Abstract
Plastid DNA, like bacterial and mitochondrial DNA, is organized into protein–DNA complexes called nucleoids. Plastid nucleoids are believed to be associated with the inner envelope in developing plastids and the thylakoid membranes in mature chloroplasts, but the mechanism for this re-localization is unknown. Here, we present the further characterization of the coiled-coil DNA-binding protein MFP1 as a protein associated with nucleoids and with the thylakoid membranes in mature chloroplasts. MFP1 is located in plastids in both suspension culture cells and leaves and is attached to the thylakoid membranes with its C-terminal DNA-binding domain oriented towards the stroma. It has a major DNA-binding activity in mature Arabidopsis chloroplasts and binds to all tested chloroplast DNA fragments without detectable sequence specificity. Its expression is tightly correlated with the accumulation of thylakoid membranes. Importantly, it is associated in vivo with nucleoids, suggesting a function for MFP1 at the interface between chloroplast nucleoids and the developing thylakoid membrane system.
INTRODUCTION
DNA-binding and membrane-anchoring proteins play an important role in condensation and spatial organization of both eukaryotic and prokaryotic genomes. In the eukaryotic nucleus, chromosome condensation is mainly achieved by binding of histones and HMG proteins to DNA. Interactions of chromatin with nuclear envelope-associated proteins and possibly an internal nuclear matrix provide spatial organization (1). In bacteria, histone-like HU proteins are involved in chromosome condensation (2,3), and association with the plasma membrane provides a spatial anchor for the genome (4). In mitochondria, the HMG-like protein Abf2p is involved in chromosome stability and packaging (5). The mitochondrial inner membrane protein Yhm2p binds DNA and is associated with mitochondrial nucleoids (6), indicative of a spatial organization of the mitochondrial chromosome via membrane attachment too.
By comparison, little is known about the packaging and spatial organization of chloroplast genomes. Plastid DNA, like bacterial and mitochondrial DNA, is organized into protein–DNA complexes termed nucleoids. An HU-like protein with DNA-compacting activity has been found in the red algae Cyanidioschyzon merolae but no homolog of this protein seems to be present in higher plants (7). Resolving the protein pattern of higher-plant chloroplast nucleoids typically shows 20–50 prominent proteins, only few of which have been identified. They include CND41 from tobacco (8), PEND from pea (9) and DPC68 (10,11) recently shown to be ferredoxin:: sulfite reductase (SiR) (12,13).
In analogy to prokaryotic and mitochondrial genomes, microscopic evidence supports the association of nucleoids with chloroplast membranes. In red algae, nucleoids are found in the chloroplast center, surrounded by thylakoids. In brown algae, a large, ring-shaped nucleoid is found within a thylakoid lamella located parallel to and just beneath the chloroplast envelope (7). In higher plants, the distribution of nucleoids changes during plastid development. In the pea proplastid, a single small nucleoid is located at the center of the plastid. Nucleoids in developing plastids are localized close to the plastid envelope. Finally, in mature chloroplasts, they are located at the center of the chloroplast, in close vicinity to the thylakoids (9,14). A similar pattern of reorganization has also been shown for wheat chloroplasts (15). Liu and Rose (16) have shown that nucleoids co-fractionate with the thylakoids and that a region of the chloroplast DNA is bound to the thylakoids.
The PEND protein is a candidate for anchoring nucleoids to the inner plastid envelope at early stages of development (9). PEND is an unusual bZIP protein that binds to plastid DNA in a sequence-specific manner and is associated with the inner envelope membrane. Its expression correlates with the early stage of plastid development at which the nucleoids are found associated with the plastid envelope (17). In contrast, no information exists about possible DNA-binding proteins associated with the thylakoid membranes and expressed in fully developed chloroplasts.
We have previously identified the nuclear-encoded protein MFP1 in tomato by its ability to bind to matrix attachment region DNA (18). MFP1 is a large coiled-coil protein with a C-terminal DNA-binding domain and a predicted N-terminal transmembrane domain. It was shown to be associated with globular structures of unknown identity at the nuclear rim of tobacco suspension culture cells (19). We show here that the MFP1-containing structures are the proplastids of tobacco suspension culture cells, which can be tightly associated with the nuclear envelope. MFP1 is targeted to plastids both in suspension culture cells and in leaves and is located in the thylakoid membranes, with the C-terminal DNA-binding domain oriented towards the stroma. MFP1 has a major DNA-binding activity in Arabidopsis chloroplasts and binds to several regions of the chloroplast DNA with equal affinity. Its expression is tightly correlated with the accumulation of thylakoid membranes, and it is in vivo associated with nucleoids. These findings present MFP1 as the first candidate for a protein involved in anchoring nucleoids to the thylakoid membrane system in fully developed chloroplasts.
MATERIALS AND METHODS
Prediction programs
For chloroplast transit peptide prediction, the programs ChloroP (20), TargetP (21) and PSORT (22) were used. The program SignalP (23,24) was used for predicting the thylakoid signal peptide. Coiled-coil domains and transmembrane helices were predicted using MultiCoil (25) and HMMTOP (26), respectively. The BLAST algorithm (27) and CDD search and the PROSITE database (28) were used for sequence homology and domain searches. Putative MAR sequences were predicted using SMARTest (http://genomatix.gsf.de).
Plant material
If not indicated otherwise, Arabidopsis ecotype Columbia was used. Seedlings were grown on MS (29) medium supplemented with B5 vitamins, 2% (w/v) sucrose and 0.8% (w/v) agar at 22°C either under continuous white light or in the dark. For chloroplast extraction, Nicotiana tabacum and Arabidopsis ecotype WS plants were grown in soil under 16 h of light and 8 h of dark at 22°C for 2 months and 6 weeks, respectively. Lycopersicon esculentum var. ‘Early Girl’ was grown under greenhouse conditions. Tobacco BY-2 suspension cells were cultured as described (30).
AtMFP1 protein expression and antibody production
For the expression of recombinant AtMFP1, a 1775 bp fragment [nucleotide 435–2184 of the AtMFP1 cDNA (31)], encoding Δ144AtMFP1, was used to avoid expression problems caused by the hydrophobic N-terminus (18). Cloning and protein expression were performed using the Affinity LIC Cloning and Protein Purification Kit (Stratagene, La Jolla, CA). Purified protein was stored in protein binding buffer (20 mM HEPES, pH 7.8, 40 mM NaCl, 0.2 mM EDTA and 1 mM DTT) with 20% glycerol at –80°C. Rabbit antiserum (OSU91) was produced by Cocalico Biologicals Inc. (Reamstown, PA).
Protein extraction from plant tissues
Total protein extracts were prepared according to Harder et al. (31) using 2 µl/mg extraction buffer [62.5 mM Tris–HCl, pH 6.8, 20% (w/v) glycerol, 4% (w/v) SDS and 1.4 M 2-mercaptoethanol] for Arabidopsis tissues and 1 µl/mg for tomato pericarp.
Immunoblots
Antibody dilutions were 1:3000 for 288 (18), 1:5000 for OSU91, 1:1000 for anti-maize OEC33 (32), 1:500 for anti-Arabidopsis ferredoxin::sulfite reductase (13), 1:250 for anti-pea Cytochrome f (33), 1:200 for anti-β-tubulin (Sigma, St Louis, MO) and 1:10 000–1:20 000 for horseradish peroxidase-coupled donkey anti-rabbit and sheep anti-mouse antibodies. Immunoblotting was done essentially as described (34). ECL detection was as described by the manufacturer (Amersham Pharmacia Biotech, Uppsala, Sweden). Membranes were stripped using Restore Western Blot Stripping Buffer (Pierce, Rockford, IL).
Chloroplast fractionation
Chloroplast isolation was performed according to Maliga et al. (35), chloroplast and thylakoid fractionations according to Peltier et al. (36). For high-salt or high-pH washes, the thylakoid pellet was incubated in 1.0 M NaCl or 0.1 M Na2CO3, pH 11.5 on ice for 30 min. Thylakoids were washed once and recovered in the original volume of lysis buffer.
