Abstract
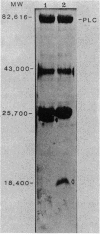
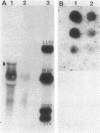
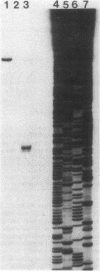
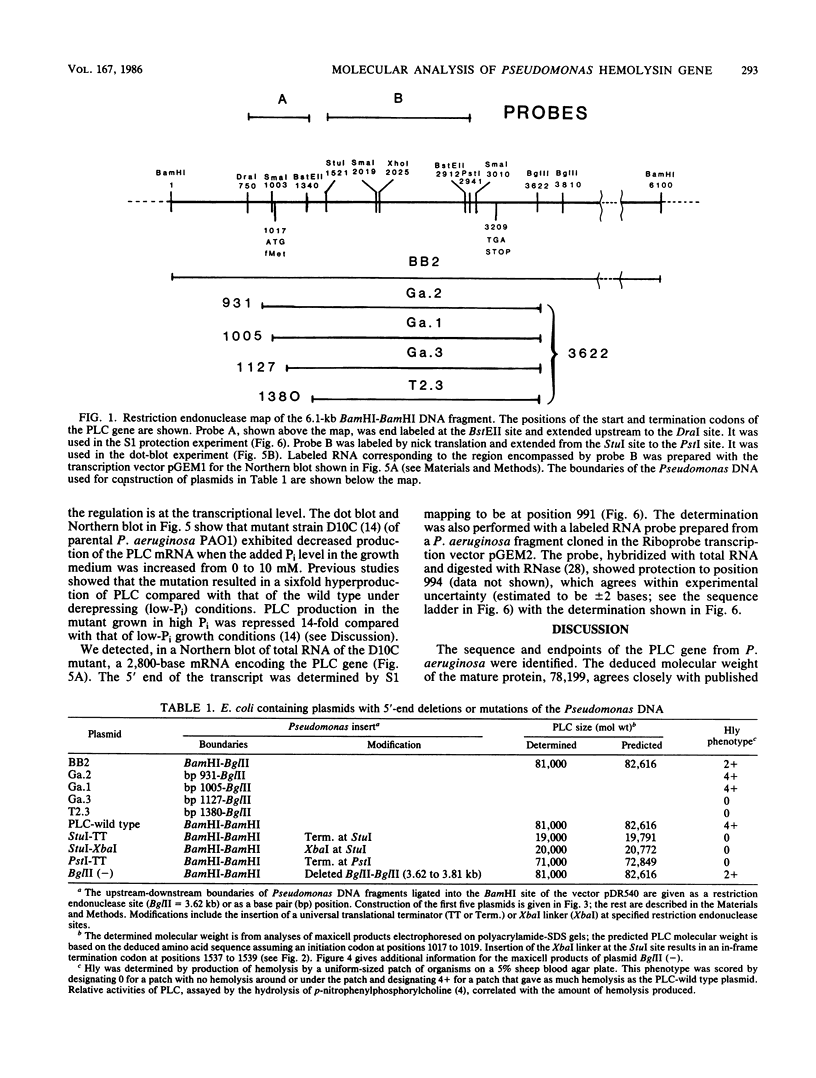
A 3.3-kilobase-pair fragment of Pseudomonas aeruginosa DNA containing the phospholipase C (heat-labile hemolysin) gene was sequenced, and the location of the gene was determined. The gene product contains at its NH2 terminus a 38-amino acid sequence which structurally resembles the signal peptides of other secreted proteins but is unusually long and positively charged (6+). The location of the translation start codon was determined by constructing a series of plasmids in which the promoter of a transcription vector was ligated to Pseudomonas DNA containing deletions at the 5' end of the gene. The plasmids were used to transform Escherichia coli, and the resulting clones were assayed for hemolysin activity. In addition, sizes of truncated proteins produced by mutants with translation terminators introduced at specific sites were analyzed in E. coli maxicells. The gene is transcribed, starting just upstream of the hemolysin gene, as an mRNA of approximately 2,800 bases. Analysis of the nucleotide sequence, analysis of mutants in maxicells, and transcriptional studies indicate that the hemolysin is part of an operon composed of two genes. Phosphate regulation of the operon is at the transcriptional level. The location of the 5' end of the transcript was determined by S1 mapping.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Berka R. M., Gray G. L., Vasil M. L. Studies of phospholipase C (heat-labile hemolysin) in Pseudomonas aeruginosa. Infect Immun. 1981 Dec;34(3):1071–1074. doi: 10.1128/iai.34.3.1071-1074.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka R. M., Vasil M. L. Phospholipase C (heat-labile hemolysin) of Pseudomonas aeruginosa: purification and preliminary characterization. J Bacteriol. 1982 Oct;152(1):239–245. doi: 10.1128/jb.152.1.239-245.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bresser J., Hubbell H. R., Gillespie D. Biological activity of mRNA immobilized on nitrocellulose in NaI. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6523–6527. doi: 10.1073/pnas.80.21.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. J., Ingram J. M., Costerton J. W. Release of alkaline phosphatase from cells of Pseudomonas aeruginosa by manipulation of cation concentration and of pH. J Bacteriol. 1970 Nov;104(2):748–753. doi: 10.1128/jb.104.2.748-753.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Coleman K., Dougan G., Arbuthnott J. P. Cloning, and expression in Escherichia coli K-12, of the chromosomal hemolysin (phospholipase C) determinant of Pseudomonas aeruginosa. J Bacteriol. 1983 Feb;153(2):909–915. doi: 10.1128/jb.153.2.909-915.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. L., Berka R. M., Vasil M. L. A Pseudomonas aeruginosa mutant non-derepressible for orthophosphate-regulated proteins. J Bacteriol. 1981 Aug;147(2):675–678. doi: 10.1128/jb.147.2.675-678.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. L., Berka R. M., Vasil M. L. Phospholipase C regulatory mutation of Pseudomonas aeruginosa that results in constitutive synthesis of several phosphate-repressible proteins. J Bacteriol. 1982 Jun;150(3):1221–1226. doi: 10.1128/jb.150.3.1221-1226.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. L., Smith D. H., Baldridge J. S., Harkins R. N., Vasil M. L., Chen E. Y., Heyneker H. L. Cloning, nucleotide sequence, and expression in Escherichia coli of the exotoxin A structural gene of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1984 May;81(9):2645–2649. doi: 10.1073/pnas.81.9.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. L., Vasil M. L. Mapping of a gene controlling the production of phospholipase C and alkaline phosphatase in Pseudomonas aeruginosa. Mol Gen Genet. 1981;183(2):403–405. doi: 10.1007/BF00270648. [DOI] [PubMed] [Google Scholar]
