Abstract
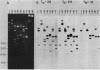
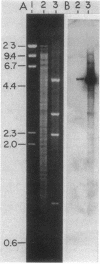
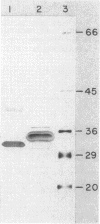
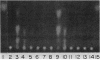
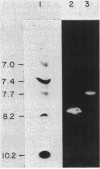
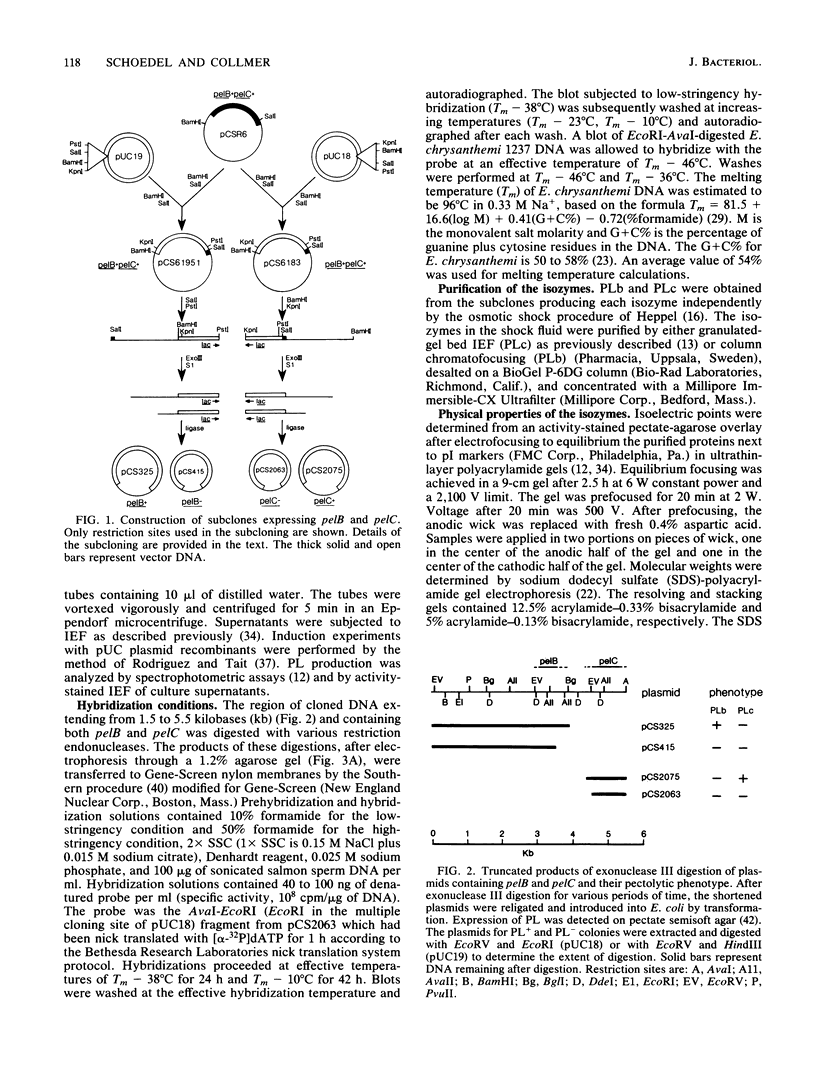
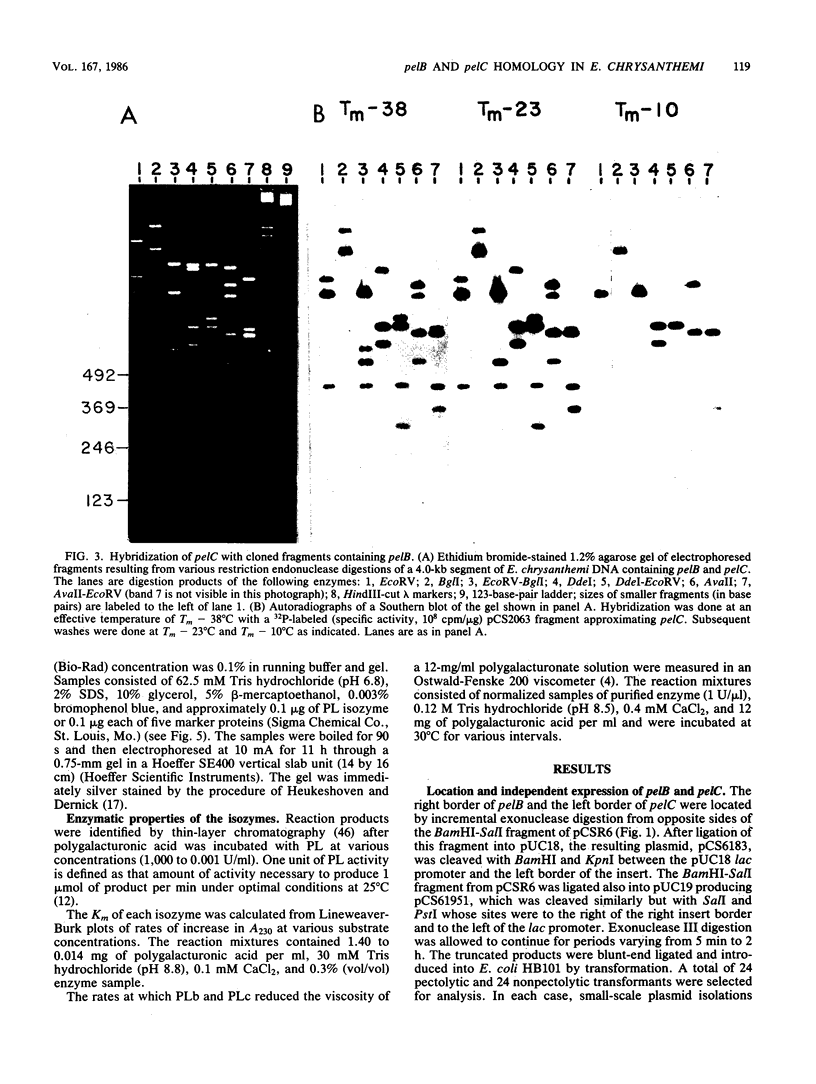
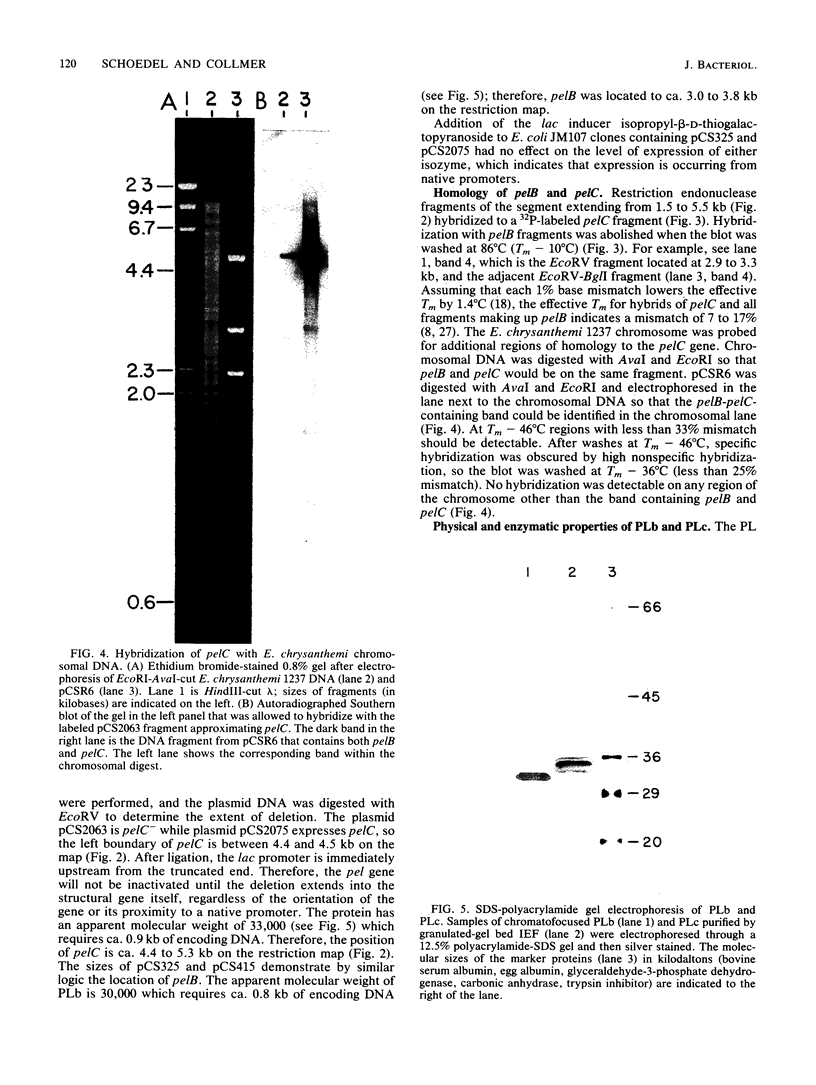
The genes for two of several pectate lyase isozymes produced by the phytopathogenic enterobacterium Erwinia chrysanthemi 1237 were subcloned and compared by DNA-DNA hybridization, and the encoded proteins were analyzed. The borders of the genes were located on a restriction map by incremental exonuclease III deletions. DNA-DNA hybridization studies revealed a low percentage of mismatch (7 to 17%) between pelB and pelC. No homology was detected between pelC and other regions of the E. chrysanthemi 1237 chromosome, in which three other isozyme genes apparently reside. The pectate lyase isozymes were readily purified by chromatofocusing or granulated-gel bed isoelectric focusing from the periplasmic shock fluids of Escherichia coli subclones. The molecular weights of PLb and PLc were 30,000 and 33,000 as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Their isoelectric points were 7.6 and 8.1, respectively, as determined by equilibrium isoelectric focusing in ultrathin polyacrylamide gels. The Km values for PLb and PLc were 0.20 and 0.32 mg/ml, respectively, with polygalacturonate as a substrate. Thin-layer chromatography of reaction products and viscometric assays revealed little difference between the two isozymes. All our data indicate that the genes are duplicates and that the proteins are isofunctional.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman D. F. Hydrolytic and trans-eliminative degradation of pectic substances by extracellular enzymes of Fusarium solani f. phaseoli. Phytopathology. 1966 Feb;56(2):238–244. [PubMed] [Google Scholar]
