Abstract
The excision repair machinery of a thermophilic bacterium has been shown to recognize and repair an expanded genetic base pair. Native Thermus aquaticus DNA polymerase will remove a mispaired natural base and replace it with a non-natural base to form an expanded base pair. In addition, DNA ligase will recognize a nick formed by polymerase between two non-natural base pairs and covalently attach the two strands, thus demonstrating complete repair of a bifurcated base-paired model duplex. These results add evidence to the idea that the cellular replication and repair machinery of an organism containing an expanded genetic alphabet could recognize and properly repair a site containing consecutive unnatural bases.
INTRODUCTION
Genetic information storage of all living organisms is derived from organized strings of just two base pairs (A:T and G:C). The simplicity of a two pair code is fascinating yet begs the question: are more than two base pairs possible? New base pairs have been explored with the hopes of one day developing synthetic organisms with additional codons encoding additional amino acids. Such synthetic organisms would have the ability to encode proteins with new chemical and physical properties. Until science can generate entirely new replication machinery, the possibilities for new life forms with expanded genetic systems will be dictated by how well nature’s machinery deals with the new alphabet.
Many efforts to expand the genetic code using a series of non-natural base pairs have been publicized. Strategies for expanding the nucleotide alphabet have included alternative hydrogen bonding schemes, or specific hydrophobic and van der Waals force interactions (1–4). Polymerase recognition and nucleotide strand incorporation have been demonstrated in all cases cited. In a more recent publication, it was shown that by combining both shape and novel hydrogen bond complementarity, one could develop an efficient and specific transcription product incorporating an unnatural base (5). There are many additional well-characterized studies involving the enzymatic recognition of unnatural base pairs in replication and transcription (6–18). Yet, in spite of all these data, there seems to be a lack of information regarding the enzymatic incorporation of consecutive non-standard bases. Another shortcoming that many of the new base pairs share is a higher than usual misincorporation rate. With this in mind, it is likely that any new base pairs that expand the genetic alphabet in living organisms will require recognition by a series of specific enzymes beyond the polymerases required to replicate them (19). When errors occur due to replication and DNA damage, the new bases will require recognition by specific excision repair enzymes.
In our simplified model of excision repair, there are three main steps: excision, replication and ligation. First, the polymerase is required to reproduce the correct copy. If an expanded genetic information system were randomly assembled with even just one additional base pair, two consecutive non-standard bases would occur on average once every nine positions. Therefore, primer extension and repair of sites containing at least two consecutively placed non-standard bases would be required. Secondly, nucleases will be required to excise incorrect nucleotides or damaged regions containing non-standard bases. In particular, the flap endonuclease-1 class enzymes (FEN-1) are believed to play a role in base excision repair, double-strand break (DSB) repair, recombination, as well as the repair of mismatches produced during the priming and synthesis of Okazaki fragments (20–22). Finally, once runs of unnatural bases are replicated and repaired, the nicked regions must be ligated to restore the covalent integrity of the repaired strand.
To investigate whether an organism has machinery necessary to repair a replication error occurring at a site containing two synthetic base pairs, a model system was created that would allow repair to be examined step wise and in unison (Fig. 1). We used our system to investigate three of the most important functions in DNA repair: polymerization, excision and ligation. These processes were studied individually as well as in combination. This model system provides standard, single-step methodology for analyzing how DNA-specific enzymes will recognize new base pairs.
Figure 1.
