Abstract
Micro-RNAs are a class of small non-coding regulatory RNAs that impair translation by imperfect base pairing to mRNAs. For analysis of their cellular function we injected different miRNA-specific DNA antisense oligonucleotides in Drosophila embryos. In four cases we observed severe interference with normal development, one had a moderate impact and six oligonucleotides did not cause detectable phenotypes. We further used the miR-13a DNA antisense oligonucleotide as a PCR primer on a cDNA library template. In this experimental way we identified nine Drosophila genes, which are characterised by 3′ untranslated region motifs that allow imperfect duplex formation with miR-13 or related miRNAs. These genes, which include Sos and Myd88, represent putative targets for miRNA regulation. Mutagenesis of the target motif of two genes followed by transfection in Drosophila Schneider 2 (S2) cells and subsequent reporter gene analysis confirmed the hypothesis that the binding potential of miR-13 is inversely correlated with gene expression.
INTRODUCTION
Micro- (mi) RNAs represent a class of regulatory small non-coding RNAs of ∼21–24 nt detected in animals and plants (1–5). Some miRNAs are conserved across kingdoms and a uniform system for their nomenclature has been recently introduced (6). miRNAs are processed from hairpin-type precursor RNA transcripts by the ribonuclease III-like enzyme Dicer (7–10). Dicer is also linked to the RNA interference (RNAi) pathway as it processes double-stranded RNA into short interfering RNA (siRNA) (reviewed in 11,12). Recent data suggest that plants may have distinct Dicer enzymes, one for the production of miRNAs and another for the generation of siRNA (13), but regardless of this potential difference, both pathways are also linked (14).
In animals, miRNAs are believed to modulate translation by binding to the 3′ untranslated regions (UTRs) of target genes. This assumption is based on two observations. First, the previously discovered small temporal (st) RNAs let-7 and lin-4 of Caenorhabditis elegans hybridise to lin-41 and lin-14 mRNA, respectively (15–17) and are now considered prototypes of miRNAs. Secondly, 3′ UTR-located sequence motifs known to mediate negative post-transcriptional regulation are complementary to some classes of miRNAs (18). Recently, more examples of target genes that are regulated by miRNAs have been described for C.elegans (19,20) and Drosophila melanogaster (21,22). Moreover, it was recently found that C.elegans contains further tiny non-coding (tnc) RNAs of similar size to miRNAs that are, however, not processed from hairpin precursors (23).
The large number of non-coding small RNAs, in combination with their small size, makes it difficult to identify loss-of-function mutants. Another complication is that several miRNA genes are redundant and occur in different loci so that mutant phenotypes are even less likely. As an alternative approach to inactivate these regulatory RNAs, we tested whether depletion of miRNAs in Drosophila embryos by injection of miRNA-specific DNA antisense oligonucleotides would cause developmental defects. We could show that some antisense RNAs interfered with normal development, whereas other antisense DNAs had no impact. We further used the same antisense DNA oligonucleotide to develop an experimental PCR strategy for identification of putative target genes that are regulated by miRNAs.
MATERIALS AND METHODS
Injections of Drosophila embryos
DNA oligonucleotides complementary to 11 miRNAs were custom synthesised (MWG Germany) purified and adjusted to a concentration of 100 µM, injected to Drosophila embryos, which were treated as described (24 and http://images.cellpress.com/supmat/cub/bb11_22Boutla_1776.pdf).
Cloning of miR-13-specific target genes
From an early (4–8 h) and late (12–24 h) cDNA expression library from Drosophila embryos (25) the DNA was pooled separately. Approximately 1 µg of each library was subjected to a PCR with anti-miDNA-13a and a primer specific for the promoter of T7 RNA polymerase (5′-TAATACGACTCACTATAGGG) present on the cloning vector. The reaction was performed in 20 mM Tris pH 8.4, 50 mM KCl, 2 mM MgCl2 with 10 cycles at an annealing temperature of 35°C, followed by 20 cycles of 50°C (temperature for extension 72°C and denaturation 94°C). The amplified DNAs were directly cloned into the pGEM®-T Easy Vector System I (Promega) and the inserts sequenced with the anti-miDNA-13a.
Expression analysis
The cDNA corresponding to the 3′ UTR of genes CG10222 and CG9498 was amplified by PCR with the DNA oligonucleotides, E134-1, CCTCTAGAGAAACTAATCGAAAATAGCCTGTGATATTGTGCATTGTATTTC, and E1328-1, CCTCTAGACATGAGTATTATAATGGTTGGGGTTCTGTGATATAAATGGAGGCTCTTC, respectively, in combination with an oligonucleotide Bam-T7, CGGGATCCTAATACGACTCACTATAGGG, specific for the promoter of T7 RNA polymerase. By using the long primers we restored the complete 3′ UTR sequence. The PCR product was cleaved with XbaI and BamHI and subcloned into the same sites of the pGL3-Basic vector (Promega), replacing the SV40 late poly(A) signal. The vector had been previously modified by the addition of the act5C promoter sequence isolated from plasmid Ract-HAdh (26) generating pact-GL3. The construction of the mutated 3′ UTR was done in the same manner but DNA oligonucleotides carrying the mutations given in Figure 3 were used instead. Recombinant DNAs were purified using the Qiagen plasmid purification system.
