Abstract
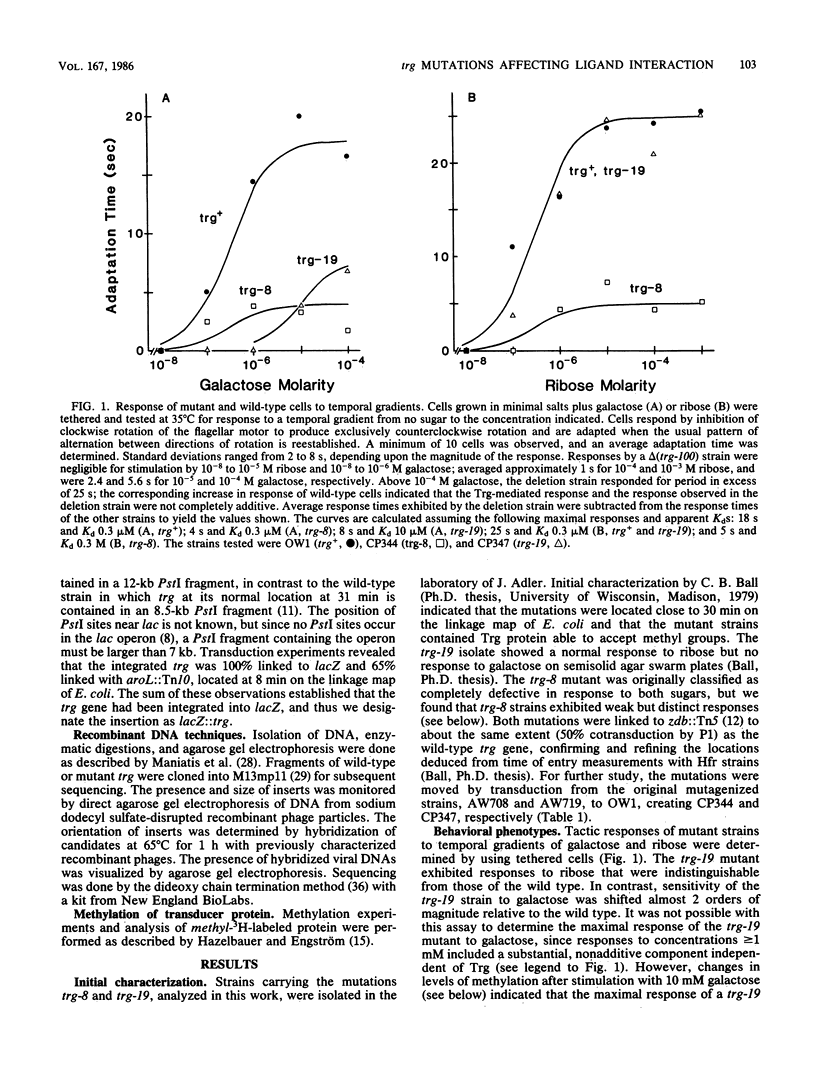
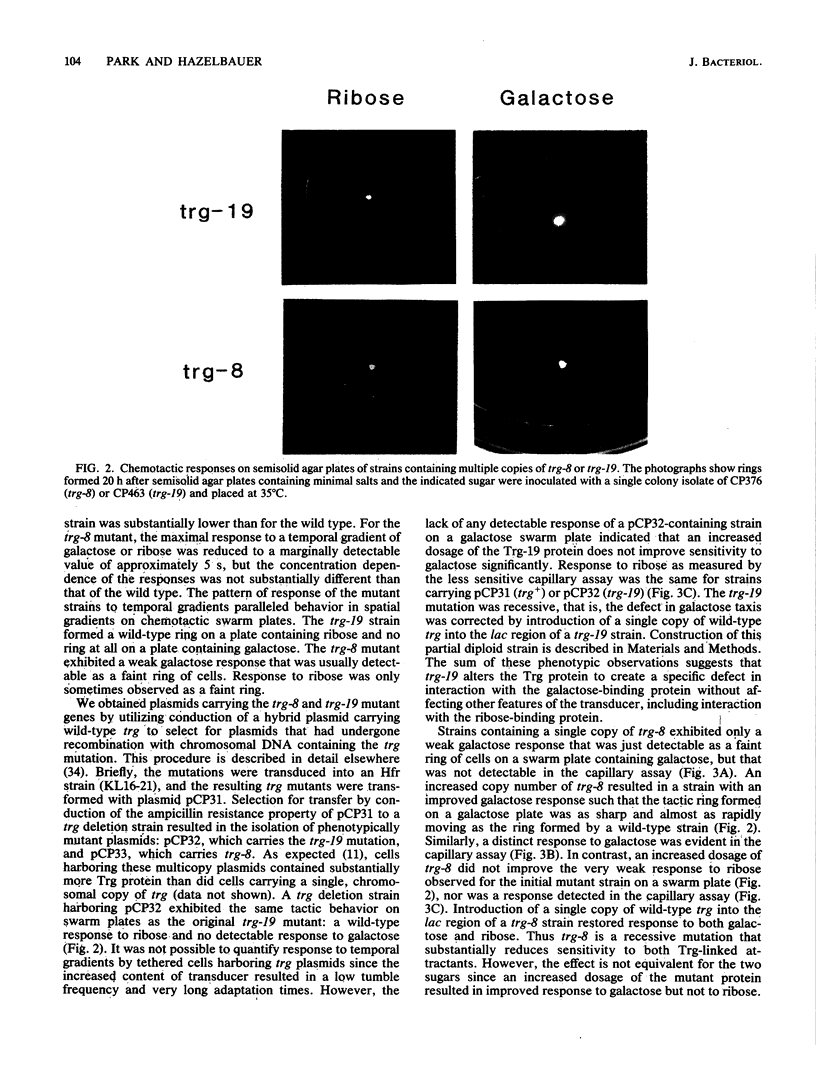
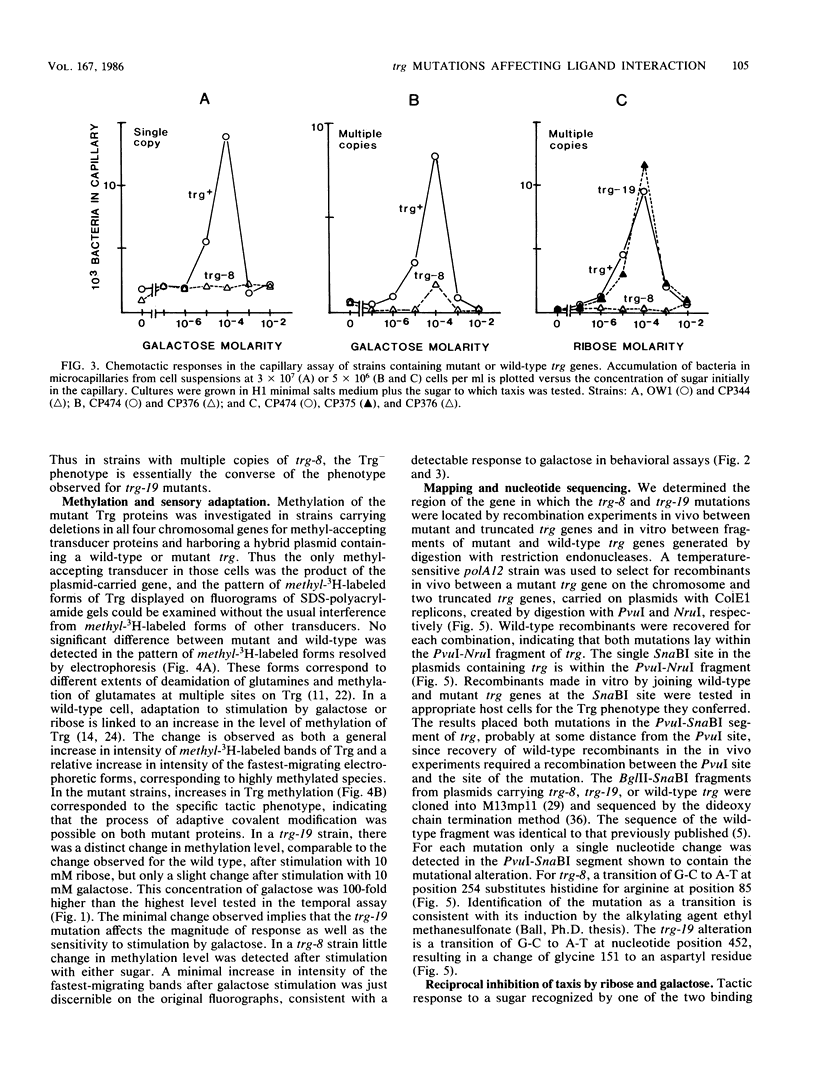
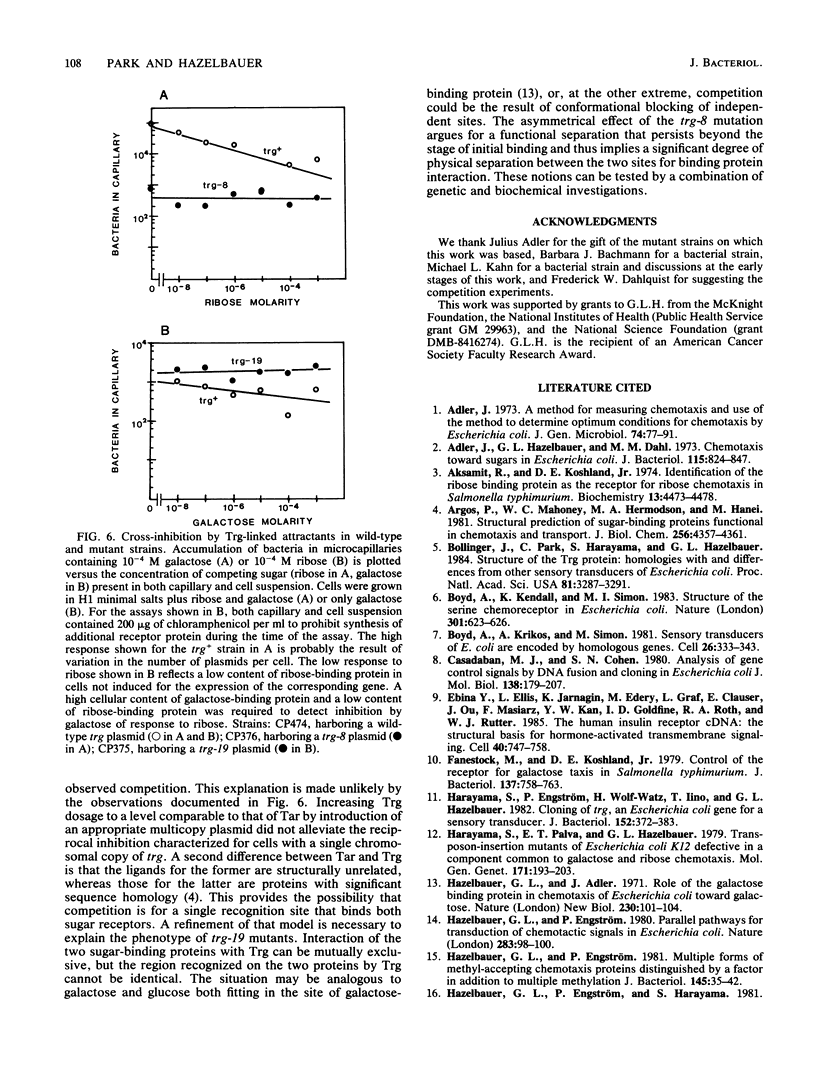
The Trg transducer mediates chemotactic response to galactose and ribose by interacting, respectively, with sugar-occupied galactose- and ribose-binding proteins. Adaptation is linked to methylation of specific glutamyl residues of the Trg protein. This study characterized two trg mutations that affect interaction with binding protein ligands but do not affect methylation or adaptation. The mutant phenotypes indicated that the steady-state activity of methyl-accepting sites is independent of ligand-binding activity. The mutation trg-8 changed arginine 85 to histidine, and trg-19 changed glycine 151 to aspartate. The locations of the mutational changes provided direct evidence for functioning of the amino-terminal domain of Trg in ligand recognition. Cross-inhibition of tactic sensitivity by the two Trg-linked attractants implies competition for a common site on Trg. However, the single amino acid substitution caused by trg-19 greatly reduced the response to galactose but left unperturbed the response to ribose. Thus Trg must recognize the two sugar-binding proteins at nonidentical sites, and the complementary sites on the respective binding proteins should differ. trg-8 mutants were substantially defective in the response to both galactose and ribose. An increase in cellular content of Trg-8 protein improved the response to galactose but not to ribose. It appears that Trg-8 protein is defective in the generation of the putative conformational change induced by ligand interaction. The asymmetry of the mutational defect implies that functional separation of interaction sites could persist beyond the initial stage of ligand binding.