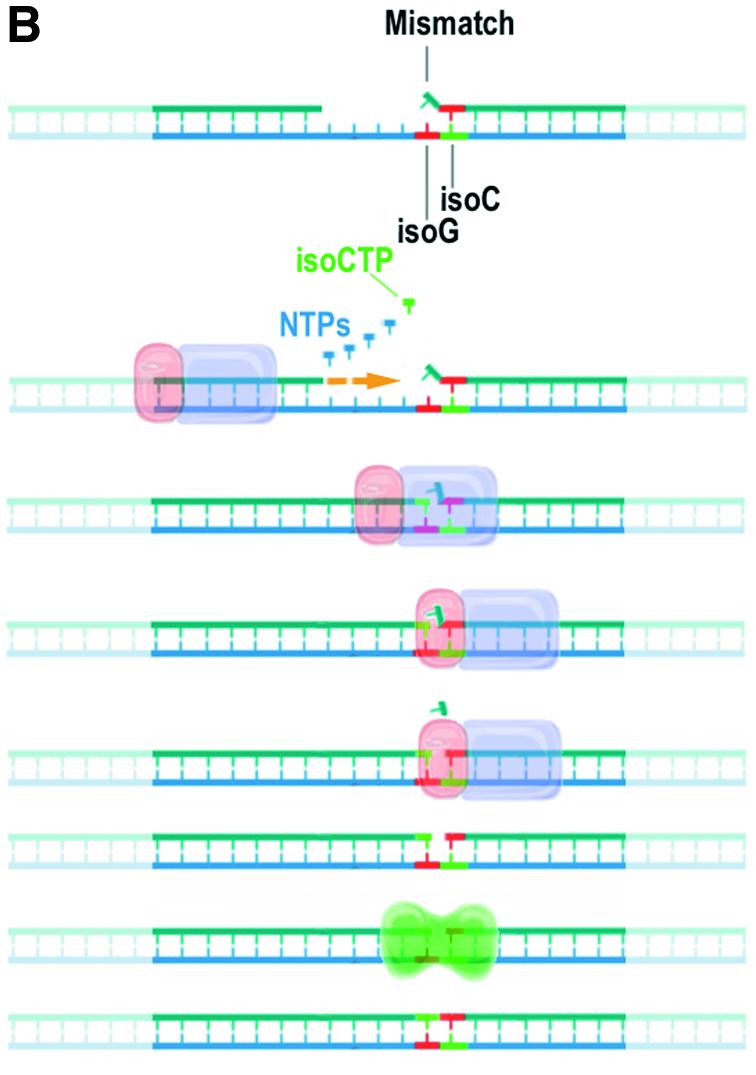
(A) Alternative bases that are structurally similar to standard nucleobases but have altered base-pairing specificity. The pattern of hydrogen bond acceptors and donors (indicated by arrows) is reversed for the non-standard base pair between 5-methyl isocytosine (isoC) and isoguanosine (isoG) (right) compared with a standard cytosine (C) guanosine (G) base pair (left). (B) Mismatch repair model of a template containing consecutive non-standard base pairs. The polymerase domain (purple) of a bacterial DNA polymerase catalyzes repair DNA synthesis by incorporation of nucleoside triphosphates. The 5′ nuclease domain (pink) of the polymerase excises a standard base (blue) mismatched with a non-natural base (isoG, red) in the template. DNA ligase (green) completes repair by formation of a covalent bond between isoC (green) and isoG (red).
MATERIALS AND METHODS
Polymerase extension
A DNA oligonucleotide primer 5′-labeled with Cy5 (5′-Cy5-GAGCAGCTCAGTGCA-3′) at 200 nM was combined with a DNA oligonucleotide template at 300 nM containing consecutive non-standard nucleotides (5′-GACCTGATCGATCGCXYTGCACTGAGCTGCTC-3′). The letters ‘X’ and ‘Y’ indicate the non-standard nucleotides 5-methyl isocytosine (isoC) and isoguanosine (isoG), respectively. KlenTaq, a nuclease-deficient, N-terminal truncation mutant of Thermus aquaticus DNA polymerase (Taq) (23) was used at 0.05 U/µl to extend the primer–template substrate in the presence of 20 µM of deoxynucleoside mixtures containing non-standard base triphosphates. Polymerase extension reactions were performed in a standardized polymerase–ligase buffer containing 10 mM bis-Tris propane–HCl pH 9.1, 40 mM potassium acetate, 2 mM magnesium chloride, 0.1 mg/ml bovine serum albumin (BSA), 5% polyethylene glycol 8000 (PEG-8000), 5 mM dithiothreitol (DTT), 5 mM NAD and 1 mM rATP. Reactions were cycled 10 times with the following conditions: 1 s at 95°C, 1 s at 50°C and 15 s at 68°C. Extension reactions were analyzed by 7 M urea, 20% PAGE and fluorescence imaging.
Nuclease cleavage
DNA oligonucleotide probes both 5′-labeled with fluorescein (FAM) and 3′-labeled with Cy3 DM120 (5′-FAM-TYTCCTGTCTGC-Cy3-T-3′) and MM257 (5′-FAM-XYTCCTGTCTGC-Cy3-T) at 200 nM were annealed to equimolar amounts of hairpin-forming target DNA oligonucleotides MM223 (5′-CACGACAGGCAGACAGGAXYGCTCACGTTTTCGTGAGC-3′) and MM271 (5′-CACG ACAGGCAGACAGGAXYGCTCACGTTTTCGTGAGCX-3′) possessing single-stranded 5′ ends complementary to the probe. Nuclease reactions were performed with full-length DNA polymerase isolated from T.aquaticus (Promega, Madison, WI) at 0.01 U/µl in the absence of nucleoside triphosphates at 55°C for 1 min. Reactions were performed in the same buffer as for polymerase extension except that nucleosides, DTT, NAD and PEG-8000 were omitted. Reactions were analyzed by 7 M urea, 20% PAGE and fluorescence imaging.