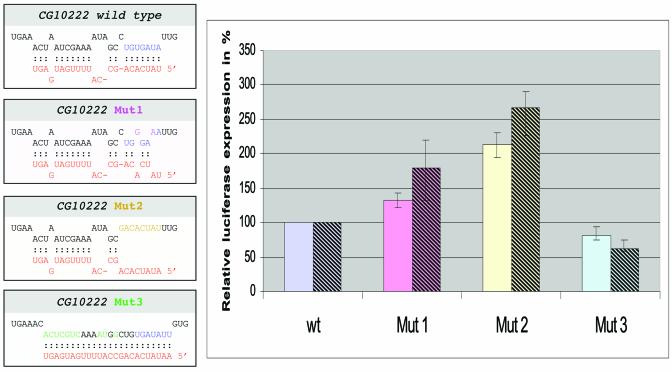
Figure 3.
Altering the miR-13 target site in the 3′ UTR of gene CG10222 and its influence on gene expression. (Left) The 3′ UTR of gene CG10222 and three mutant constructs with its predicted base pairing with miR-13b. The 3′ UTRs were fused to a pact-GL3 reporter construct and transfected into S2 cells. Transfection efficiency was first normalised to a reporter plasmid expressing LacZ. (Right) Relative expression level in percent with respect to the wild-type 3′ UTR of gene CG1022, which was set to 100%. Average expression levels and standard deviations obtained from six independent experiments with 1800 (left bars) or 450 ng (right bars) are given. The relative effect of the mutations is stronger with the lower amount of transfected plasmid.
Two micrograms of plasmid DNA was transfected using the calcium phosphate method into one million Drosophila Schneider 2 (S2) cells as described (27). The DNA contained either 1800 ng of the pGL3-based luciferase fusion construct plus 200 ng of a LacZ expressing plasmid or 450 ng of the former plus 200 ng of the latter, supplemented with 1350 ng of pBluescript-SK (Stratagene). The cells were incubated in Shields and Sang M3 insect medium (Sigma) supplemented with 10% fetal bovine serum (Gibco) and 50 mg/l gentamycin (Fluka) at 25°C for 48 h. After cells were collected, luciferase activity was determined according to the Promega protocol.
RESULTS AND DISCUSSION
Injection of micro antisense DNAs in Drosophila embryos
We were interested to test whether depletion of specific miRNAs would cause developmental defects, as is the case for loss-of-function mutations of lin-4 and let-7 in C.elegans (15–17). From the originally described Drosophila miRNAs (1), we selected 11 sequences that are expressed in the early embryo. DNA oligonucleotides of antisense polarity and covering the entire miRNA sequence were synthesised (anti-miDNA). For each DNA, approximately 200 Drosophila embryos were injected from the posterior side at a concentration where DNA oligonulceotides do not cause any unspecific effects (∼100 pl/embryo of a 100 µM solution) (24). We anticipated that the high amount of perfectly matching anti-miDNA would be sufficient to titrate out the free single-stranded form of miRNAs, thus preventing them from hybridising to their target RNAs. Forty-eight hours after injection, the cuticle phenotypes of the embryos were analysed. Buffer-injected controls showed a viability rate of 60–70%, which is slightly lower than the rate of ∼80–90% in untreated animals. A variety of developmental defects were seen after the injection of four anti-miDNAs specific for miR-1 and miR-3 and the two related RNAs miR-2a and miR-13a. The frequency of occurrence was ∼35% for anti-miDNA-1 and anti-miDNA-3 and 60–65% for anti-miDNA-13a and anti-miDNA-2a. Simultaneously, the viability dropped to 30–35%. The defects observed after injection with anti-miDNA-1 and anti-miDNA-3 were highly variable (data not shown), yet different to non-viable buffer-injected controls, which mostly exhibited head defects (Fig. 1A and B). In contrast, specific defects were seen after injection with anti-miDNA-2a and anti-miDNA-13a (Fig. 1C–F). Most embryos exhibited defects in the head and posterior abdominal segments, including cuticle holes and denticle belt malformations. Yet, the overall body pattern was not altered and both anterior-posterior and dorso-ventral polarity were clearly visible. The phenotypes between anti-miDNA-2a and anti-miDNA-13a were indistinguishable, reflecting the close relationship of the two targeted miRNAs (Fig. 2A). Despite a size difference of 1 nt, both miRNAs are identical in the 11 5′-terminal nucleotides along with two further motifs of 5 and 3 nt. In view of the consistency of the induced phenotypes, we conclude that the related miRNAs miR-2a and miR-13a act on the same target genes, together with the related miR-2b and miR-13b, which each differ by just 1 nt, respectively, in position 5 from the 3′ end. These four miRNAs form a functional sub-group of small RNAs, henceforth called miRs-2/13.