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol. 1973 Jan;74(1):77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- Adler J., Hazelbauer G. L., Dahl M. M. Chemotaxis toward sugars in Escherichia coli. J Bacteriol. 1973 Sep;115(3):824–847. doi: 10.1128/jb.115.3.824-847.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksamit R. R., Koshland D. E., Jr Identification of the ribose binding protein as the receptor for ribose chemotaxis in Salmonella typhimurium. Biochemistry. 1974 Oct 22;13(22):4473–4478. doi: 10.1021/bi00719a001. [DOI] [PubMed] [Google Scholar]
- Argos P., Mahoney W. C., Hermodson M. A., Hanei M. Structural prediction of sugar-binding proteins functional in chemotaxis and transport. J Biol Chem. 1981 May 10;256(9):4357–4361. [PubMed] [Google Scholar]
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Boyd A., Krikos A., Simon M. Sensory transducers of E. coli are encoded by homologous genes. Cell. 1981 Nov;26(3 Pt 1):333–343. doi: 10.1016/0092-8674(81)90202-6. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Ebina Y., Ellis L., Jarnagin K., Edery M., Graf L., Clauser E., Ou J. H., Masiarz F., Kan Y. W., Goldfine I. D. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell. 1985 Apr;40(4):747–758. doi: 10.1016/0092-8674(85)90334-4. [DOI] [PubMed] [Google Scholar]