Ligation
A DNA oligonucleotide probe both 3′-labeled with Cy3 and containing a 5′ phosphoryl isoG (5′-PO4-YTCCTGTCTGC-Cy3-T-3′) at 20 nM was annealed to a 100-fold excess of hairpin-forming target DNA oligonucleotides MM223, MM271 and MM224 (5′-CACGACAGGCAGACAGGAXAGCTCACGTTTTCGTGAGCT-3′). Ligation reactions were performed in the same buffer as for polymerase extension except that the magnesium chloride concentration was increased to 10 mM. Reactions containing T4 ligase at 1.2 Weiss U/µl were incubated at 37°C for 60 min. Reactions containing Taq ligase at 8 U/µl were cycled 20 times between 1 s at 85°C and 3 min at 55°C. A DNA oligonucleotide 3′-labeled with Cy3 corresponding to the predicted reaction product was synthesized chemically and used as a size marker for electrophoresis. Reactions were analyzed by 7 M urea, 20% PAGE and fluorescence imaging.
Complete repair
DNA oligonucleotide probes both 5′-labeled with FAM and 3′-labeled with Cy3 DM120 and MM257 at 200 nM were annealed to equimolar amounts of hairpin-forming target DNA oligonucleotide MM223. Complete repair and control reactions were performed for 60 min at 37°C in the same buffer as for polymerase extension with combinations of either KlenTaq (nuclease minus) or Taq DNA polymerase at 0.2 U/µl; and T4 ligase at 0.6 U/µl or Taq ligase at 4 U/µl; and in the presence or absence of 10 µM 5-methyl isoCTP without other nucleosides except rATP. Repair reactions were also performed in the presence of 10 µM of the four standard nucleoside triphosphates, isoCTP and isoGTP. Reactions were analyzed by 7 M urea, 20% PAGE and fluorescence imaging.
RESULTS
Polymerase extension
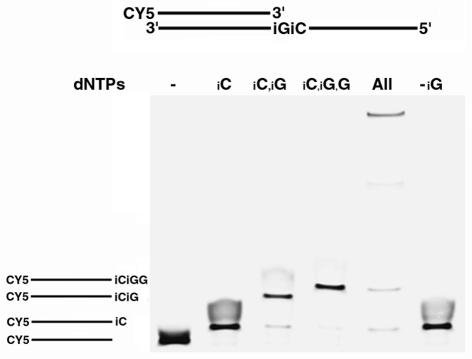
In vivo DNA synthesis of a non-natural nucleotide alphabet may require the incorporation of multiple non-standard bases, and in many cases these alternative bases will be strung together consecutively. Therefore, we first examined replication at a site containing two consecutive non-standard base pairs. We used gel electrophoresis and fluorescence analysis to demonstrate that Taq DNA polymerase is capable of polymerizing through and past the non-natural sites (Fig. 2). To demonstrate specificity, differing combinations of deoxynucleoside triphosphates were added to the PCRs. The results of the experiment demonstrate specific incorporation of isoG opposite isoC, incorporation of isoC opposite isoG, consecutive non-standard base incorporation as well as read through beyond the non-standard bases. In order to demonstrate preferential incorporation of isoG opposite isoC, we covalently labeled the 8 position of isoG with a number of modifiers that change the electrophoretic mobility. From these experiments, isoG was found to preferentially incorporate opposite isoC (data not shown).
Figure 2.
Polymerase extension through consecutive non-standard bases. A cyanine 5 (CY5) 5′-labeled DNA primer hybridized to a complementary DNA template was extended by Taq DNA polymerase in the presence of various non-standard and standard deoxynucleoside triphosphates. Extension products were resolved by denaturing polyacrylamide gel electrophoresis and detected by fluorescence imaging. Deoxynucleoside triphosphate combinations are indicated on top of each lane. Lane 1, no triphosphate (–); lane 2, d-isoCTP (iC); lane 3, d-isoCTP + d-isoGTP (iC,iG); lane 4, d-isoCTP + d-isoGTP + dGTP (iC,iG,G); lane 5, d-isoCTP + d-isoGTP + dNTP, where N = A, C, G, T (All); lane 6, d-isoCTP + dNTP (-iG). Reaction products are diagrammed on the left and are lined up with their respective band size.