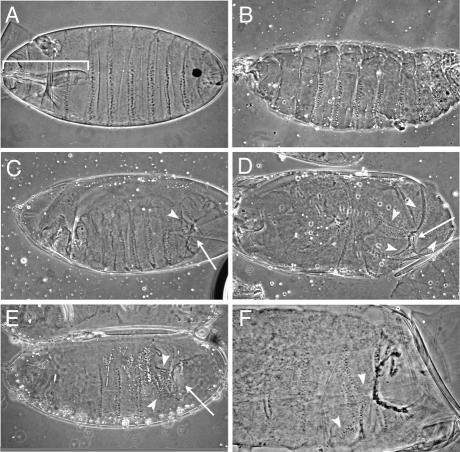
Figure 1.
Developmental defects due to injection of DNA oligonucleotides complementary to miRNAs. All embryos are oriented with anterior to the left and ventral down. (A) Wild-type embryo just before hatching (buffer- injected control). Note the wild-type mouth parts (bracket) that are missing in all other embryos. (B) Example of non-specific developmental defects in buffer-injected embryos (loss/deformation of mouth parts). (C–F) Anti-mi-DNA injected embryos. Embryos injected with anti-miDNA-2a (C and D) or anti-miDNA-13a (E and F) consistently produce abdominal defects, such as cuticle holes (arrows) and disorganised denticle belts (arrowheads), seen clearly in the higher magnification detail in (F).
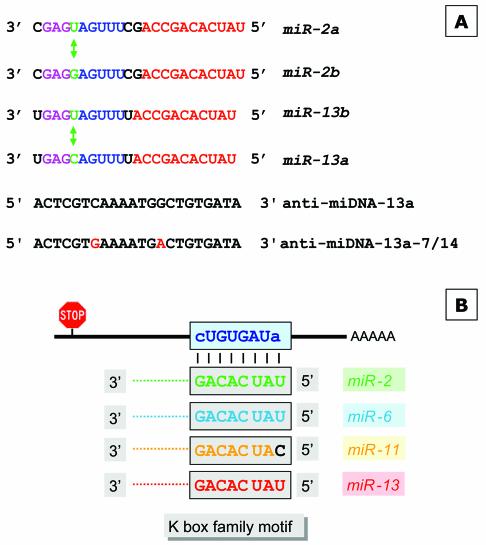
Figure 2.
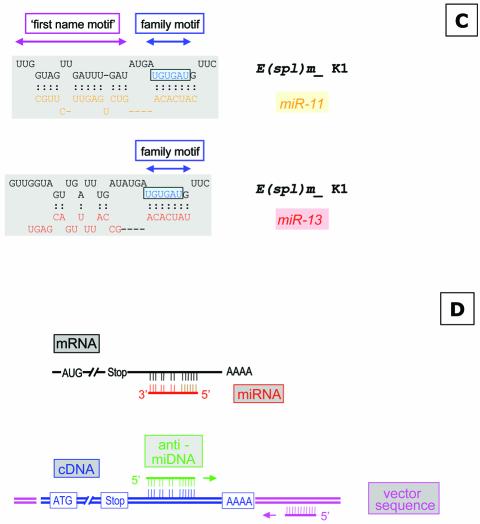
Schematic overview of miRNAs specific for interaction with the K box and the DNA antisense oligonucleotides used for injection or target gene identification. (A) The sequences of miR-2a, miR-2b, miR-13a and miR-13b are given; identical sequence elements are color-coded. The nucleotide which discriminates variant a and b of each miRNA are indicated as well. The anti-miDNA oligonucleotides used for injection are given and the positions of the mutations introduced to anti-miDNA-13a-7/14 are indicated. (B) Schematic interaction of the miRNAs with the K box consensus sequence as described (18). (C) Examples of base pairing interaction of a K box motif. The K1 box of E (spl) mδ is able to form an interaction with miR-11 within the family and the first name motif, while miR-13 can interact only via the family motif. (D) Strategy to identify target genes that are negatively regulated by miRNAs. (Top) The schematic map of a mRNA and the binding of a miRNA at the 3′ UTR (similar to B and C). Many miRNAs may pair perfectly via the family domain at their 5′ terminus, while the base pairing in the 3′ part of the miRNA is not continuous. Thus, the antisense DNA oligonucleotide will base pair perfectly via its 3′ domain allowing the initiation of a PCR on a cloned cDNA template (bottom), supported by additional base pairing in the 5′ domain. The second primer is specific for a vector sequence.