Thermolysin treatment
Intact chloroplasts in 1× HSM buffer (50 mM HEPES, 0.33 M sorbitol, 8.4 mM methionine) were centrifuged at 1000 g for 5 min at 4°C and the pellet was resuspended in lysis buffer (50 mM Tris–HCl, 5 mM MgCl2, pH 8.0). After incubation for 20 min on ice to rupture chloroplast membranes, thylakoids were recovered by centrifugation at 1000 g for 5 min at 4°C. The pellet was washed and resuspended in the same volume of thermolysin buffer (10 mM Tris–HCl, 10 mM CaCl2 and 0.2 M NaOH, pH 8.0) or thermolysin buffer plus 1% Triton X-100, with or without 100 µg/ml thermolysin (Type X protease; Sigma Aldrich, St Louis, MO). After incubation for 0, 2, 10 and 30 min at room temperature, the reactions were stopped by adding 5 µl of 0.5 M EDTA.
Fractionation of tobacco BY-2 cells and protein extraction
BY-2 protoplasts, nuclei and nuclear matrix were prepared as described (37). Plastids of BY-2 cells were isolated as described by Kapoor and Sugiura (38). BY-2 total protein was isolated from sedimented protoplasts by incubating for 10 min at 70°C in extraction buffer (see Protein extraction from plant tissues). Nuclei, nuclear matrix and plastid samples were concentrated by centrifugation at 4°C for 10 min at 3000 g and dissolved for 10 min at 70°C in extraction buffer prior to sample preparation for SDS–PAGE.
Isolation of chloroplast nucleoids
Nucleoid isolation from chloroplasts of N.tabacum was carried out essentially as described (10,11). Briefly, intact chloroplasts from 100 g of tobacco leaves were pelleted and resuspended in 30 ml of nucleoid isolation buffer [17% (w/v) sucrose, 20 mM Tris–HCl, 0.5 mM EDTA, 1.2 mM spermidine, 7 mM 2-mercaptoethaol, 1 mM PMSF, 5 µg/ml antipain, 5 µg/ml leupeptin, 1 µg/ml pepstatin, 1 µg/ml aprotinin, 1 µg/ml chymostatin, pH 7.6]. A 1/20 volume of 20% (v/v) Nonidet P-40 was added and stirred at 4°C for 30 min. The clarified solution was centrifuged for 10 min at 3000 g at 4°C. The supernatant was centrifuged at 48 000 g for 40 min at 4°C. This supernatant (S1) was kept on ice. The pellet was resuspended in 30 ml of nucleoid isolation buffer/2% Nonidet P-40 with a syringe and centrifuged at 48 000 g for 40 min at 4°C. The pellet was resuspended in 200 µl of nucleoid isolation buffer/2% Nonidet P-40, brought to 33% glycerol, and stored at –80°C. Two milliliters of the supernatant (S2) were precipitated with 10% TCA and the pellet was dissolved in 50 µl of protein loading buffer.
‘South-western’ DNA-binding experiments
‘South-western’ DNA-binding experiments were performed essentially as described (18). Binding reactions were performed overnight at 4°C with 35–50 ng/ml PCR probes and 50 μg/ml (Fig. 6C) or 10 µg/ml (Fig. 6D) sheared herring sperm DNA. For single-stranded DNA, the probes were boiled for 5 min and cooled on ice immediately before the binding reactions.
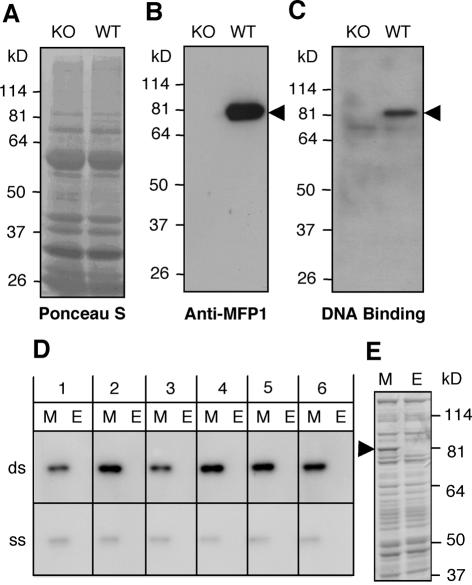
Figure 6.
AtMFP1 has a major DNA-binding activity in Arabidopsis leaf chloroplasts. (A–C) Total chloroplast protein from wild-type Arabidopsis (WT) and a T-DNA insertion line (KO) were separated by 10% SDS–PAGE and transferred to a nitrocellulose membrane. (A) Ponceau S staining. (B) Immunoblot with anti-AtMFP1 antibody OSU91. (C) ‘South-western’ blot probed with the Drosophila histone H1/H3 spacer MAR in the presence of 50 µg/ml sheared herring sperm DNA. Arrowheads in (B) and (C) show the expected position of AtMFP1. Molecular mass markers are given on the left. (D) ‘South-western’ DNA-binding experiment with recombinant AtMFP1. Escherichia coli lysate containing Δ144AtMFP1 (M) or E.coli control lysate (E) were resolved by 10% SDS–PAGE, blotted to nitrocellulose membranes and incubated with radiolabeled DNA fragments as indicated, in the presence of 10 µg/ml sheared herring sperm DNA. (Top) Double-stranded DNA probes (ds); (bottom) single stranded DNA probes (ss). Probes were (see Materials and Methods for details): 1, OriA fragment (48% AT); 2, OriB fragment (67% AT); 3, petA ORF (62% AT); 4, predicted chloroplast MAR (74% AT); 5, Drosophila histone H1/H3 spacer MAR (75% AT); 6, AtMFP1 gene fragment (56% AT). (E) Coomassie brilliant blue staining of lanes M and E shown in (D). The arrowhead indicates the position of Δ144AtMFP1.
Total RNA purification and RNA blot
Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Fifteen micrograms of total RNA were separated on a formaldehyde gel, transferred to Hybond-NX (Amersham Pharmacia Biotech) and hybridized essentially as described (34).
Southern dot blot of chloroplast and nucleoid DNA
DNA was extracted from tobacco chloroplasts with genomic DNA extraction buffer (0.2 M Tris–HCl, 0.25 M NaCl, 25 mM EDTA, 0.5% SDS, pH 7.5), followed by phenol/chloroform extraction and ethanol precipitation. Chloroplast nucleoids were extracted once with phenol/chloroform and ethanol-precipitated. Approximately 1 µg of denatured DNA (as estimated by ethidium bromide staining) was spotted on Hybond-NX, hybridized and subjected to autoradiography at room temperature for 24 h.
PCR probes
The PCR fragments listed in Table 1 were synthesized as probes for ‘South-western’ blots, RNA blots and DNA dot blots. Radioactive PCR was carried out with 2 µl of 2.5 mM d(CTG)TP, 0.8 mM dATP and 7 µl of [γ-32P]ATP (3000 Ci/mmol) under standard conditions in a final volume of 25 µl. The product was adjusted to 200 µl with dH2O and purified through Sephadex G-25.
Table 1. PCR probes used in this study.
| Fragment (size) | Primers (5′–3′) | Template | Experiment | Reference |
|---|---|---|---|---|
| AtMFP1 ORF (1.8 kb) | GACGACGACAAGTATGCACTCGCTCGGCAAGAT | pGEM-T-AtMFP1 | RNA blot | (31) |
| GGAACAAGACCCGTTCAAGAACTGGTACTGCTCTTTC | ||||
| OriA (0.8 kb) | GGATCCCCTTTTAC GCCC | MAB 17 | ‘South-western’/Southern dot blot | (68) |
| CGTTGTACTGCGCTCTCCA | 69) | |||
| OriB (0.8 kb) | GTTTCGATGCAACAACAAGATGTTATT | MAB 17 | ‘South-western’ | (68) |
| ACTAATAAATTTTGTATATAATTGGTCATATAATCGTGG | (69) | |||
| petA (1.0 kb) | CCATCGATATGCAAACTAGAAATACCTT | MAB 17 | ‘South-western’/Southern dot blot | (68) |
| CGGGATCCCTAAAAATTCATTTCGGATAA | 69) | |||
| ‘Chloroplast MAR’ (0.8 kb) | GAATGTATTAAATACGTATGAAGGGAGACT | MAH2 | ‘South-western’ | (68) |
| GACTGTAAATTCATGACCTTTTTTGAATGA | ||||
| Drosophila histone H1/H3 spacer MAR (0.66 kb) | GTAAAACGACGGCCAGT | pBluescript II SK+/0.66 MAR | ‘South-western’ | (70) |
| GGAAACAGCTATGACCATG | ||||
| AtMFP1 internal fragment (0.67 kb) | GACGACGACAAGGCTAGCGAACTTGCACTGGGAG | pGEM-T-AtMFP1 | ‘South-western’/Southern dot blot | (31) |
| GGAACAAGACCCGTTCAAGAACTGGTACTGCTCTTTC | ||||
| psbA (1.1 kb) | CCATCGATATGACTGCAATTTTAGAGAG | MAH2 | Southern dot blot | (68) |
| CGGGATCCTATCCATTTGTAGATGGAGC | ||||
| atpI (0.75 kb) | CCATCGATATGAATGTTTTATCATGTTC | MAH2 | Southern dot blot | (68) |
| CGGGATCC TTAATGATGACCTTCCATAG | 69) | |||
| accD (1.5 kb) | CATGCCATGGATGGAAAAATCGTGGTTCAA | MAB 17 | Southern dot blot | (68) |
| GACTAGTTTAATTTGTGTTCAAAGGAAA |
Ballistic transient transformation
Transient transformation of BY-2 cells was performed as described previously (19).