- Fahnestock M., Koshland D. E., Jr Control of the receptor for galactose taxis in Salmonella typhimurium. J Bacteriol. 1979 Feb;137(2):758–763. doi: 10.1128/jb.137.2.758-763.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Engström P., Wolf-Watz H., Iino T., Hazelbauer G. L. Cloning of trg, a gene for a sensory transducer in Escherichia coli. J Bacteriol. 1982 Oct;152(1):372–383. doi: 10.1128/jb.152.1.372-383.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Palva E. T., Hazelbauer G. L. Transposon-insertion mutants of Escherichia coli K12 defective in a component common to galactose and ribose chemotaxis. Mol Gen Genet. 1979 Mar 20;171(2):193–203. doi: 10.1007/BF00270005. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Adler J. Role of the galactose binding protein in chemotaxis of Escherichia coli toward galactose. Nat New Biol. 1971 Mar 24;230(12):101–104. doi: 10.1038/newbio230101a0. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Engström P. Multiple forms of methyl-accepting chemotaxis proteins distinguished by a factor in addition to multiple methylation. J Bacteriol. 1981 Jan;145(1):35–42. doi: 10.1128/jb.145.1.35-42.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G. L., Engström P. Parallel pathways for transduction of chemotactic signals in Escherichia coli. Nature. 1980 Jan 3;283(5742):98–100. doi: 10.1038/283098a0. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Harayama S. Mutants in transmission of chemotactic signals from two independent receptors of E. coli. Cell. 1979 Mar;16(3):617–625. doi: 10.1016/0092-8674(79)90035-7. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Harayama S. Sensory transduction in bacterial chemotaxis. Int Rev Cytol. 1983;81:33–70. doi: 10.1016/s0074-7696(08)62334-7. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Mesibov R. E., Adler J. Escherichia coli mutants defective in chemotaxis toward specific chemicals. Proc Natl Acad Sci U S A. 1969 Dec;64(4):1300–1307. doi: 10.1073/pnas.64.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehry M. R., Dahlquist F. W. Adaptation in bacterial chemotaxis: CheB-dependent modification permits additional methylations of sensory transducer proteins. Cell. 1982 Jul;29(3):761–772. doi: 10.1016/0092-8674(82)90438-x. [DOI] [PubMed] [Google Scholar]
- Kehry M. R., Dahlquist F. W. The methyl-accepting chemotaxis proteins of Escherichia coli. Identification of the multiple methylation sites on methyl-accepting chemotaxis protein I. J Biol Chem. 1982 Sep 10;257(17):10378–10386. [PubMed] [Google Scholar]
- Kehry M. R., Engström P., Dahlquist F. W., Hazelbauer G. L. Multiple covalent modifications of Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1983 Apr 25;258(8):5050–5055. [PubMed] [Google Scholar]
- Koiwai O., Hayashi H. Studies on bacterial chemotaxis. IV. Interaction of maltose receptor with a membrane-bound chemosensing component. J Biochem. 1979 Jul;86(1):27–34. [PubMed] [Google Scholar]
- Kondoh H., Ball C. B., Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Conley M. P., Boyd A., Berg H. C., Simon M. I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Maloy S. R., Nunn W. D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981 Feb;145(2):1110–1111. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mowbray S. L., Foster D. L., Koshland D. E., Jr Proteolytic fragments identified with domains of the aspartate chemoreceptor. J Biol Chem. 1985 Sep 25;260(21):11711–11718. [PubMed] [Google Scholar]