Injection of the residual seven anti-miDNAs (specific for miR-4, miR-5, miR-6, miR-7, miR-8, miR-9, miR-11) had no impact on viability (∼60%), which was comparable with that of buffer-injected controls. Only a small fraction (∼10–20%) of embryos injected with anti-miDNA-11 showed a mild form of the defect seen when miRs-2/13 was targeted, namely disorganised posterior segments, but no cuticle disruptions. This lack of interfering activity suggested that either the residual six miRNAs were not accessible to their anti-miDNA, for example, because they were part of a multi-component ribonucleoprotein (RNP) complex (28), or that the hybridisation to anti-miDNA did not cause dramatic effects. We cannot rule out the possibility that some miRNAs may have redundant functions, so that depletion of one specific sequence might be tolerated.
In order to confirm that phenotypic defects were caused by functional inactivation of miRs-2/13, we altered the anti-miDNA-13a in positions 7 (C to G) and 14 (G to A), which corresponds to positions 16 and 9 in miR-13a (Fig. 2A). The two nucleotide changes were selected to prevent efficient interaction of the antisense DNA oligonucleotide with mi-R13a or any other miRNAs of the miRs-2/13 family. No contiguous base pairing was possible, but only three short helices of 6, 6 and 8 bp, respectively. Injection of anti-miDNA-13a-7/14 did not cause any developmental defect—the viability and phenotype of embryos were no different than the buffer-injected controls (data not shown). Though we have no direct proof that the injected anti-miDNA-2 or anti-miDNA-13 interacts with the miRs-2/13 in the embryo, the observation that the developmental defects correlate with the predicted ability of the injected DNA to hybridise with miRs-2/13 strongly supports the hypothesis that these miRNAs play an important regulatory role in embryonic development. In this context, it is important to note that the depletion strategy allowed us to inactivate simultaneously a group of miRNAs that occur in the Drosophila genome in at least seven genes in four loci. Therefore, antisense DNA oligonucleotides provide a specific way to assign miRNA function, as an alternative to recently described examples, where mutant phenotypes in human and Drosophila could be attributed to the deletion of specific miRNA genes (21,22,29).
Identification of target genes negatively regulated by miR-13
To obtain an insight on how miRs-2/13 might regulate embryonic development, we set out to identify putative target mRNAs. Recently, target genes were identified for some plant miRNAs, based on computational methods (4,30). miR-39 (also called miR-171) of Arabidopsis thaliana is completely complementary to the mRNA of three Scarecrow-like (SCL) transcription factors (4,5,30,31). In accordance with the perfect complementarity, plant miRNA can cleave their mRNA targets via the siRNA pathway (31,32). Also, non-plant single-stranded short RNAs, including miRNAs, may be incorporated into the RNA-induced silencing complex (RISC) and if a complementary target RNA is available they may enter the RNAi pathway (33,34). Rhoades et al. (30) predicted for a total of 14 miRNAs collectively, 49 plant mRNA targets when they allowed some mismatches. The occurrence of hits in annotated sequences was significantly higher than in randomised sequences. In this context, it is of interest that a similar analysis for animal miRNAs, including Drosophila miRNAs, did not reveal target genes (30), suggesting that animal miRNAs work by a less stringent interaction, which makes it unlikely that miRNAs enter the RNAi pathway. On the other hand, it was shown that siRNAs with deliberately introduced mutations to make them only partially complementary to sites in 3′ UTRs of reporter genes, may function as miRNAs by translational repression (35).
To identify putative miR-13 target genes we used a combined bioinformatics and molecular approach. The basis for our strategy is the previous observation that miRNAs have two domains. Lai pointed out (18) that the 5′ terminal domains of Drosophila’s miR-2a, miR-2b, miR-6, miR-11, miR-13a and miR-13b are related and contain a sequence element that is complementary to the consensus sequence of the K box (36), a 3′ UTR sequence motif that mediates negative post- transcriptional regulation (Fig. 2B), while other miRNAs have similarities to boxes called Brd and GY (37). Thus, the first 6–8 nt at their 5′ terminus characterise the miRNA family and can therefore be considered the ‘family name’ of miRNAs. The domain further downstream distinguishes miRNAs of the same family and characterises a specific ‘first name’ (Fig. 2C). As pointed out earlier, some miRNAs share similarity not only in their family domain but also in the 3′-terminal domain, for example, miR-13a, miR-13b, miR-2a and miR-2b. In order to interact specifically with a miRNA, an mRNA needs to have the equivalent matching two-part target motif, for example, the K box plus some additional matching sequences further upstream in the mRNA. We inspected 18 known K boxes for their potential interaction with different miRNAs of the K box family, which includes miRs-2/13, miR-6 and miR-11, and found that in many cases there is a clear preference for interaction with a specific miRNA (see Supplementary Material which lists the potential interactions of four types of miRNAs with K boxes sequences located in the 3′ UTR of various genes of the Enhancer of split gene complex). For example, the first K box (K1) of the E(spl)mδ transcription unit is able to interact only with miR-11 (Fig. 2C), while the second K box (K2) in the same 3′ UTR can interact to some extent only with miR-13a. Similarly, there are K boxes with preference for miR-6 [E(spl)mγ] or miR-2b [E(spl)m7], but other K boxes may interact with two or even three miRNAs. For example, E(spl) m4 seems to interact almost equally well with miR-2b, miR-11 and miR-13. These examples suggest a potentially very complex regulation of translation by miRNAs. A specific miRNA may influence several targets, and conversely the target RNA may require a specific miRNA or a group of related miRNAs.