Nuclease cleavage
An important question in any replication system is how will the machinery handle mistakes? For example, if the polymerase placed a standard base opposite a non-standard base, the machinery should be capable of repairing the mistake. To begin to investigate the question of repair, we built model systems using two oligonucleotides containing multiple non-standard bases, added the 5′ nuclease of Taq polymerase and observed cleavage activity (Fig. 3). The 5′ nuclease of Taq is a member of a family of excision repair enzymes termed FEN-1 enzymes. FEN-1 enzymes are specific endonucleases that cleave single-stranded DNA or RNA at a bifurcated site of a base-paired duplex (24). The FEN-1 family includes nucleases from all cellular life forms including Eubacteria as well as Archaea and Eukarya. This large family of structure-specific nucleases is believed to be involved in DNA replication, excision repair, DSB repair and recombination. Although family members possess similar enzymatic activity, there is little sequence similarity across the kingdoms (25,26).
Figure 3.
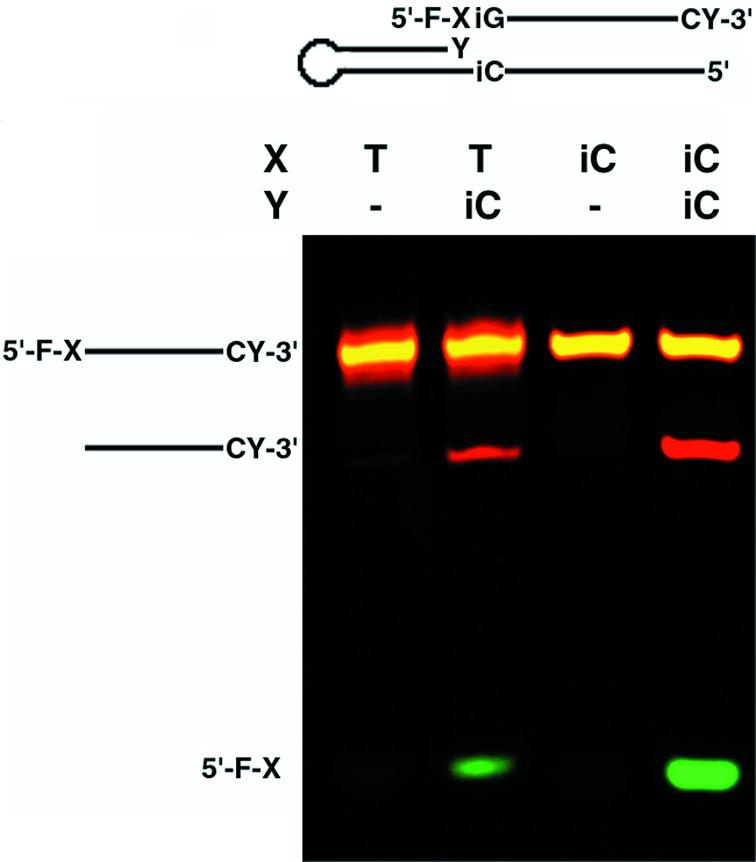
A model system demonstrating FEN-1 nuclease cleavage of a bifurcated site. DNA both 5′-labeled with fluorescein (F) and 3′-labeled with cyanine 3 (CY) was annealed to equimolar amounts of complementary hairpin-forming DNA. The 3′ ends of the hairpins either contained an isoC (iC) or no base (–) at position Y. The complementary base in the hairpin opposite position Y was isoG. The annealed substrates were treated with T.aquaticus DNA polymerase containing the 5′ FEN-1 domain in the absence of nucleoside triphosphates. Cleavage products were resolved by denaturing PAGE and detected by fluorescence imaging.
Model systems designed to analyze the activity of these nucleases as they relate to repair have previously been generated and used to probe enzyme activity and specificity (27,28). We developed a similar model system containing either an isoC:isoG match or a T:isoG mismatch. Our models represent a bifurcation product recognized in lagging strand replication and in excision repair, respectively. These model systems allow for the testing of how the FEN-1 enzymes recognize non-standard bases. Particularly, the mismatched model identifies the ability of the enzyme to remove a natural base opposite a non-natural base, required for non-natural base incorporation and repair. The isoG:T non-complementary mismatch was chosen since there are data that show that an isoG can direct polymerase misincorporation of T (2,15). In addition, this mispair has the apparent tendency to adopt Watson–Crick geometry (29), unlike the most stable natural mispair G:T that adopts wobble geometry. Therefore, it seemed appropriate to use this mismatch in the model.