- Hacker J., Hughes C. Genetics of Escherichia coli hemolysin. Curr Top Microbiol Immunol. 1985;118:139–162. doi: 10.1007/978-3-642-70586-1_8. [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Poole K., Benz R. Outer membrane protein P of Pseudomonas aeruginosa: regulation by phosphate deficiency and formation of small anion-specific channels in lipid bilayer membranes. J Bacteriol. 1982 May;150(2):730–738. doi: 10.1128/jb.150.2.730-738.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Determination of the transcription initiation site and identification of the protein product of the regulatory gene xylR for xyl operons on the TOL plasmid. J Bacteriol. 1985 Sep;163(3):863–869. doi: 10.1128/jb.163.3.863-869.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the promoter region of the xylDEGF operon on TOL plasmid of Pseudomonas putida. Gene. 1984 Sep;29(3):323–330. doi: 10.1016/0378-1119(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Kurioka S., Liu P. V. Effect of the hemolysin of Pseudomonas aeruginosa on phosphatides and on phospholipase c activity. J Bacteriol. 1967 Feb;93(2):670–674. doi: 10.1128/jb.93.2.670-674.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lory S., Tai P. C. Characterization of the phospholipase C gene of Pseudomonas aeruginosa cloned in Escherichia coli. Gene. 1983 Apr;22(1):95–101. doi: 10.1016/0378-1119(83)90068-9. [DOI] [PubMed] [Google Scholar]
- Lory S., Tai P. C., Davis B. D. Mechanism of protein excretion by gram-negative bacteria: Pseudomonas aeruginosa exotoxin A. J Bacteriol. 1983 Nov;156(2):695–702. doi: 10.1128/jb.156.2.695-702.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N., Lehrbach P. R., Reineke W., Timmis K. N. Transcription of the TOL plasmid toluate catabolic pathway operon of Pseudomonas putida is determined by a pair of co-ordinately and positively regulated overlapping promoters. EMBO J. 1984 Nov;3(11):2461–2466. doi: 10.1002/j.1460-2075.1984.tb02156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Minton N. P., Atkinson T., Bruton C. J., Sherwood R. F. The complete nucleotide sequence of the Pseudomonas gene coding for carboxypeptidase G2. Gene. 1984 Nov;31(1-3):31–38. doi: 10.1016/0378-1119(84)90192-6. [DOI] [PubMed] [Google Scholar]
- Oliver D. Protein secretion in Escherichia coli. Annu Rev Microbiol. 1985;39:615–648. doi: 10.1146/annurev.mi.39.100185.003151. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Poncz M., Solowiejczyk D., Ballantine M., Schwartz E., Surrey S. "Nonrandom" DNA sequence analysis in bacteriophage M13 by the dideoxy chain-termination method. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4298–4302. doi: 10.1073/pnas.79.14.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Export of protein in bacteria. Microbiol Rev. 1984 Dec;48(4):290–298. doi: 10.1128/mr.48.4.290-298.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D. R., Bennett G. N. Construction and analysis of in vivo activity of E. coli promoter hybrids and promoter mutants that alter the -35 to -10 spacing. Gene. 1982 Dec;20(2):231–243. doi: 10.1016/0378-1119(82)90042-7. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K. Vectors for gene cloning in Pseudomonas and their applications. Curr Top Microbiol Immunol. 1982;96:31–45. doi: 10.1007/978-3-642-68315-2_3. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy T. J., Benson S. A., Emr S. D. Mechanisms of protein localization. Microbiol Rev. 1983 Sep;47(3):313–344. doi: 10.1128/mr.47.3.313-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson M. W., Hayden C. Secretion of phospholipase C by Pseudomonas aeruginosa. Infect Immun. 1979 Aug;25(2):558–564. doi: 10.1128/iai.25.2.558-564.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil M. L., Berka R. M., Gray G. L., Nakai H. Cloning of a phosphate-regulated hemolysin gene (phospholipase C) from Pseudomonas aeruginosa. J Bacteriol. 1982 Oct;152(1):431–440. doi: 10.1128/jb.152.1.431-440.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil M. L., Berka R. M., Gray G. L., Pavlovskis O. R. Biochemical and genetic studies of iron-regulated (exotoxin A) and phosphate-regulated (hemolysin phospholipase C) virulence factors of Pseudomonas aeruginosa. Antibiot Chemother (1971) 1985;36:23–39. doi: 10.1159/000410469. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wretlind B., Pavlovskis O. R. Genetic mapping and characterization of Pseudomonas aeruginosa mutants defective in the formation of extracellular proteins. J Bacteriol. 1984 Jun;158(3):801–808. doi: 10.1128/jb.158.3.801-808.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]