Guided by these considerations of miRNA/target interaction via two motifs we developed a strategy to identify target genes from a cDNA library (Fig. 2D) that are regulated by miR-13. The key point was that the anti-miDNA would match perfectly at its 3′ end (corresponding to the more constant family name of the miRNA) to a target sequence allowing initiation of a PCR. The strategy cannot avoid the amplification of cDNAs that have an accidental match with the six 3′ terminal nucleotides of the anti-miDNA. However, we anticipated that genuine miR-13 targets would have additional matches in the residual domain and would thus be amplified preferentially. We subjected pooled DNA from a cDNA library originating either from early embryos (4–8 h) or from late embryos (12–24 h) to this PCR amplification procedure using anti-miDNA-13a together with a vector-specific DNA oligonucleotide as primers. The PCR products were cloned and after analysing the size of the inserts, 10 clones from each library were sequenced. Comparison with the Drosophila sequence data showed that all cDNA inserts originated from priming of anti-miDNA-13a. With two exceptions, all cloned sequences showed perfect matches with the six to nine 3′-terminal nucleotides of the primer. Approximately half (9/20) of the sequences were the result of occurrence of the miR-13 family motif outside of a 3′ UTR region, e.g. far upstream of the stop codon. We observed that two potential binding sites for miR-13 were in exons close to exon–intron junctions. These were not considered further.
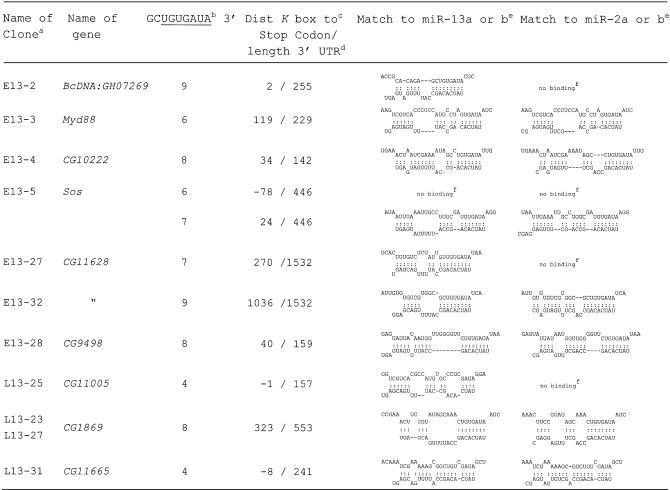
The genes that contained sequences with potential target motifs for miR-13 and miR-2 within or close to the 3′ UTR are listed in Table . With two exceptions (CG11005 and CG11665) they contain a K box consensus motif in their 3′ UTR, which is where the PCR product was primed from. The sequences were analysed with the aid of the Mfold program (38) for the potential to form RNA duplexes with miR-13a or miR-13b and also with the related miR-2a or miR-2b. All cloned motifs showed a potential interaction with either miR-13a or miR-13b. As expected, in most cases the same motif would also allow interaction with miR-2a or miR-2b. Cases where interaction was limited to the K box only and did not extend to flanking nucleotides, are denoted ‘no binding’. In general, the interactions detected in our novel putative target genes seemed to be at least as good as that of previously characterised K box genes (see Supplementary Material). The Sos gene exhibited a second K box consensus sequence 78 nt upstream of the stop codon. Unlike the K box motif identified by cloning (24 nt downstream of the stop codon) the upstream motif shows no specific interaction, neither with miR-13a and miR-13b nor with miR-2a and miR-2b, nor any other known miRNA, so that the occurrence of the upstream K box is likely to be accidental. A more careful inspection of the 3′ UTR of the target genes revealed in several cases additional potential binding sites, lacking a consensus K box.
Table 1. Identified Drosophila genes, their 3′ UTR motifs and potential interaction with miR-13 and miR-2.
aClones originating from an early and late cDNA library are denoted with E13 and L13, respectively.
bNumber of nucleotides matching to the 3′ end of the outlined 9 nt sequence element, wherein the K box is underlined.
cNumber of nucleotides between the last nucleotide of the stop codon and the 3′ terminal A of the K box and matching to the 5′ U of miR-13. Negative numbers indicate that the miR-13 target site is located upstream of the stop codon or overlapping with it (CG1105).
dThe given length of the 3′ UTR is based on sequence data from the cDNA clones; it indicates the number of nucleotides between the stop codon and poly(A).
eThe structures have been calculated with the Mfold program (38) by artificially joining the two RNAs via a oligo A or C; the sequences of miR-13a and miR-13b refer to table 1 of Lagos-Quintana et al. (1).
fNo binding, only interaction of the two RNAs via the K box.