To follow the excision reaction, the model substrates were labeled on both 5′ and 3′ ends with two separate fluorescent markers that could be individually resolved via gel electrophoresis and fluorescence imaging (Fig. 3). The flap endonucleases have been previously demonstrated to have varying activities toward matched versus mismatched substrates and typically cleave the 5′ flap leaving a nick (24). The flap can be created by polymerase 3′ extension of the correct base. Extension would force the mismatched base out of the way, leaving a bifurcated structure ready for specific nuclease activity to take place as would be the case when X = T and Y = isoC in our model. During initial rates, our results demonstrate that the flap substrate created by an isoC:isoG match is preferred over the T:isoG mismatch, which is further preferred over the non-flap substrates. It seems clear from the data that the FEN-1 enzyme has the ability to recognize the structure even though it contains non-standard bases on either side of the cleavage site.
Ligation
To look at the final step in this excision repair model, we tested Taq and T4 ligase for their ability to covalently attach a 3′ 5-methyl isoC or thymidine to a 5′-isoG when the bases were at the ends of two DNA strands hybridized to their complementary sequences. DNA ligases are essential for DNA replication as they catalyze the joining of Okazaki fragments during lagging strand synthesis and the nicks formed during DNA repair and recombination (30,31). Therefore, it is essential that ligase not only recognizes single non-natural base pairs but, as with the other enzymes, the ligase needs to recognize two non-natural bases and ligate them together to produce consecutive non-standard base pairs. Using a model system similar to that used to test the nuclease activity, covalent attachment by ligase of multiple non-natural bases was confirmed (Fig. 4). Where a gap was present between the strands, no ligation was observed. Our data suggest that both the T4 and Taq ligases can efficiently join consecutive non-standard bases, although our Taq ligase preparation was not as efficient as the T4 ligase (data not shown). The inefficient ligation activity of Taq ligase towards all-natural base substrates has been previously reported (32).
Figure 4.
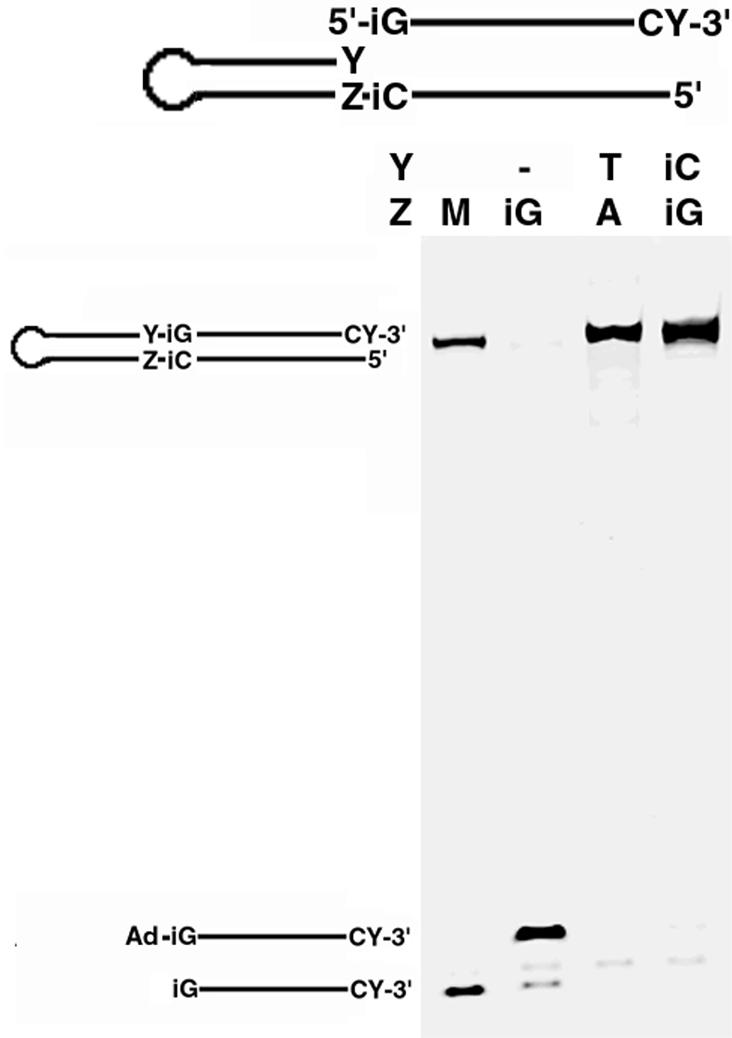
Ligation between consecutive non-standard bases. A 3′-labeled single-stranded DNA containing a 5′ phosphoryl isoguanosine (iG) was annealed to the complementary 5′ single-stranded ends of one of three DNA hairpins. The hairpins used all differed at position Y. The annealed substrates were treated with T4 DNA ligase under appropriate conditions. Ligation products including the 5′ adenylated iG probe intermediate (Ad-iG) indicated on the left of the gel were resolved by denaturing PAGE and detected by fluorescence imaging. A mock product was synthesized chemically and run along with the unligated probe as a size marker (M).