- Ordal G. W., Adler J. Isolation and complementation of mutants in galactose taxis and transport. J Bacteriol. 1974 Feb;117(2):509–516. doi: 10.1128/jb.117.2.509-516.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordal G. W., Adler J. Properties of mutants in galactose taxis and transport. J Bacteriol. 1974 Feb;117(2):517–526. doi: 10.1128/jb.117.2.517-526.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Transfer of chromosomal mutations to plasmids via Hfr-mediated conduction. J Bacteriol. 1986 Jan;165(1):312–314. doi: 10.1128/jb.165.1.312-314.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Strange P. G., Koshland D. E., Jr Receptor interactions in a signalling system: competition between ribose receptor and galactose receptor in the chemotaxis response. Proc Natl Acad Sci U S A. 1976 Mar;73(3):762–766. doi: 10.1073/pnas.73.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C., Bogonez E., Wang E. A., Koshland D. E., Jr Sites of methyl esterification on the aspartate receptor involved in bacterial chemotaxis. J Biol Chem. 1983 Aug 25;258(16):9608–9611. [PubMed] [Google Scholar]
- Terwilliger T. C., Koshland D. E., Jr Sites of methyl esterification and deamination on the aspartate receptor involved in chemotaxis. J Biol Chem. 1984 Jun 25;259(12):7719–7725. [PubMed] [Google Scholar]
- Ullrich A., Coussens L., Hayflick J. S., Dull T. J., Gray A., Tam A. W., Lee J., Yarden Y., Libermann T. A., Schlessinger J. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. 1984 May 31-Jun 6Nature. 309(5967):418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wang E. A., Koshland D. E., Jr Receptor structure in the bacterial sensing system. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]