Mutations of the miR-13-responsive domain in reporter constructs and their influence on translation efficiency
In order to test for the functional relevance of the identified structures given in Table , we selected gene CG10222, which is characterised by a relatively short 3′ UTR, for a more detailed analysis. We transferred its 3′ UTR just downstream of the stop codon of an actin promoter-driven luciferase reporter gene. In addition, we introduced three types of mutations to the potential target motif for miR-13a and miR-13b (Fig. 3). All of those mutations would also influence binding to miR-2a and miR-2b. The first variant contained two point mutations in the 3′ UTR, which weaken the interaction with the K box family motif of miR-13a. The second type of mutation destroys the family motif completely and the third construct converts the target motif into a domain that matches perfectly to miR-13a. These luciferase-fusion constructs were transfected into S2 cells, which had been shown earlier to express miR-13a (1). Luciferase activity was determined and always normalised to a co-transfected plasmid expressing LacZ. The normalised luciferase values varied slightly between individual experiments depending on the conditions of the S2 cells. For easier comparison between individual experiments we determined expression levels of the mutated sequences in relation to the fusion constructs of the corresponding wild-type sequence of gene CG10222, which was set to 100%, respectively. Figure 3 summarises six transfection experiments performed with a high concentration of DNA. Mutation type 1, that weakened binding to miR-13, increased the expression level of luciferase by 25%. Mutation 2, which completely destroyed the K box motif, resulted in a further increase of expression to approximately twice the level of the wild-type sequence. In contrast, the conversion into a perfect miR-13 binding site (mutation 3) reduced luciferase activity by ∼18%. This pattern is consistent with the reduced or increased binding potential of the target motif for miR-13 and/or miR-2. A mutation similar to type 1 also increased expression in CG9498 by 25% (data not shown).
The observed difference in gene expression of the CG10222 wild-type 3′ UTR and its mutated form (non-matching to miR-13) was only approximately a factor of two. This observation allows several interpretations, which are not mutually exclusive. First, miRNAs may be used to ‘fine tune’ expression levels. This would be in agreement with the observation that 7 of the 11 tested anti-miDNAs did not cause any noticeable defects in vivo; however, it might not explain the severe defects seen after out-titration of miRs-2/13. Secondly, the 3′ UTR of the miRs-2/13 responsive gene CG10222 might contain a second binding motif for another miRNA, similar to lin-41 of C.elegans that provides binding sites for miRNAs let-7 and lin-4. We inspected the 3′ UTR of gene CG10222 and indeed we could identify a further binding site without a canonical K box motif that potentially could interact with miR-6, miR-11 and miR-13. A third possibility is that the moderate effect is caused by too high a concentration of the reporter target RNA. The transfection experiment makes use of the endogenous concentration of miRs-2/13 in S2 cells. Therefore, it is possible that the expressed luc/CG10222 reporter mRNA was present in a molar excess. If that were the case, most of the target RNAs would be free of miRNAs, so that the introduced mutations should have even less or no effect at all. Although we could not control expression levels in individual cells, we repeated the experiment using 25% of the plasmid DNA encoding the luc/CG10222 fusion constructs for transfection (Fig. 3, right bars). For the lower concentration we observed stronger influences of the mutations, which included a stronger inhibition (to 61%) for the matching miR-13 motif (mut3) and a stronger de-repression (∼180 and ∼260%) when the motif was weakened or destroyed (mut1 and mut2). This can be expected if the intracellular concentration of target RNA is reduced making a larger percentage of it susceptible to regulation by the endogenous miRNA. It should be added that we cannot completely rule out that the introduced sequence changes may influence mRNA stability, especially since K boxes seem to work by a combination of translational repression and influencing mRNA stability (18,36). However, that would not explain the repression observed with the matching mutation (mut3), where the K box is left intact.