Complete repair
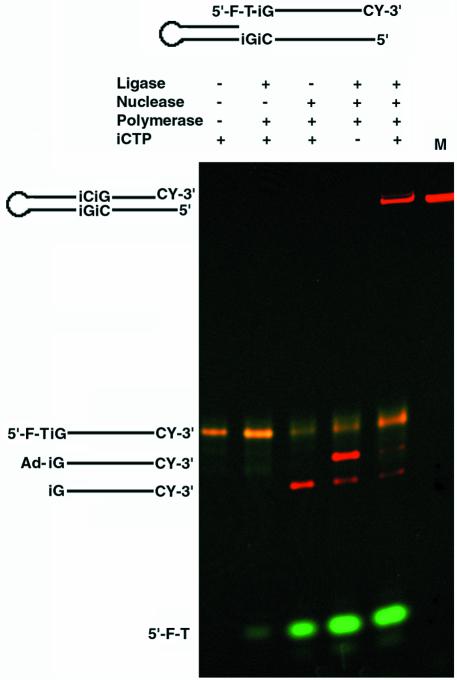
After demonstrating the repair steps individually, we tested a system in which all three steps could occur consecutively. The single-step model systems used above were combined to demonstrate that full repair of a bifurcated site containing unnatural bases and base pairs could be accomplished in a single reaction. In our model, complete repair required all three enzymes and the non-standard nucleotide (Fig. 5). First, when we omitted the FEN-1 nuclease activity from the reactions, ligase-specific products were not observed. This argues that the 5′ nuclease domain of Taq polymerase was involved in the removal of the bifurcated strand. The nuclease, polymerase and isoCTP were all required to form the nick substrate for DNA ligase. Ligase itself was also required for formation of the complete repair product generated by the polymerase and its substrate isoCTP. The isoCTP is required to form the nick. In its absence, nucleotide is not incorporated by polymerase opposite isoG, leaving a gap. A gap is not a substrate for complete ligation as we demonstrated in Figure 4. These data show that the activities of all three enzymes (polymerase, nuclease and ligase) as well as the nucleotide triphosphate are requirements to fully repair the bifurcated site. Given that the site of repair is flanked by two non-standard base pairs, and that repair required the replacement of a standard base with a non-standard base, the experiment demonstrates that these enzymes may be capable of functioning within a living system to create an in vivo expanded genetic information system.
Figure 5.
Complete repair of the mismatched bifurcated model system at a site containing consecutive non-standard DNA bases. A single-stranded DNA both 5′-labeled with fluorescein (F) and 3′-labeled with cyanine 3 (CY) was annealed to equimolar amounts of complementary hairpin-forming DNA. The 5′ thymidine (T) of the single-stranded DNA was mispaired with an isoguanosine (iG) in the hairpin. Repair reactions contained combinations of ligase, polymerase, FEN-1 nuclease and 5-methyl isocytosine triphosphate (iCTP) as indicated above the lanes. Complete repair reaction product, cleavage intermediate, 5′-phosphoryl probe and ligation intermediate adenylated probe (Ad-iG) indicated on the left of the gel were resolved by denaturing PAGE and detected by fluorescence imaging. A mock repair product was synthesized chemically and used as a size marker (M).
DISCUSSION
Here we demonstrate further evidence that it may be possible to use an expanded genetic alphabet to create synthetic organisms. First, we confirm that DNA polymerase accurately places non-standard nucleotides opposite their cognate bases, and we add to earlier findings by demonstrating that two non-standard bases can be incorporated consecutively. Secondly, we find that DNA ligase can recognize and ligate a site between two non-standard bases. Thirdly, we show evidence that sequences containing these non-naturally occurring bases are substrates for a repair nuclease. This work builds upon other findings where enzymes and cellular systems recognize and process non-standard bases. In one particular past example, the RecA protein catalyzed strand exchange between DNA oligomers containing isoG and isoC (21). The rate and efficiency of the exchange were comparable with those of a reaction where the substrate was all natural. It is important to note, however, that DNA replication and repair are catalyzed by an extremely wide variety of processing enzymes, of which only three were mentioned here. The genetic program that rectifies the errors or damage that occur includes mismatch repair, transcription-coupled repair, excision repair, DSB repair, checkpoints and recombination (19,33). Future exploration of these processes will allow for a more complete understanding of how these bases will be accepted by nature.