Fluorescent dyes and imaging
BY-2 cells were incubated with MitoTracker Red CMXRos (Molecular Probes, Eugene, OR) at a final concentration of 100 nM for 5–10 min, centrifuged at 300 g for 5 min and resuspended in dye-free medium. DNA staining with SYTO 82 Orange was performed as described (30). Chloroplast nucleoids were stained with 3 µg/ml DAPI for 5 min. Digitized confocal images of GFP fluorescence in the green channel and MitoTracker Red or SYTO 82 Orange in the red channel were acquired on a PCM 2000/Nikon Eclipse E600 confocal laser scanning microscope (Nikon Bioscience Confocal Systems, Melville, NY) as described previously (30). DAPI fluorescence was examined by epifluorescence microscopy.
Isolation of T-DNA insertion line
A T-DNA insertion line of AtMFP1 in Arabidopsis WS-2 was obtained by screening the lines provided by the University of Wisconsin Arabidopsis Knockout Facility (39). The AtMFP1-specific primers MFP1-F (5′-TTT CCG AGG TCA CAA GTC AGT ACA CAA AG-3′) and MFP1-R (5′-TTT TTG GTG CTA GCT GAA AAC TCC TTG AG-3′) were used. The insertion site was located to intron 3 of AtMFP1 by cloning and sequencing the PCR product. An individual heterozygote plant was identified and obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH). The genotype of its progeny was determined by PCR and homozygote mutants were chosen for further analysis.
RESULTS
We have previously shown that tomato MFP1 (LeMFP1) is associated with globular structures at the nuclear rim (19). Subsequently, a study by Köhler and Hanson (40) alerted us to plastids being associated with the nuclear rim in tobacco suspension culture cells in a pattern resembling that of LeMFP1. To test if the observed structures are plastids, we performed a dual labeling experiment with LeMFP1–GFP and MitoTracker, which stains chloroplasts and mitochondria in plant cells (41). Figure 1 shows that the structures labeled with LeMFP1–GFP (Fig. 1A) are also labeled by MitoTracker (Fig. 1B). The distribution of red and green fluorescence in the merge (Fig. 1C) indicates that LeMFP1–GFP is localized inside the organelles.
Figure 1.
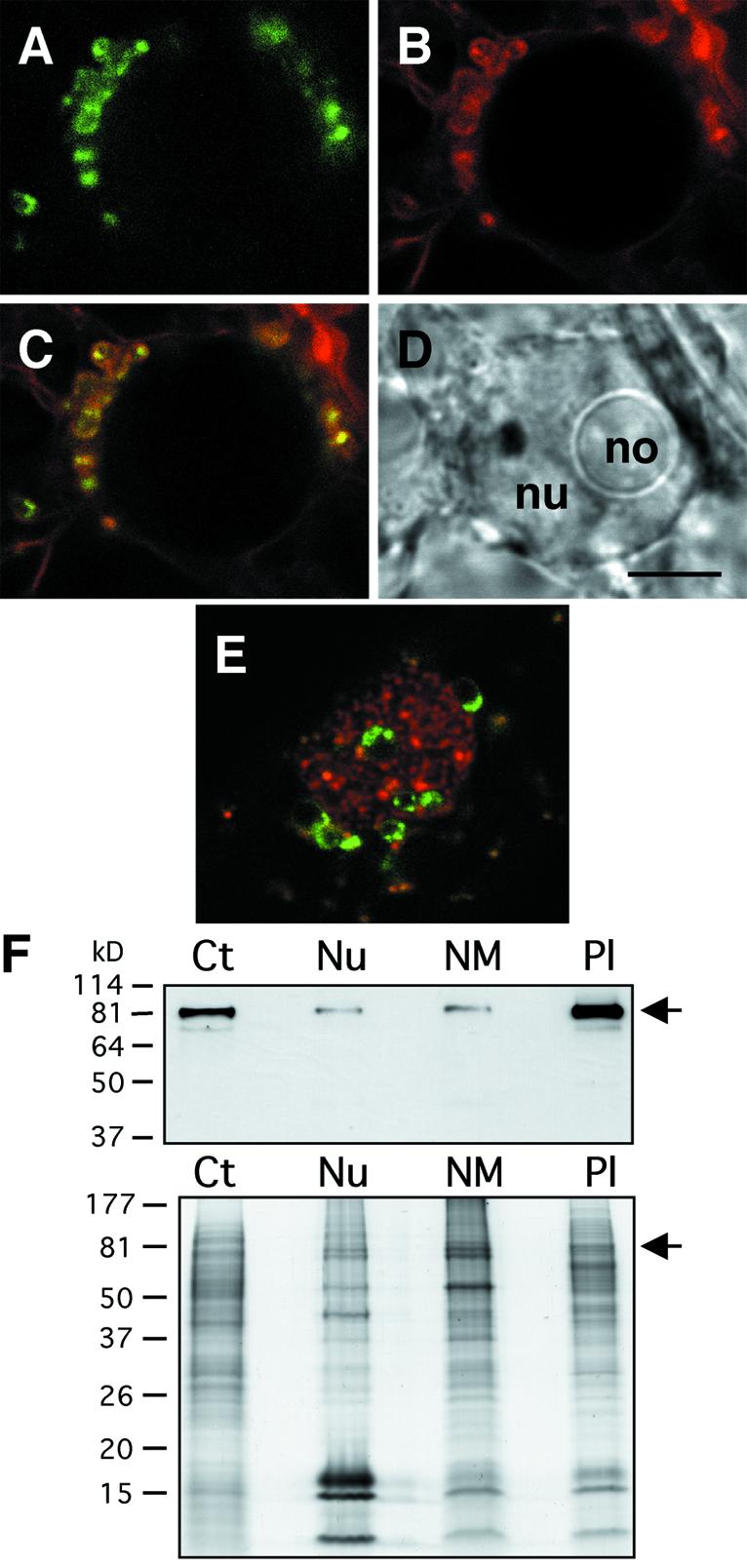
Localization of LeMFP1 in BY-2 plastids. (A–D) Nucleus of a BY-2 cell transiently transformed with LeMFP1–GFP and counter-stained with MitoTracker. (A) GFP fluorescence signal. (B) MitoTracker fluorescence signal. (C) Merge of (A) and (B). (D) Transmitted light image. Nu, nucleus; no, nucleolus. (E) Nucleus of BY-2 cell counter-stained for nucleic acids with SYTO 82 Orange. Bar in (D), 5 µm for (A–E). (F) (Top) BY-2 cell fractions were separated by SDS–PAGE and probed in an immunoblot with the anti-LeMFP1 antibody 288. (Bottom) Coomassie brilliant blue staining of a replica gel. Ct, total cell extract; Nu, nuclear fraction; NM, nuclear matrix fraction; Pl, plastid fraction. Arrows on the right indicate the expected position of MFP1. Protein mass markers are indicated on the left in kilodaltons (kDa).
