1987) Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem., 56, 63–87. [DOI] [PubMed] [Google Scholar]
- Hollander N., Selvaraj,P. and Springer,T.A. (1988) Biosynthesis and function of LFA-3 in human mutant cells deficient in phosphatidylinositol-anchored proteins. J. Immunol., 141, 4283–4290. [PubMed] [Google Scholar]
- Ilgoutz S.C., Zawadzki,J.L., Ralton,J.E. and McConville,M.J. (1999) Evidence that free GPI glycolipids are essential for growth of Leishmania mexicana. EMBO J., 18, 2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbach T. et al. (2000) Deficiency of dolichol-phosphate-mannose synthase-1 causes congenital disorder of glycosylation type Ie. J. Clin. Invest., 105, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J.W. and Schutzbach,J.S. (1985) Activation of dolichyl-phospho-mannose synthase by phospholipid. Eur. J. Biochem., 153, 41–48. [DOI] [PubMed] [Google Scholar]
- Kim S., Westphal,V., Srikrishna,G., Mehta,D.P., Peterson,S., Filiano,J., Karnes,P.S., Patterson,M.C. and Freeze,H.H. (2000) Dolichol phosphate mannose synthase (DPM1) mutations define congenital disorder of glycosylation Ie (CDG-Ie). J. Clin. Invest., 105, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Ohishi,K. and Takeda,J. (1997) GPI-anchor synthesis in mammalian cells: genes, their products and a deficiency. J. Biochem., 122, 251–257. [DOI] [PubMed] [Google Scholar]
- Kornfeld R. and Kornfeld,S. (1985) Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem., 54, 631–664. [DOI] [PubMed] [Google Scholar]
- Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lennon G.G., Auffray,C., Polymeropoulos,M. and Soares,M.B. (1996) Molecular analysis of genomes and their expression. Genomics, 33, 151–152. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Tomita,S., Watanabe,R., Ohishi,K. and Kinoshita,T. (1998) DPM2 regulates biosynthesis of dolichol phosphate-mannose in mammalian cells: correct subcellular localization and stabilization of DPM1 and binding of dolichol phosphate. EMBO J., 17, 4920–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki R., Yamamoto,A. and Tashiro,Y. (1994) Microsomal aldehyde dehydrogenase is localized to the endoplasmic reticulum via its carboxyl-terminal 35 amino acids. J. Cell Biol., 126, 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazhari-Tabrizi R., Eckert,V., Blank,M., Mueller,R., Mumberg,D., Funk,M. and Schwarz,R.T. (1996) Cloning and functional expression of glycosyltransferases from parasitic protozoans by heterologous complementation in yeast: the dolichol phosphate mannose synthase from Trypanosoma brucei brucei.Biochem. J., 316, 853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Inoue,N., Watanabe,R., Takahashi,M., Takeda,J., Stevens,V.L. and Kinoshita,T. (1997) Expression cloning of PIG-L, a candidate N-acetylglucosaminyl-phosphatidylinositol deacetylase. J. Biol. Chem., 272, 15834–15840. [DOI] [PubMed] [Google Scholar]
- Nozaki M., Ohishi,K., Yamada,N., Kinoshita,T., Nagy,A. and Takeda,J. (1999) Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab. Invest., 79, 293–299. [PubMed] [Google Scholar]
- Ohishi K., Kurimoto,Y., Inoue,N., Endo,Y., Takeda,J. and Kinoshita,T. (1996) Cloning and characterization of the murine GPI anchor synthesis gene Pigf, a homologue of the human PIGF gene. Genomics, 34, 340–346. [DOI] [PubMed] [Google Scholar]
- Orlean P., Albright,C. and Robbins,P.W. (1988) Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem., 263, 17499–17507. [PubMed] [Google Scholar]
- Schutzbach J.S., Zimmerman,J.W. and Forsee,W.T. (1993) The purification and characterization of recombinant yeast dolichyl-phosphate-mannose synthase. Site-directed mutagenesis of the putative dolichol recognition sequence. J. Biol. Chem., 268, 24190–24196. [PubMed] [Google Scholar]
- Sugita Y., Ito,K., Shiozuka,K., Suzuki,H., Gushima,H., Tomita,M. and Masuho,Y. (1994) Recombinant soluble CD59 inhibits reactive haemolysis with complement. Immunology, 82, 34–41. [PMC free article] [PubMed] [Google Scholar]
- Tomita S., Inoue,N., Maeda,Y., Ohishi,K., Takeda,J. and Kinoshita,T. (1998) A homologue of Saccharomyces cerevisiae Dpm1p is not sufficient for synthesis of dolichol-phosphate-mannose in mammalian cells. J. Biol. Chem., 273, 9249–9254. [DOI] [PubMed] [Google Scholar]
- Ware F.E. and Lehrman,M.A. (1996) Expression cloning of a novel suppressor of the Lec15 and Lec35 glycosylation mutations of Chinese hamster ovary cells. J. Biol. Chem., 271, 13935–13938. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Inoue,N., Westfall,B., Taron,C.H., Orlean,P., Takeda,J. and Kinoshita,T. (1998) The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J., 17, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman J.W., Specht,C.A., Cazares,B.X. and Robbins,P.W. (1996) The isolation of a Dol-P-Man synthase from Ustilago maydis that functions in Saccharomyces cerevisiae.Yeast, 12, 765–771. [DOI] [PubMed] [Google Scholar]