Finally, the work presented here is not limited to the creation of synthetic organisms. Indeed, the three enzymes shown here to recognize the isoC:isoG base pair also have utility in basic research and commercial diagnostics. Recently we reported that by using the polymerase and nuclease activity of Taq polymerase, a simple real-time PCR system could be constructed to quantitate and genotype nucleic acids (34). Work is in progress to show how new high-throughput directional cloning systems can be developed using isoC during PCR. We have also used reporter-modified isoGTP to demonstrate that during PCR, labels can be incorporated site specifically. The method allows for an easy way to place reporters on the 3′ ends of DNA amplicons and RNA transcripts. Additionally, a six base PCR system has been developed that should allow non-cross-reacting codes to be placed internally within PCR amplicons, allowing for quick deconvolution of multiplexed reactions. Also, six base PCR can be used for the production of aptamers with covalently attached functional groups allowing for the selection of better catalysts.
We view the science of additional base pairs to be a true paradigm shift in nucleic acid research. Certainly, if naturally occurring DNA replication, repair and recombination enzymes recognize these additional base pairs, the work involved in creating an expanded genetic code within a living organism becomes easier. Also an expanded genetic information system that is recognized by DNA repair enzymes may allow for the development of new and improved therapeutics and/or diagnostic systems.
REFERENCES
- 1.McMinn D.L., Ogawa,A.K., Wu,Y., Liu,J. and Romesberg,F.E. (1999) Efforts toward expansion of the genetic alphabet: DNA polymerase recognition of a highly stable, self-pairing hydrophobic base. J. Am. Chem. Soc., 121, 11585–11586. [Google Scholar]
- 2.Switzer C.Y., Moroney,S.E. and Benner,S.A. (1989) Enzymatic incorporation of a new base pair into DNA and RNA. J. Am. Chem. Soc., 111, 8322–8323. [Google Scholar]
- 3.Tae E.L., Wu,Y., Xia,G., Schultz,P.G. and Romesberg,F.E. (2001) Efforts toward expansion of the genetic alphabet: replication of DNA with three base pairs. J. Am. Chem. Soc., 123, 7439–7440. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y., Ogawa,A.K., Berger,M., McMinn,D.L., Schultz,P.G. and Romesberg,F.E. (2000) Efforts toward expansion of the genetic alphabet: optimization of interbase hydrophobic interactions. J. Am. Chem. Soc., 122, 7621–7632. [Google Scholar]
- 5.Ohtsuki T., Kimoto,M., Ishikawa,M., Mitsui,T., Hirao,I. and Yokoyama,S. (2001) Unnatural base pairs for specific transcription. Proc. Natl Acad. Sci. USA, 98, 4922–4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitsui T., Kitamura,A., Kimoto,M., To,T., Sato,A., Hirao,I. and Yokoyama,S. (2003) An unnatural hydrophobic base pair with shape complementarity between pyrrole-2-carbaldehyde and 9-methylimidazo[(4,5)-b]pyridine. J. Am. Chem. Soc., 125, 5298–5307. [DOI] [PubMed] [Google Scholar]
- 7.Yu C., Henry,A.A., Romesberg,F.E. and Schultz,P.G. (2002) Polymerase recognition of unnatural base pairs. Angew. Chem. Int. Ed., 41, 3841–3844. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y., Fa,M., Tae,E.L., Schultz,P.G. and Romesberg,F.E. (2002) Enzymatic phosphorylation of unnatural nucleosides. J. Am. Chem. Soc., 124, 14626–11430. [DOI] [PubMed] [Google Scholar]
- 9.Berger M., Luzzi,S.D., Henry,A.A. and Romesberg,F.E. (2002) Stability and selectivity of unnatural DNA with five-membered-ring nucleobase analogues. J. Am. Chem. Soc., 124, 1222–1226. [DOI] [PubMed] [Google Scholar]
- 10.Matsuda S., Henry,A.A., Schultz,P.G. and Romesberg,F.E. (2003) The effect of minor-groove hydrogen-bond acceptors and donors on the stability and replication of four unnatural base pairs. J. Am. Chem. Soc., 125, 6134–6139. [DOI] [PubMed] [Google Scholar]
- 11.Lutz M.J., Horlacher,J. and Benner,S.A. (1998) Recognition of a non-standard base pair by thermostable DNA polymerases. Bioorg. Med. Chem. Lett., 8, 1149–1152. [DOI] [PubMed] [Google Scholar]
- 12.Lutz M.J., Horlacher,J. and Benner,S.A. (1998) Recognition of 2′-deoxyisoguanosine triphosphate by HIV-1 reverse transcriptase and mammalian cellular DNA polymerases. Bioorg. Med. Chem. Lett., 8, 499–504. [DOI] [PubMed] [Google Scholar]