Previously, we had shown that MFP1 is present in nuclei and in a nuclear matrix fraction of tobacco suspension culture cells (19). The association of plastids with the nuclear envelope suggested that this localization could be due to contamination of these preparations with plastid material. In individual cases, we found plastids actually embedded in the nucleus (Fig. 1E). It has been shown that BY-2 cell nuclei contain major grooves and invaginations (42), which might explain the presence of organelles in the center of nuclei, and therefore the appearance of their proteins in nuclear matrix fractions. We determined the relative amount of MFP1 by immunoblotting after fractionating BY-2 cells into nuclei, nuclear matrix and plastids. Figure 1F shows that MFP1 is significantly enriched in the plastid fraction of these white, heterotrophic cells, while the amount in nuclei and the nuclear matrix fraction is comparably lower, indicating that the majority of MFP1 is clearly associated with plastids.
MFP1 is associated with thylakoid membranes
Previously, the amino acid sequence of Arabidopsis MFP1 (31) had been predicted by PSORT and PredictNLS to localize to the endoplasmic reticulum and contain a C-terminal NLS. We re-analyzed the sequence using the programs ChloroP and TargetP, which predicted an N-terminal chloroplast transit peptide with a cleavage site at amino acid 42. In the virtually processed protein, SignalP detected a twin-arginine (RR) signal peptide characteristic of thylakoid import with a cleavage site at amino acid 95 (Fig. 2A).
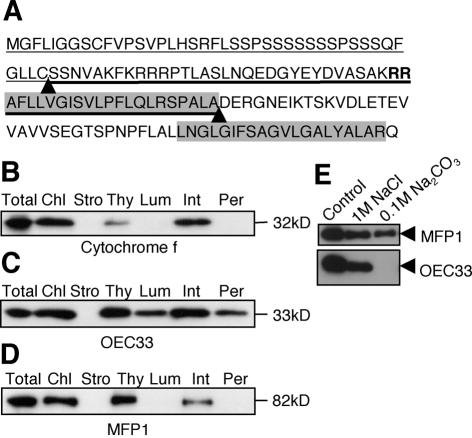
Figure 2.
MFP1 is associated with thylakoid membranes. (A) N-terminal sequence of AtMFP1. The predicted transit peptide for chloroplast import is underlined with a thin line. The predicted signal sequence for thylakoid import is underlined with a thick line. The twin arginine motif is marked in bold. The two predicted cleavage sites are indicated by arrowheads. The two hydrophobic domains previously identified are shaded in gray. (B–D) Fractionation of tobacco leaf chloroplasts. (B) Immunoblot probed with anti-Cytochrome f. (C) Immunoblot with anti-OEC33. (D) Immunoblot with the anti-LeMFP1 antibody 288. Total, total protein extract from tobacco leaves; Chl, total chloroplast protein; Stro, stroma and envelope fraction; Thy, thylakoid fraction; Lum, lumenal fraction; Int, internal membrane fraction; Per, peripheral protein fraction. Molecular masses are indicated on the right. (E) High-salt and high-pH washes of thylakoid fraction. (Top) Immunoblot probed with anti-LeMFP1 antibody 288. (Bottom) Immunoblot probed with anti-OEC33.
To confirm the predicted localization, we isolated and fractionated leaf chloroplasts from N.tabacum. Cytochrome f was used as marker for thylakoid integral membrane proteins (Fig. 2B). Oxygen evolving complex protein 33 (OEC33), a peripheral protein on the lumen side, fractionated between lumen, integral and stroma-peripheral fractions (Fig. 2C) as previously reported (43). Figure 2D shows that MFP1 is clearly associated with the thylakoid fraction. It remains associated with the membrane fraction after treatment with either 1 M NaCl or 0.1 M Na2CO3, pH 11.5 (Fig. 2E), which dissociate peripheral membrane proteins (44).
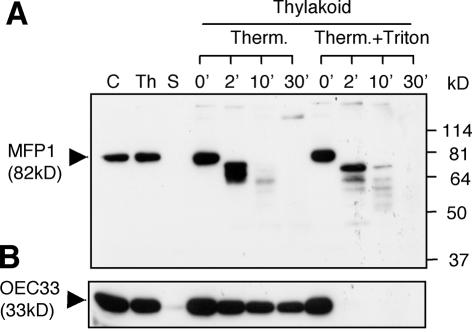
Most proteins containing the RR motif are imported into the lumen (45,46). However, plastid fusion/protein translocation factor (Pftf), which is inserted into the thylakoid membrane via an N-terminal transmembrane domain with the C-terminus oriented towards the stroma, contains the RR motif and utilizes the ΔpH-dependent pathway (47). To determine if MFP1 has the same topology, thermolysin proteolysis in the absence or presence of Triton X-100 was performed (Fig. 3). Proteins on the stroma side are degradable under both conditions, while lumen proteins will only be accessible to thermolysin after Triton X-100 treatment (48). Figure 3 shows that MFP1 is accessible to thermolysin in the absence of Triton X-100, while lumenal OEC33 is only minutely degraded under the same conditions.
Figure 3.
Topology of the MFP1–thylakoid association. The thylakoid fraction was incubated with thermolysin in a time course as indicated. Proteins were resolved by 10% SDS–PAGE and probed by immunoblotting with the anti-LeMFP1 antibody 288 (A) or anti-OEC33 (B). C, total chloroplast protein; Th, thylakoid protein fraction; S, stroma and envelope protein fraction; Therm., thermolysin; 0, 2, 10, 30, incubation times in minutes. Molecular masses in kilodaltons (kDa) are indicated on the right in (A).
Together, these data indicate that MFP1 is imported into chloroplasts and associated with thylakoid membranes. The major portion of the protein is present on the stroma side and accessible to proteases. This is consistent with a topology where the N-terminal transmembrane domain of the processed protein is inserted into the thylakoid membrane and the C-terminal DNA-binding domain is facing the stroma.
MFP1 expression in Arabidopsis is tissue specific and light regulated
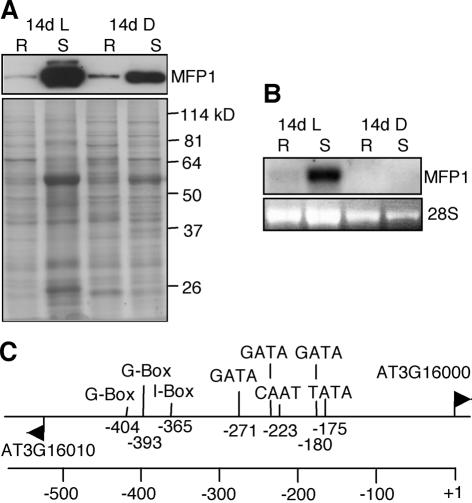
The anti-AtMFP1 antibody OSU91 recognized a single band of 82 kDa in light- and dark-grown Arabidopsis seedlings (Fig. 4A). AtMFP1 accumulates to high levels in shoots of light-grown seedling. A significantly lower amount of protein was detected in roots of both light- and dark-grown seedlings, and an intermediate amount in dark-grown shoots. RNA blot analysis indicated that protein abundance is primarily regulated at the RNA level (Fig. 4B).
Figure 4.
AtMFP1 expression is tissue specific and light regulated. (A) AtMFP1 protein accumulation in roots (R) and shoots (S) of 14-day-old light-grown seedlings (14d L) and 14-day-old dark-grown seedlings (14d D). (Top) An immunoblot probed with the anti-AtMFP1 antibody OSU91. (Bottom) The Coomassie brilliant blue staining of a replica gel (8%). Molecular masses in kilodaltons (kDa) are indicated on the right. (B) RNA blot analysis of AtMFP1 mRNA accumulation with total RNA from roots (R) and shoots (S) of 14-day-old light-grown seedlings (14d L) and 14-day-old dark-grown seedlings (14d D). (Top) RNA blot probed with the AtMFP1 cDNA. (Bottom) Ethidium bromide staining of 28S rRNA as loading control. (C) Predicted core promoter elements and cis-acting elements in the intergenic region between AT3G16010 and AT3G16000 (AtMFP1). Only elements predicted on the top strand (upstream of AtMFP1) are shown. Numbers indicate positions upstream of the AtMFP1 start codon.