Recently, it was found that the proapoptotic hid gene of Drosophila is under the control of the temporally and spatially regulated bantam miRNA (21). Five potential binding sites of the miRNA to the 3′ UTR of the target gene were reported. We subjected these binding sites to the Mfold analysis (38), like the genes that we had identified. This revealed that the interactions listed in Table are at least as strong as those reported for the bantam/hid pair. It is also noteworthy that the bantam miRNA exhibited a good match via its 5′ terminal nucleotides (family motif). Based on this analogy and the consistent effects seen in our analysis of the CG10222 3′ UTR, we conclude that the nine genes listed in Table represent good candidates to be negatively regulated by miRs-2/13. Little is known about the cellular role of CG10222 and CG9498 so that any consideration as to why these genes are under the control of miR-13 would be speculative. However, these two and some of the residual seven genes identified are most likely to be only a subset of the genes that are regulated by miRs-2/13, since our identified gene set seems far from saturated, because only two out of nine genes were recovered twice. Yet our putative target gene sample includes genes that encode a wide variety of products in terms of structure, presumed subcellular localisation and putative function. The only well studied gene is Sos, which encodes a Ras guanidine nucleotide exchange factor (GEF) (39). Another two genes appear to encode signalling molecules: Myd88 encodes a DEATH domain protein that is known to interact with the family of Toll receptors. CG11628 encodes a GEF for another GTP-binding protein, ARF, which is implicated in regulating vesicle traffic. Four genes are apparently involved in metabolic processes: CG10222 has similarity to ATPases, CG11005 has a short-chain dehydrogenase motif, CG1869 may be a chitinase and CG11665 is probably a monocarboxylic acid transporter. Finally, two genes, CG9498 and BcDNA:GH07269, encode completely novel proteins. It is noteworthy that we identified in a surprisingly small number of clones several potential target genes. Genes CG1869 and CG11628 were identified twice—the latter by two different motifs in its long 3′ UTR.
In conclusion, we could show that antisense DNA oligonucleotides specific for miRNAs can be used to inactivate miRNA function. Moreover, we have demonstrated that such oligonucleotides can be used for an experimental strategy to identify target genes that are post-transcriptionally regulated by miRNAs. A modification of the strategy would involve RNA oligonucleotides to initiate reverse transcription on a mRNA target. This would allow ‘authentic’ RNA–RNA base pairing and mimic the interaction of miRNA and target mRNA. Any of the two strategies can be used in a variety of systems to identify targets for miRNAs or other forms of tncRNAs. This will be of great importance as it has been demonstrated that more than half the cases of human B-cell chronic lymphocytic leukaemias (B-CLL) are correlated with the loss of a 30 kb fragment on human chromosome 13 encoding genes for miR-15 and miR-16 or the down-regulation of their expression (29). It will be interesting to see whether target genes will have the same tissue specificity as observed for their regulatory miRNAs (21,40).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Nikos Giagztoglou, Giorgos Vrenztos and Kostis Koumpanakis for providing instructions on the transfection assays and Ioannis Livadaras for injecting Drosophila embryos. We further thank Drs Mina Tsagris, Kriton Kalantidis and Michalis Averof for helpful suggestions. This work was supported by a grant from the General Secretariat for Research and Technology of the Hellenic Ministry of Development (contract PENED 01ED325).
REFERENCES
- 1.Lagos-Quintana M., Rauhut,R., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 2.Lau N.C., Lim,L.P., Weinstein,E.G. and Bartel,D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–62. [DOI] [PubMed] [Google Scholar]
- 3.Lee R.C. and Ambros,V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862–864. [DOI] [PubMed] [Google Scholar]
- 4.Llave C., Kasschau,K.D., Rector,M.A. and Carrington,J.C. (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell, 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinhart B.J., Weinstein,E.G., Rhoades,M.W., Bartel,B. and Bartel,D.P. (2002) MicroRNAs in plants. Genes Dev., 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambros V., Bartel,B., Bartel,D.P., Burge,C.B., Carrington,J.C., Chen,X., Dreyfuss,G., Eddy,S.R., Griffiths-Jones,S., Marshall,M. et al. (2003) A uniform system for microRNA annotation. RNA, 9, 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 8.Hutvagner G., Mclachlan,J., Pasquinelli,A.E., Balint,E., Tuschl,T. and Zamore,P.D. (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science, 293, 834–838. [DOI] [PubMed] [Google Scholar]
- 9.Grishok A., Pasquinelli,A.E., Conte,D., Li,N., Parrish,S., Ha,I., Baillie,D.L., Fire,A., Ruvkun,G. and Mello,C.C. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y., Jeon,K., Lee,J.T., Kim,S. and Kim,V.N. (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J., 21, 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutvagner G. and Zamore,P.D. (2002) RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev., 12, 225–232. [DOI] [PubMed] [Google Scholar]