- Bertheau Y., Madgidi-Hervan E., Kotoujansky A., Nguyen-The C., Andro T., Coleno A. Detection of depolymerase isoenzymes after electrophoresis or electrofocusing, or in titration curves. Anal Biochem. 1984 Jun;139(2):383–389. doi: 10.1016/0003-2697(84)90022-8. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Campbell J. H., Lengyel J. A., Langridge J. Evolution of a second gene for beta-galactosidase in Escherichia coli. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1841–1845. doi: 10.1073/pnas.70.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A. K., Starr M. P. Donor strains of the soft-rot bacterium Erwinia chrysanthemi and conjugational transfer of the pectolytic capacity. J Bacteriol. 1977 Dec;132(3):862–869. doi: 10.1128/jb.132.3.862-869.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer A., Schoedel C., Roeder D. L., Ried J. L., Rissler J. F. Molecular cloning in Escherichia coli of Erwinia chrysanthemi genes encoding multiple forms of pectate lyase. J Bacteriol. 1985 Mar;161(3):913–920. doi: 10.1128/jb.161.3.913-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer A., Whalen C. H., Beer S. V., Bateman D. F. An exo-poly-alpha-D-galacturonosidase implicated in the regulation of extracellular pectate lyase production in Erwinia chrysanthemi. J Bacteriol. 1982 Feb;149(2):626–634. doi: 10.1128/jb.149.2.626-634.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijsegem F., Toussaint A., Schoonejans E. In vivo cloning of the pectate lyase and cellulase genes of Erwinia chrysanthemi. EMBO J. 1985 Mar;4(3):787–792. doi: 10.1002/j.1460-2075.1985.tb03698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G., Reeve E. C. A third beta-galactosidase in a strain of Klebsiella that possesses two lac genes. J Bacteriol. 