AtMFP1 is a single gene in Arabidopsis (AT3G16000), with a predicted 540 bp of non-coding sequence upstream of the ATG and downstream of the next, divergently oriented open reading frame (ORF) (AT3G16010). Analyzing this putative promoter region for cis-elements involved in light-activation of plant gene expression (49), two G-boxes, one I-box and three additional GATA elements were found close to and upstream of the predicted CAAT and TATA boxes (Fig. 4C). Consistent with the expression pattern, this promoter architecture is highly suggestive of a ‘classic’ nuclear, light-activated gene such as RBCS or LHCII (50).
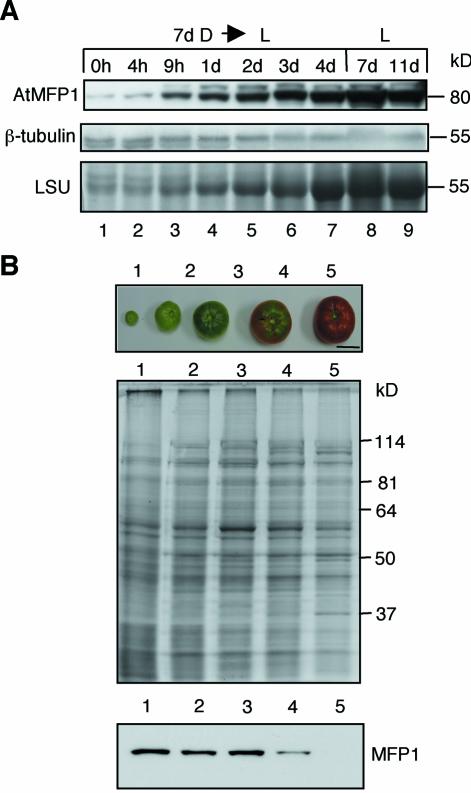
MFP1 protein accumulation parallels chloroplast development
To investigate if MFP1 protein accumulation parallels chloroplast development, we determined the level of AtMFP1 during greening of etiolated Arabidopsis seedlings. Seven-day-old dark-grown seedlings were placed into light, and were assayed at different time points for AtMFP1 abundance in total shoot protein (Fig. 5A). AtMFP1 accumulation was first observed after 9 h and reached the level of light-grown seedlings after 3 days. This paralleled the accumulation of the large subunit of Rubisco (Fig. 5A) and the visually observed greening of the cotyledons (data not shown).
Figure 5.
MFP1 protein abundance correlates with chloroplast development. (A) AtMFP1 protein abundance was analyzed during greening of etiolated, 7-day-old Arabidopsis seedlings (7d D→L). Seedlings were placed into light for the indicated amount of time and total shoot protein extracts were prepared. Controls are seedlings grown for 7 or 11 days in constant light (L, 7d; L, 11d). (Top) Immunoblot with anti-AtMFP1 antibody OSU91. (Middle) Immunoblot with anti-β-tubulin as loading control. (Bottom) Accumulation of the large subunit of Rubisco (LSU), as detected by Coomassie staining after 10% SDS–PAGE. Molecular masses are indicated on the right. (B) MFP1 protein abundance during tomato fruit development. Lycopersicon esculentum var. ‘Early Girl’ fruit were harvested at the stages shown in the top panel. Total pericarp protein extracts were separated by 10% SDS–PAGE (middle) and probed by immunoblotting with anti-LeMFP1 288 (bottom). Bar in the top panel, 2.5 cm. Molecular mass markers are indicated on the right.
To test if MFP1 accumulation also correlates with the presence of photosynthetic membranes in a different system, we investigated its abundance during tomato fruit development. Young, green tomato fruits contain chloroplasts that are similar to leaf chloroplasts. During fruit ripening, the photosynthetic membranes break down and the chromoplasts accumulate carotenoids and lycopene (51,52). Figure 5B shows that LeMFP1 accumulation remains constant until the onset of ripening, when the protein level begins to decline (Fig. 5B, lane 4). Subsequently, it drops to an undetectable level coincident with the transition from chloroplasts to chromoplasts (Fig. 5B, lane 5). Therefore, MFP1 abundance not only matches the abundance of photosynthetic membranes during greening, but also during their degradation in fruit development.
DNA-binding activity of AtMFP1
We tested the DNA-binding activity of endogenous, chloroplast-localized and recombinant AtMFP1. Chloroplasts were isolated from rosette leaves of wild-type plants and a T-DNA insertion line (see Materials and Methods). Figure 6A shows the protein profile of the two chloroplast preparations. MFP1 is detected in an immunoblot in wild-type, but not the T-DNA insertion line (Fig. 6B). Figure 6C shows a ‘South-western’ DNA-binding experiment with a Drosophila MAR (which had been previously shown to bind recombinant LeMFP1) in the presence of sheared herring-sperm DNA as competitor (18). The data show a DNA-binding activity of 82 kDa, the size of AtMFP1, which is present in wild-type but not in the T-DNA insertion line. A second, minor DNA-binding activity of ∼70 kDa was detected, which was not influenced by the T-DNA insertion. Its size would be consistent with Arabidopsis sulfite reductase (see Fig. 7).
Figure 7.
MFP1 is associated with chloroplast nucleoids. (A) Epi fluorescence microscopy after DAPI staining of 1 µl of the final nucleoid fraction shown in lane Nc in (C). (B) Dot blot hybridization of 1 µg each of pGEM-AtMFP1 (row 1), total chloroplast DNA (row 2) and nucleoid DNA (row 3). Probes for the five chloroplast DNA fragments psbA, petA, atpI, accD and oriA are described in Materials and Methods. The AtMFP1 ORF was used as the hybridization control. (C) Immunoblot analysis of OEC33, Cytochrome f (Cyto f), ferredoxin::sulfite reductase (SiR) and MFP1 in different fractions of the nucleoid isolation. Chl, total chloroplast extract; S1, first 48 000 g supernatant; S2, second 48 000 g supernatant; Nc, nucleoid fraction. Ten microliters of the final fractions were loaded in each lane (see Materials and Methods). A replica gel (10%) stained with Coomassie brilliant blue is shown on the right. Molecular mass markers are indicated on the right for the replica gel.
Figure 6D shows that recombinant Δ144AtMFP1 binds in vitro with comparable affinity to six DNA fragments of similar size and different AT content in the presence of sheared herring sperm DNA as competitor. In contrast, binding was significantly weaker when the probes were denatured. Approximately equal binding was also observed when three restriction-digested BAC clones (which together represent the complete Arabidopsis chloroplast genome) were used separately as probes in ‘South-western’ blots with recombinant Δ144AtMFP1 (data not shown). Together, these data indicate that AtMFP1 is a prominent DNA-binding protein in mature Arabidopsis chloroplasts and that it binds to different double-stranded DNA fragments without detectable sequence specificity.
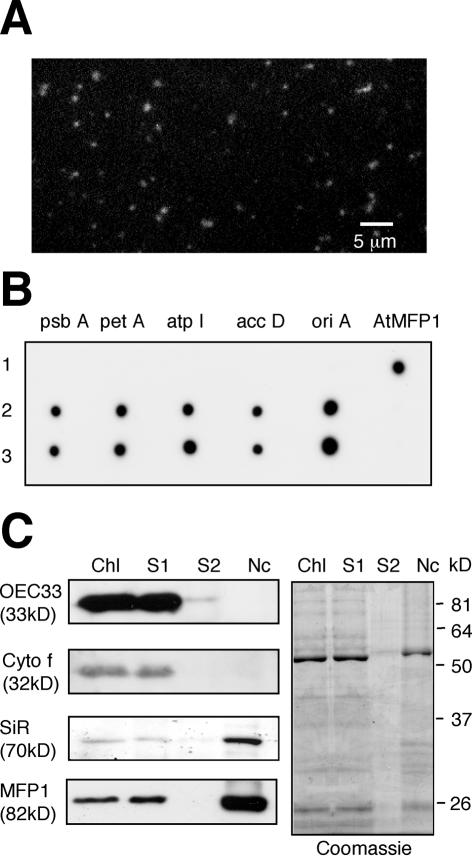
MFP1 is associated with chloroplast nucleoids
To investigate if MFP1 is associated with nucleoids in mature chloroplasts, we purified nucleoids and probed in an immunoblot for the presence of MFP1. A nucleoid fraction was isolated from tobacco chloroplasts and confirmed by DAPI staining (Fig. 7A). In addition, nucleoid DNA was probed in a dot-blot experiment for the presence of five regions of the chloroplast genome. Figure 7B shows that the signals were comparable with those obtained with total chloroplast DNA, confirming the presence of the chloroplast genome in the nucleoid fraction.