- 12.Tijsterman M., Ketting,R.F. and Plasterk,R.H. (2002) The genetics of RNA silencing. Annu. Rev. Genet., 36, 489–519. [DOI] [PubMed] [Google Scholar]
- 13.Finnegan E.J., Margis,R. and Waterhouse,P.M. (2003) Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol., 13, 236–240. [DOI] [PubMed] [Google Scholar]
- 14.Boutet S., Vazquez,F., Liu,J., Beclin,C., Fagard,M., Gratias,A., Morel,J.B., Crete,P., Chen,X. and Vaucheret,H. (2003) Arabidopsis HEN1. A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol., 13, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee R.C., Feinbaum,R.L. and Ambros,V. (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 16.Pasquinelli A.E., Reinhart,B.J., Slack,F., Martindale,M.Q., Kuroda,M.I., Maller,B., Hayward,D.C., Ball,E.E., Degnan,B., Muller,P. et al. (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature, 408, 86–89. [DOI] [PubMed] [Google Scholar]
- 17.Reinhart B.J., Slack,F.J., Basson,M., Pasquinelli,A.E., Bettinger,J.C., Rougvie,A.E., Horvitz,H.R. and Ruvkun,G. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 403, 901–906. [DOI] [PubMed] [Google Scholar]
- 18.Lai E.C. (2002) Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genet., 30, 363–364. [DOI] [PubMed] [Google Scholar]
- 19.Abrahante J.E., Daul,A.L., Li,M., Volk,M.L., Tennessen,J.M., Miller,E.A. and Rougvie,A.E. (2003) The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by MicroRNAs. Dev. Cell, 4, 625–637. [DOI] [PubMed] [Google Scholar]
- 20.Lin S.Y., Johnson,S.M., Abraham,M., Vella,M.C., Pasquinelli,A., Gamberi,C., Gottlieb,E. and Slack,F.J. (2003) The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable MicroRNA target. Dev. Cell, 4, 639–650. [DOI] [PubMed] [Google Scholar]
- 21.Brennecke J., Hipfner,D.R., Stark,A., Russell,R.B. and Cohen,S.M. (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell, 113, 25–36. [DOI] [PubMed] [Google Scholar]
- 22.Xu P., Vernooy,S.Y., Guo,M. and Hay,B.A. (2003) The Drosophila MicroRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol., 13, 790–795. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V., Lee,R.C., Lavanway,A., Williams,P.T. and Jewell,D. (2003) MicroRNAs and other tiny endogenous RNAs in C. elegans.Curr. Biol., 13, 807–818. [DOI] [PubMed] [Google Scholar]
- 24.Boutla A., Delidakis,C., Livadaras,I., Tsagris,M. and Tabler,M. (2001) Short 5′-phosphorylated double-stranded RNAs induce RNA interference in Drosophila. Curr. Biol., 11, 1776–1780. [DOI] [PubMed] [Google Scholar]
- 25.Brown N.H. and Kafatos,F.C. (1988) Functional cDNA libraries from Drosophila embryos. J. Mol. Biol., 203, 425–437. [DOI] [PubMed] [Google Scholar]
- 26.Swevers L., Cherbas,L., Cherbas,P. and Iatrou,K. (1996) Bombyx EcR (BmEcR) and Bombyx USP (BmCF1) combine to form a functional ecdysone receptor. Insect Biochem. Mol. Biol., 26, 217–221. [DOI] [PubMed] [Google Scholar]
- 27.Eastman D.S., Slee,R., Skoufos,E., Bangalore,L., Bray,S. and Delidakis,C. (1997) Synergy between suppressor of Hairless and Notch in regulation of Enhancer of split m gamma and m delta expression. Mol. Cell. Biol., 17, 5620–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mourelatos Z., Dostie,J., Paushkin,S., Sharma,A., Charroux,B., Abel,L., Rappsilber,J., Mann,M. and Dreyfuss,G. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev., 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calin G.A., Dumitru,C.D., Shimizu,M., Bichi,R., Zupo,S., Noch,E., Aldler,H., Rattan,S., Keating,M., Rai,K. et al. (2002) Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA, 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhoades M.W., Reinhart,B.J., Lim,L.P., Burge,C.B., Bartel,B. and Bartel,D.P. (2002) Prediction of plant microRNA targets. Cell, 110, 513–520. [DOI] [PubMed] [Google Scholar]
- 31.Llave C., Xie,Z., Kasschau,K.D. and Carrington,J.C. (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science, 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- 32.Tang G., Reinhart,B.J., Bartel,D.P. and Zamore,P.D. (2003) A biochemical framework for RNA silencing in plants. Genes Dev., 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez J., Patkaniowska,A., Urlaub,H., Luhrmann,R. and Tuschl,T. (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell, 110, 563. [DOI] [PubMed] [Google Scholar]
- 34.Hutvagner G. and Zamore,P.D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- 35.Doench J.G., Petersen,C.P. and Sharp,P.A. (2003) siRNAs can function as miRNAs. Genes Dev., 17, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai E.C., Burks,C. and Posakony,J.W. (1998) The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of enhancer of split complex transcripts. Development, 125, 4077–4088. [DOI] [PubMed] [Google Scholar]
- 37.Lai E.C. and Posakony,J.W. (1997) The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development, 124, 4847–4856. [DOI] [PubMed] [Google Scholar]
- 38.Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raabe T. (2000) The sevenless signaling pathway: variations of a common theme. Biochim. Biophys. Acta, 1496, 151–163. [DOI] [PubMed] [Google Scholar]
- 40.Lagos-Quintana M., Rauhut,R., Yalcin,A., Meyer,J., Lendeckel,W. and Tuschl,T. (2002) Identification of tissue-specific MicroRNAs from mouse. Curr. Biol., 12, 735–739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.