1977 Oct;132(1):219–223. doi: 10.1128/jb.132.1.219-223.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley P. M., Israel M. A., Law M. F., Martin M. A. A rapid method for detecting and mapping homology between heterologous DNAs. Evaluation of polyomavirus genomes. J Biol Chem. 1979 Jun 10;254(11):4876–4883. [PubMed] [Google Scholar]
- Keen N. T., Dahlbeck D., Staskawicz B., Belser W. Molecular cloning of pectate lyase genes from Erwinia chrysanthemi and their expression in Escherichia coli. J Bacteriol. 1984 Sep;159(3):825–831. doi: 10.1128/jb.159.3.825-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A., Gorini L. Similarity of genes argF and argI. Nature. 1975 Aug 21;256(5519):621–624. doi: 10.1038/256621a0. [DOI] [PubMed] [Google Scholar]
- Kotoujansky A., Diolez A., Boccara M., Bertheau Y., Andro T., Coleno A. Molecular cloning of Erwinia chrysanthemi pectinase and cellulase structural genes. EMBO J. 1985 Mar;4(3):781–785. doi: 10.1002/j.1460-2075.1985.tb03697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Matthysse A. G. Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J Bacteriol. 1983 May;154(2):906–915. doi: 10.1128/jb.154.2.906-915.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Ow D. W., Ausubel F. M. Regulation of nitrogen metabolism genes by nifA gene product in Klebsiella pneumoniae. Nature. 1983 Jan 27;301(5898):307–313. doi: 10.1038/301307a0. [DOI] [PubMed] [Google Scholar]
- Reverchon S., Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Cloning of genes encoding pectolytic enzymes from a genomic library of the phytopathogenic bacterium, Erwinia chrysanthemi. Gene. 1985;35(1-2):121–130. doi: 10.1016/0378-1119(85)90164-7. [DOI] [PubMed] [Google Scholar]