The immunoblots in Figure 7C follow the abundance of four chloroplast proteins during nucleoid fractionation. OEC33 and Cytochrome f were present in the total chloroplast extract (Fig. 7C, lane Chl) and the 48 000 g supernatant (Fig. 7C, lane S1), but not in the concentrated wash (Fig. 7C, lane S2) or the final nucleoid pellet (Fig. 7C, lane Nc). This controls for the absence of non-specific thylakoid proteins in the nucleoid fraction. In contrast, ferredoxin::sulfite reductase/DPC68 (SiR), a major component of purified chloroplast nucleoids (10,12,13), was enriched in the nucleoid fraction. Like SiR, MFP1 co-fractionated with the 48 000 g pellet, indicating a direct physical interaction of MFP1 with nucleoids in mature leaf chloroplasts.
DISCUSSION
MFP1 was originally identified as a DNA-binding protein with affinity for matrix attachment regions (18) and was subsequently shown to be associated with nuclei and the nuclear matrix in tobacco suspension culture cells (19). We have demonstrated here that the primary location of MFP1 in both suspension culture cells and in plants is in plastids and associated with the thylakoid membranes. MFP1 can also be clearly identified in nuclei and the nuclear matrix and a convincing NLS is predicted at the C-terminus of MFP1 from Arabidopsis, tomato and tobacco. At present, we cannot rule out that there is a second fraction of MFP1 which is associated with the nuclear matrix and which might correspond to the onion MFP1-like protein found in discrete locations in the nuclear matrix, which resemble replication factories (53,54). However, the MFP1–GFP signal that we have found in speckles at the nuclear rim (19), corresponds to proplastids associated with the nucleus in tobacco suspension cultured cells. Further work at the electron microscopy level using intact tissue sections could address if there is a separate pool of intranuclear MFP1. A precedence for a protein with such a dual cellular address is the yeast inner nuclear envelope protein Trm1p, which is also located in mitochondria (55).
Alternatively, the nuclear portion of MFP1 might be due to a contamination of this fraction with plastid material. The close association of plastids with the nuclear rim and cases of embedded plastids into nuclear material shown here might lead to a co-fractionation of insoluble plastid material with the nuclear matrix fraction from suspension culture cells. It should be pointed out that the isolation procedures widely used to prepare the nuclear matrix from plants were originally developed for animal cells, which do not contain plastids.
MFP1 has the features of an unusual TAT pathway target
Analysis of the N-terminal sequence of AtMFP1 indicated that the protein contains a chloroplast transit peptide, a hydrophobic signal peptide, and a hydrophobic membrane anchor. The signal peptide is of the twin arginine type, indicative of a substrate of the ΔpH-dependent or TAT pathway (46). While most substrates of this pathway known to date enter the thylakoid lumen, there are two examples of thylakoid membrane-anchored proteins, which are TAT-pathway substrates, Pftf and the Rieske Fe/S protein, which utilizes the TAT pathway, but lacks the twin-arginine motif and a cleavable signal peptide (56). Our data are consistent with MFP1 being inserted into the thylakoid membrane with a short N-terminal domain located in the lumen and the long coiled-coil domain located in the stroma. This would require the second hydrophobic domain to act as a transmembrane domain. Its length of 20 amino acids is sufficient and the resistance of MFP1 to the extraction with 1 M NaCl indicates its integral association with the lipid bilayer (44,47). The TAT pathway might therefore function for more membrane associated, stroma-exposed proteins than previously expected.
MFP1 is associated with both nucleoids and thylakoid membranes in mature leaf chloroplasts
For more than 30 years, there have been indications that in mature chloroplasts DNA is associated with thylakoid membranes. Woodcock and Fernandez-Moran (57) noticed in their electron microscopy studies of spinach chloroplast DNA that the DNA fibers are frequently associated with and extrude from the grana lamella. Lindbeck and Rose (58) demonstrated that chloroplast DNA is associated with thylakoid vesicles. Liu and Rose (16) identified a region of the chloroplast genome that remains bound to thylakoid membrane vesicles after restriction digestion. Cytological studies in wheat and pea showed a developmental re-localization of nucleoids from the plastid periphery to the interior (9,15,59). While all these studies indicated an association of nucleoids with the thylakoids in mature chloroplast, the molecular nature of this interaction has not been understood.
We have shown here that MFP1 is associated with thylakoid membranes, with the coiled-coil DNA-binding domain of the protein located on the stroma surface and that it has a major DNA-binding activity in mature Arabidopsis chloroplasts. Most importantly, MFP1 is in vivo associated with nucleoids similar to SiR/DCP68, a known component of chloroplast nucleoids (12). This suggests that MFP1 is acting at the interface between nucleoids and the thylakoid membranes in mature chloroplasts. The expression pattern of MFP1 is tightly correlated with the accumulation of photosynthetic membranes and reciprocal to that of PEND, the other known membrane-associated DNA-binding protein in chloroplasts. This is consistent with a model in which nucleoids are associated with the plastid envelope at the early stages of plastid development when PEND is expressed, and relocate to the thylakoids in mature chloroplasts, when PEND is absent but MFP1 is abundant. We did not find a difference, however, in the pattern of DAPI-stained nucleoids associated with isolated thylakoids from wild-type and MFP1 knock-out Arabidopsis plants. In addition, we found no significant difference in abundance of SiR in the two thylakoid fractions (data not shown). This might be due to functional redundancy among MFP1-like chloroplast proteins or point to a function of MFP1 other than physical association (see below).
MFP1 is a unique long coiled-coil protein from plants
Proteins with extended coiled-coil domains have been mostly described in animals and yeast, where many of them function as structural proteins. The most prominent example of DNA-binding coiled-coil proteins are the nuclear lamins, which are involved in connecting chromatin with the inner nuclear envelope (1). While no canonical nuclear lamins appear to exist in plants, MFP1 is somewhat similar to this protein class in being a long coiled-coil protein that is anchored to membranes and binds DNA. Sato (60) suggests that during evolution the plastid genetic machinery has sequentially lost many of the original ‘prokaryotic’ DNA-organizing proteins, such as HUs, and has instead acquired nuclear ‘eukaryotic’ types of proteins. The bZIP protein PEND with its similarity to nuclear transcription factors is an example of the latter class. While MFP1 is structurally similar to ‘eukaryotic’ long coiled-coil proteins, no homologs have been found in the fully sequenced genomes of Cynechocystis and Nostoc (data not shown). MFP1 therefore appears to be another example in favor of Sato’s hypothesis of discontinuous evolution of the plastid ‘chromatin’ by incorporation of nuclear proteins.
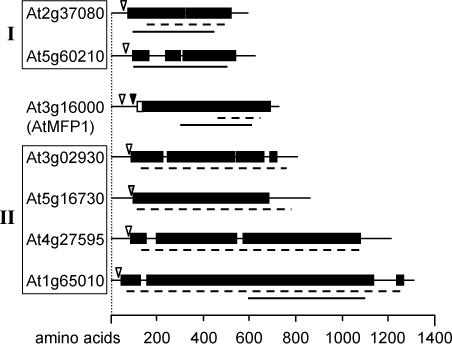
We asked whether other chloroplast-located long coiled-coil proteins are encoded by the Arabidopsis genome. In a computational screen of the translated genome with the algorithm MultiCoil and subsequent search for putative targeting signals, we found six additional predicted chloroplast proteins with a coiled-coil domain similar in length and type to that of MFP1. They can be grouped into two families of homologous sequences (Fig. 8). The coiled-coil domains of MFP1 and both families are rich in glutamic acid residues and show weak similarity to the coiled-coil segments of mammalian and prokaryotic SMC proteins (61,62). However, in contrast to MFP1, no thylakoid targeting signals and transmembrane domains were predicted for these proteins. In Arabidopsis, MFP1 is therefore unique in its combination of domains.
Figure 8.
MFP1-like proteins in Arabidopsis. Proteins are drawn to scale as bar diagrams. (Closed boxes) Coiled-coil domains; (open boxes) putative transmembrane helices. Cleavage sites for chloroplast targeting signal and TAT pathway signal peptides as predicted by ChloroP and SignalP are indicated by open and closed arrowheads, respectively. Broken lines underneath the diagrams mark glutamic acid-rich regions; closed lines indicate the position of domains with similarity to SMC proteins. Proteins showing statistically significant sequence similarity were grouped together (group I, 36% identical; group II, 37–54% identical).