- 13.Lutz M.J., Held,H.A., Hottiger,M., Hubscher,U. and Benner,S.A. (1996) Differential discrimination of DNA polymerase for variants of the non-standard nucleobase pair between xanthosine and 2,4-diaminopyrimidine, two components of an expanded genetic alphabet. Nucleic Acids Res., 24, 1308–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horlacher J., Hottiger,M., Podust,V.N., Hubscher,U. and Benner,S.A. (1995) Recognition by viral and cellular DNA polymerases of nucleosides bearing bases with nonstandard hydrogen bonding patterns. Proc. Natl Acad. Sci. USA, 92, 6329–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Switzer C.Y., Moroney,S.E. and Benner,S.A. (1993) Enzymatic recognition of the base pair between isocytidine and isoguanosine. Biochemistry, 32, 10489–10496. [DOI] [PubMed] [Google Scholar]
- 16.Bain J.D., Switzer,C., Chamberlin,A.R. and Benner,S.A. (1992) Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature, 356, 537–539. [DOI] [PubMed] [Google Scholar]
- 17.Piccirilli J.A., Krauch,T., Moroney,S.E. and Benner,S.A. (1990) Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature, 343, 33–37. [DOI] [PubMed] [Google Scholar]
- 18.Hirao I., Ohtsuki,T., Fujiwara,T., Mitsui,T., Yokogawa,T., Okuni,T., Nakayama,H., Takio,K., Yabuki,T., Kigawa,T. et al. (2002) An unnatural base pair for incorporating amino acid analogs into proteins. Nat. Biotechnol., 20, 177–182. [DOI] [PubMed] [Google Scholar]
- 19.Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 20.Harrington J.J. and Lieber,M.R. (1994) The characterization of a mammalian DNA structure-specific endonuclease. EMBO J., 13, 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieber M.R. (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays, 19, 233–240. [DOI] [PubMed] [Google Scholar]
- 22.Wu X., Wilson,T.E. and Lieber,M.R. (1999) A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl Acad. Sci. USA, 96, 1303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes W.M. (1992) The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene, 112, 29–35. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser M.W., Lyamicheva,N., Ma,W., Miller,C., Neri,B., Fors,L. and Lyamichev,V.I. (1999) A comparison of eubacterial and archaeal structure-specific 5′-exonucleases. J. Biol. Chem., 274, 21387–21394. [DOI] [PubMed] [Google Scholar]
- 25.Hosfield D.J., Mol,C.D., Shen,B. and Tainer,J.A. (1998) Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell, 95, 135–146. [DOI] [PubMed] [Google Scholar]
- 26.Karanjawala Z.E., Shi,X., Hsieh,C.L. and Lieber,M.R. (2000) The mammalian FEN-1 locus: structure and conserved sequence features. Microb. Comp. Genomics, 5, 173–177. [DOI] [PubMed] [Google Scholar]
- 27.Murante R.S., Rust,L. and Bambara,R.A. (1995) Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem., 270, 30377–30383. [DOI] [PubMed] [Google Scholar]
- 28.Harrington J.J. and Lieber,M.R. (1995) DNA structural elements required for FEN-1 binding. J. Biol. Chem., 270, 4503–4508. [DOI] [PubMed] [Google Scholar]
- 29.Roberts C., Bandaru,R. and Switzer,C. (1997) Theoretical and experimental study of isoguanine and isocytosine: base pairing in an expanded genetic system. J. Am. Chem. Soc., 119, 4640–4649. [Google Scholar]
- 30.Timson D.J., Singleton,M.R. and Wigley,D.B. (2000) DNA ligases in the repair and replication of DNA. Mutat. Res., 460, 301–318. [DOI] [PubMed] [Google Scholar]
- 31.Lehman I.R. (1974) DNA ligase: structure, mechanism and function. Science, 186, 790–797. [DOI] [PubMed] [Google Scholar]
- 32.Housby J.N., Thorbjarnardottir,S.H., Jonsson,Z.O. and Southern,E.M. (2000) Optimised ligation of oligonucleotides by thermal ligases: comparison of Thermus scotoductus and Rhodothermus marinus DNA ligases to other thermophilic ligases. Nucleic Acids Res., 28, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedberg E.C. (2003) DNA damage and repair. Nature, 421, 436–440. [DOI] [PubMed] [Google Scholar]
- 34.Moser M.J., Marshall,D.J., Grenier,J.K., Kieffer,C.D., Killeen,A.A., Ptacin,J.L., Richmond,C.S., Roesch,E.B., Scherrer,C.W., Sherrill,C.B. et al. (2003) Exploiting the enzymatic recognition of an unnatural base pair to develop a universal genetic analysis system. Clin. Chem., 49, 407–414. [DOI] [PubMed] [Google Scholar]