- Ried J. L., Collmer A. Activity stain for rapid characterization of pectic enzymes in isoelectric focusing and sodium dodecyl sulfate-polyacrylamide gels. Appl Environ Microbiol. 1985 Sep;50(3):615–622. doi: 10.1128/aem.50.3.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M., Anilionis A. Evolution of the bacterial genome. Annu Rev Microbiol. 1978;32:519–560. doi: 10.1146/annurev.mi.32.100178.002511. [DOI] [PubMed] [Google Scholar]
- Roberts T. M., Lauer G. D. Maximizing gene expression on a plasmid using recombination in vitro. Methods Enzymol. 1979;68:473–482. doi: 10.1016/0076-6879(79)68036-9. [DOI] [PubMed] [Google Scholar]
- Ros J., Aguilar J. Genetic and structural evidence for the presence of propanediol oxidoreductase isoenzymes in Escherichia coli. J Gen Microbiol. 1984 Mar;130(3):687–692. doi: 10.1099/00221287-130-3-687. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Squires C. H., De Felice M., Devereux J., Calvo J. M. Molecular structure of ilvIH and its evolutionary relationship to ilvG in Escherichia coli K12. Nucleic Acids Res. 1983 Aug 11;11(15):5299–5313. doi: 10.1093/nar/11.15.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr M. P., Chatterjee A. K., Starr P. B., Buchanan G. E. Enzymatic degradation of polygalacturonic acid by Yersinia and Klebsiella species in relation to clinical laboratory procedures. J Clin Microbiol. 1977 Oct;6(4):379–386. doi: 10.1128/jcm.6.4.379-386.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zink R. T., Chatterjee A. K. Cloning and expression in Escherichia coli of pectinase genes of Erwinia carotovora subsp. carotovora. Appl Environ Microbiol. 1985 Mar;49(3):714–717. doi: 10.1128/aem.49.3.714-717.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipkas D., Riley M. Proposal concerning mechanism of evolution of the genome of Escherichia coli. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1354–1358. doi: 10.1073/pnas.72.4.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]