Interestingly, a short coiled-coil DNA-binding, plastid-localized protein has been recently described in the resurrection plant Craterostigma plantagineum (63). CpPTP is a 200 amino acid protein with an N-terminal chloroplast import signal and a C-terminal approximately 100 amino acids long coiled-coil domain, which non-specifically binds DNA. CpPTP is induced by drought and not detectable in unstressed plants. It has no homolog in the Arabidopsis genome. Interestingly, a CpPTP–GFP fusion protein localizes in a spot-like pattern in tobacco chloroplasts, very similar to MFP1. While the function of CpPTP is presently not known, the authors propose a structural role in protecting chloroplast DNA from dehydration. If this is confirmed, it will be worth investigating whether CpPTP in C.plantagineum functions by replacing DNA-binding, coiled-coil proteins such as MFP1 under extreme drought stress, and whether this mechanism is absent from desiccation-sensitive plants such as Arabidopsis.
What are the function and selective advantage of nucleoid–thylakoid association?
The MFP1 T-DNA insertion line described here has no detectable protein accumulation, but shows no visible phenotype under standard laboratory conditions (data not shown). It is therefore possible that the MFP1-like coiled-coil proteins provide functional redundancy despite the lack of true sequence homologs of MFP1 in Arabidopsis. In addition, there are several possible scenarios for the function of MFP1, the disruption of which would lead to no drastic phenotypes. For example, the expression of several chloroplast genes coding for PSI and PSII subunits is fine-tuned by the redox state of plastoquinone (64–66). It is possible that the presently unknown signal transduction chain leading to this regulation is facilitated by a short physical distance between the chloroplast gene expression machinery and the photosynthetic membranes and that MFP1 is involved in providing this connection. Alternatively (or additionally), MFP1 might be involved in facilitating the co-translational membrane insertion and assembly of thylakoid proteins such as the D1 protein (67) and Cytochrome f (33) by contributing to short distances between transcription, translation and protein insertion.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Peter Buchenau for alerting us to the study by Köhler and Hanson. We thank Drs Alice Barkan, Klaas van Wijk and Jens Schwenn for kindly providing antisera against OEC33, Cytochrome f and SiR, respectively, Drs Ling Xiong and Richard Sayre for help with the chloroplast fractionation, and Dr Biao Ding for generous user time on his confocal microscope. This work was supported by a grant from the National Science Foundation (MCB-0079577) to I.M.
REFERENCES
- 1.Gruenbaum Y., Wilson,K.L., Harel,A., Goldberg,M. and Cohen,M. (2000) Review: nuclear lamins—structural proteins with fundamental functions. J. Struct. Biol., 129, 313–323. [DOI] [PubMed] [Google Scholar]
- 2.Rouviere-Yaniv J. and Gros,F. (1975) Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc. Natl Acad. Sci. USA, 72, 3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouviere-Yaniv J., Yaniv,M. and Germond,J.E. (1979) E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell, 17, 265–274. [DOI] [PubMed] [Google Scholar]
- 4.Norris V., Turnock,G. and Sigee,D. (1996) The Escherichia coli enzoskeleton. Mol. Microbiol., 19, 197–204. [DOI] [PubMed] [Google Scholar]
- 5.Diffley J.F. and Stillman,B. (1991) A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA, 88, 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho J.H., Ha,S.J., Kao,L.R., Megraw,T.L. and Chae,C.B. (1998) A novel DNA-binding protein bound to the mitochondrial inner membrane restores the null mutation of mitochondrial histone Abf2p in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 5712–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi T., Takahara,M., Miyagishima,S.Y., Kuroiwa,H., Sasaki,N., Ohta,N., Matsuzaki,M. and Kuroiwa,T. (2002) Detection and localization of a chloroplast-encoded HU-like protein that organizes chloroplast nucleoids. Plant Cell, 14, 1579–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakano T., Murakami,S., Shoji,T., Yoshida,S., Yamada,Y. and Sato,F. (1997) A novel protein with DNA binding activity from tobacco chloroplast nucleoids. Plant Cell, 9, 1673–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato N., Albrieux,C., Joyard,J., Douce,R. and Kuroiwa,T. (1993) Detection and characterization of a plastid envelope DNA-binding protein which may anchor plastid nucleoids. EMBO J., 12, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon G.C., Ward,L.N., Case,C.I. and Heinhorst,S. (1999) The 68 kDa DNA compacting nucleoid protein from soybean chloroplasts inhibits DNA synthesis in vitro. Plant Mol. Biol., 39, 835–845. [DOI] [PubMed] [Google Scholar]
- 11.Nemoto Y., Kawano,S., Nakamura,S., Mita,T., Nagata,T. and Kuroiwa,T. (1988) Studies on plastid-nuclei (nucleoids) in Nicotiana tabacum L.I. Isolation of proplastid-nuclei from cultured cells and identification of proplastid-nuclear protein. Plant Cell Physiol., 29, 167–177. [Google Scholar]
- 12.Sato N., Nakayama,M. and Hase,T. (2001) The 70-kDa major DNA-compacting protein of the chloroplast nucleoid is sulfite reductase. FEBS Lett., 487, 347–350. [DOI] [PubMed] [Google Scholar]
- 13.Chi-Ham C.L., Keaton,M.A., Cannon,G.C. and Heinhorst,S. (2002) The DNA-compacting protein DCP68 from soybean chloroplasts is ferredoxin:sulfite reductase and co-localizes with the organellar nucleoid. Plant Mol. Biol., 49, 621–631. [DOI] [PubMed] [Google Scholar]
- 14.Sato N., Rolland,N., Block,M.A. and Joyard,J. (1999) Do plastid envelope membranes play a role in the expression of the plastid genome? Biochimie, 81, 619–629. [DOI] [PubMed] [Google Scholar]
- 15.Miyamura S., Nagata,T. and Kuroiwa,T. (1986) Quantitative fluorescence microscopy on dynamic changes of plastid nucleoids during wheat development. Protoplasma, 133, 66–72. [Google Scholar]
- 16.Liu J.W. and Rose,R.J. (1992) The spinach chloroplast chromosome is bound to the thylakoid membrane in the region of the inverted repeat. Biochem. Biophys. Res. Commun., 184, 993–1000. [DOI] [PubMed] [Google Scholar]
- 17.Sato N., Ohshima,K., Watanabe,A., Ohta,N., Nishiyama,Y., Joyard,J. and Douce,R. (1998) Molecular characterization of the PEND protein, a novel bZIP protein present in the envelope membrane that is the site of nucleoid replication in developing plastids. Plant Cell, 10, 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meier I., Phelan,T., Gruissem,W., Spiker,S. and Schneider,D. (1996) MFP1, a novel plant filament-like protein with affinity for matrix attachment region DNA. Plant Cell, 8, 2105–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gindullis F. and Meier,I. (1999) Matrix attachment region binding protein MFP1 is localized in discrete domains at the nuclear envelope. Plant Cell, 11, 1117–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emanuelsson O., Nielsen,H. and von Heijne,G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci., 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emanuelsson O., Nielsen,H., Brunak,S. and von Heijne,G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol., 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- 22.Nakai K. and Horton,P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci., 24, 34–36. [DOI] [PubMed] [Google Scholar]
- 23.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst., 8, 581–599. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen H., Brunak,S. and von Heijne,G. (1999) Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng., 12, 3–9. [DOI] [PubMed] [Google Scholar]
- 25.Wolf E., Kim,P.S. and Berger,B. (1997) MultiCoil: A program for predicting two- and three-stranded coiled coils. Protein Sci., 6, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusnady G.E. and Simon,I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics, 17, 849–850. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falquet L., Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murashige T. and Skoog,F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant, 103, 473–479. [Google Scholar]
- 30.Rose A. and Meier,I. (2001) A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc. Natl Acad. Sci. USA, 98, 15377–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harder P.A., Silverstein,R.A. and Meier,I. (2000) Conservation of matrix attachment region-binding filament-like protein 1 among higher plants. Plant Physiol., 122, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voelker R. and Barkan,A. (1995) Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J., 14, 3905–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohl T. and van Wijk,K.J. (2001) In vitro reconstitution of insertion and processing of cytochrome f in a homologous chloroplast translation system. J. Biol. Chem., 276, 35465–35472. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- 35.Maliga P., Klessig,D., Cashmore,A., Gruissem,W. and Varner,J. (1995) Methods in Plant Molecular Biology: A Laboratory Course Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- 36.Peltier J.B., Friso,G., Kalume,D.E., Roepstorff,P., Nilsson,F., Adamska,I. and van Wijk,K.J. (2000) Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell, 12, 319–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall G. Jr, Allen,G.C., Loer,D.S., Thompson,W.F. and Spiker,S. (1991) Nuclear scaffolds and scaffold-attachment regions in higher plants. Proc. Natl Acad. Sci. USA, 88, 9320–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapoor S. and Sugiura,M. (1999) Identification of two essential sequence elements in the nonconsensus type II PatpB-290 plastid promoter by using plastid transcription extracts from cultured tobacco BY-2 cells. Plant Cell, 11, 1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krysan P.J., Young,J.C. and Sussman,M.R. (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell, 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köhler R.H. and Hanson,M.R. (2000) Plastid tubules of higher plants are tissue-specific and developmentally regulated. J. Cell Sci., 113, 81–89. [DOI] [PubMed] [Google Scholar]
- 41.Nebenführ A., Gallagher,L.A., Dunahay,T.G., Frohlick,J.A., Mazurkiewicz,A.M., Meehl,J.B. and Staehelin,L.A. (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol., 121, 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collings D.A., Carter,C.N., Rink,J.C., Scott,A.C., Wyatt,S.E. and Allen,N.S. (2000) Plant nuclei can contain extensive grooves and invaginations. Plant Cell, 12, 2425–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisk D.G., Walker,M.B. and Barkan,A. (1999) Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J., 18, 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J.M., Cho,J.H., Kang,S.G., Jang,H.J., Pih,K.T., Piao,H.L., Cho,M.J. and Hwang,I. (1998) A dynamin-like protein in Arabidopsis thaliana is involved in biogenesis of thylakoid membranes. EMBO J., 17, 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson C., Woolhead,C. and Edwards,W. (2000) Transport of proteins into and across the thylakoid membrane. J. Exp. Bot., 51, 369–374. [DOI] [PubMed] [Google Scholar]
- 46.Robinson C., Thompson,S.J. and Woolhead,C. (2001) Multiple pathways used for the targeting of thylakoid proteins in chloroplasts. Traffic, 2, 245–251. [DOI] [PubMed] [Google Scholar]
- 47.Summer E.J., Mori,H., Settles,A.M. and Cline,K. (2000) The thylakoid delta pH-dependent pathway machinery facilitates RR-independent N-tail protein integration. J. Biol. Chem., 275, 23483–23490. [DOI] [PubMed] [Google Scholar]
- 48.Walker M.B., Roy,L.M., Coleman,E., Voelker,R. and Barkan,A. (1999) The maize tha4 gene functions in Sec-independent protein transport in chloroplasts and is related to hcf106, tatA and tatB. J. Cell Biol., 147, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higo K., Ugawa,Y., Iwamoto,M. and Korenaga,T. (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res., 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilmartin P.M., Sarokin,L., Memlink,J. and Chua,N.-H. (1990) Molecular light switches for plant genes. Plant Cell, 2, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bathgate B., Purton,M., Grierson,D. and Goodenough,P. (1985) Plastid changes during the conversion of chloroplasts to chromoplasts in ripening tomatoes. Planta, 165, 197–204. [DOI] [PubMed] [Google Scholar]
- 52.Thelander M., Narita,J. and Gruissem,W. (1986) Plastid differentiation and pigment biosynthesis during tomato fruit ripening. Curr. Topics Plant Biochem. Physiol., 5, 128–141. [Google Scholar]
- 53.Samaniego R., Yu,W., Meier,I. and Moreno Diaz de la Espina,S. (2001) Characterisation and high-resolution distribution of a matrix attachment region-binding protein (MFP1) in proliferating cells of onion. Planta, 212, 535–546. [DOI] [PubMed] [Google Scholar]
- 54.Moreno Diaz De La Espina S., Samaniego,R., Yu,W. and De La Torre,C. (2003) Intermediate filament proteins with nuclear functions: NuMA, lamin-like proteins and MFP1. Cell. Biol. Int., 27, 233–235. [DOI] [PubMed] [Google Scholar]
- 55.Rose A.M., Belford,H.G., Shen,W.C., Greer,C.L., Hopper,A.K. and Martin,N.C. (1995) Location of N2,N2-dimethylguanosine-specific tRNA methyltransferase. Biochimie, 77, 45–53. [DOI] [PubMed] [Google Scholar]
- 56.Molik S., Karnauchov,I., Weidlich,C., Herrmann,R.G. and Klosgen,R.B. (2001) The Rieske Fe/S protein of the cytochrome b6/f complex in chloroplasts: missing link in the evolution of protein transport pathways in chloroplasts? J. Biol. Chem., 276, 42761–42766. [DOI] [PubMed] [Google Scholar]
- 57.Woodcock C.L. and Fernandez-Moran,H. (1968) Electron microscopy of DNA conformations in spinach chloroplasts. J. Mol. Biol., 31, 627–631. [DOI] [PubMed] [Google Scholar]
- 58.Lindbeck A.G. and Rose,R.J. (1990) Thylakoid-bound chloroplast DNA from spinach is enriched for replication forks. Biochem. Biophys. Res. Commun., 172, 204–210. [DOI] [PubMed] [Google Scholar]
- 59.Sato N., Misumi,O., Shinada,Y., Sasaki,M. and Yoine,M. (1997) Dynamics of localization and protein composition of plastid nuceloids in light-grown pea seedlings. Protoplasma, 200, 163–173. [Google Scholar]
- 60.Sato N. (2001) Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci., 6, 151–155. [DOI] [PubMed] [Google Scholar]
- 61.Ball J.A.R. and Yokomori,K. (2001) The structural maintenance of chromosomes (SMC) family of proteins in mammals. Chromosome Res., 9, 85–96. [DOI] [PubMed] [Google Scholar]
- 62.Soppa J. (2001) Prokaryotic structural maintenance of chromosomes (SMC) proteins: distribution, phylogeny and comparison with MukBs and additional prokaryotic and eukaryotic coiled-coil proteins. Gene, 278, 253–264. [DOI] [PubMed] [Google Scholar]
- 63.Phillips J.R., Hilbricht,T., Salamini,F. and Bartels,D. (2002) A novel abscisic acid- and dehydration-responsive gene family from the resurrection plant Craterostigma plantagineum encodes a plastid-targeted protein with DNA-binding activity. Planta, 215, 258–266. [DOI] [PubMed] [Google Scholar]
- 64.Allen J.F. and Pfannschmidt,T. (2000) Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Phil. Trans. R. Soc. Lond. B Biol. Sci., 355, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfannschmidt T., Nilsson,A., Tullberg,A., Link,G. and Allen,J.F. (1999) Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. IUBMB Life, 48, 271–276. [DOI] [PubMed] [Google Scholar]
- 66.Pfannschmidt T., Schütze,K., Brost,M. and Oelmüller,R. (2001) A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J. Biol. Chem., 276, 36125–36130. [DOI] [PubMed] [Google Scholar]
- 67.Zhang L., Paakkarinen,V., van Wijk,K.J. and Aro,E.M. (1999) Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem., 274, 16062–16067. [DOI] [PubMed] [Google Scholar]
- 68.Sato S., Nakamura,Y., Kaneko,T., Asamizu,E. and Tabata,S. (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res., 6, 283–290. [DOI] [PubMed] [Google Scholar]
- 69.Kunnimalaiyaan M. and Nielsen,B.L. (1997) Fine mapping of replication origins (ori A and ori B) in Nicotiana tabacum chloroplast DNA. Nucleic Acids Res., 25, 3681–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.von Kries J.P., Buhrmester,H. and Strätling,W.H. (1991) A matrix/scaffold attachment region binding protein: identification, purification and mode of binding. Cell, 64, 123–135. [DOI] [PubMed] [Google Scholar]