Abstract
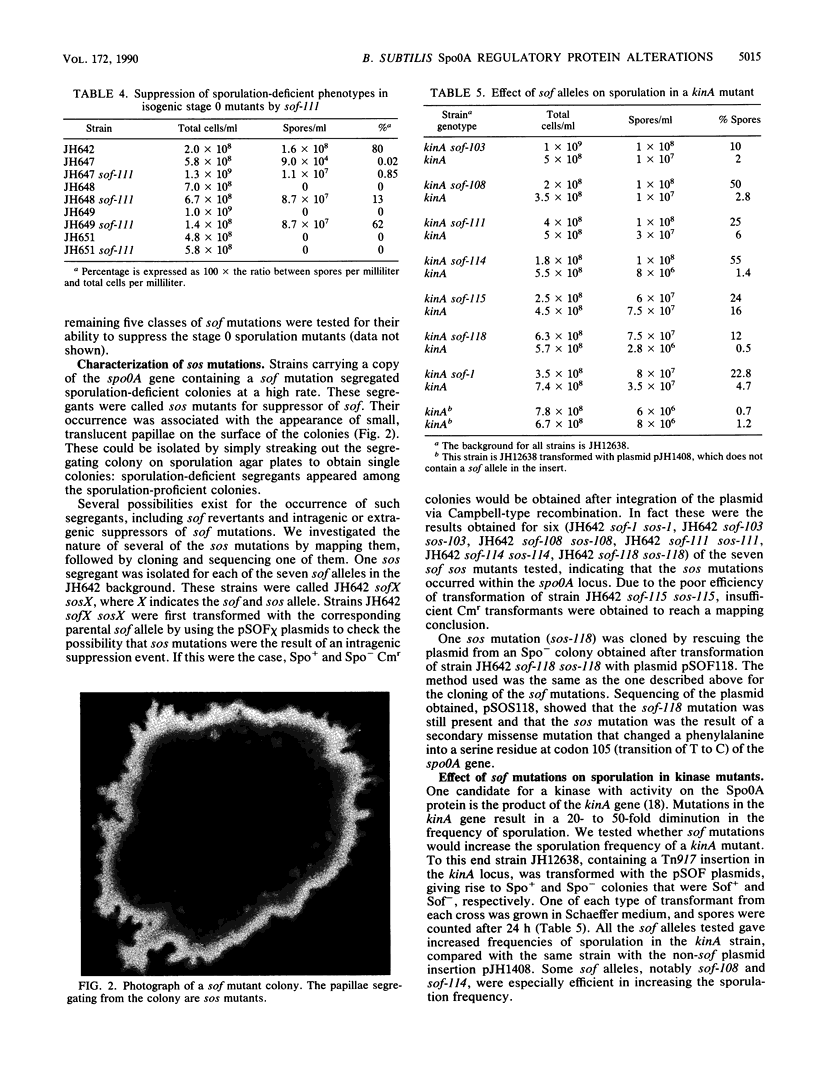
Secondary site mutations that restore sporulation to sporulation-defective spo0F or spo0B deletion mutants were found to reside in the spo0A gene. Sequence analysis of 23 such sof mutants showed that the sof mutations fell into six classes of missense codon changes, primarily in the conserved amino-terminal domain of the response regulator Spo0A protein. Changes were observed in codons 12, 14, 60, 92, and 121. The residues affected were predominantly located in the potential turn regions at one end of the amino-terminal conserved domain on the same topological face as the active site aspartate residues. The ability of sof mutations to suppress deficiencies in the transmitter kinases, KinA and KinB, of two-component regulatory systems was tested. All of the sof mutations suppressed the sporulation deficiency of kinA mutants but only two classes among five tested suppressed kinB mutations. sof mutants segregated Spo- colonies at high frequency. Five of these Spo- mutants were found to result from mutations in the spo0A locus that reversed the effect of the sof mutatation. One of these was sequenced and found to have the original sof mutation and a new mutation, sos, at codon 105. The accumulation of sos mutations in sof strains suggested that the sof mutations have a subtle, yet deleterious, effect on the growth of the cell. The results suggested that the sof mutations increase the avidity for or reactivity with transmitter kinases in an allele-specific manner, although in some cases it is possible that the sof mutations obviate the need for phosphorylation to activate the Spo0A protein. An alternative hypothesis is presented in which the sof mutations play the role of bypass mutations for kinases.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewski C., Savelli B., Stragier P. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol. 1990 Jan;172(1):86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm S. P., Staal S. P., Hoch J. A. Phenotypes of pleiotropic-negative sporulation mutants of Bacillus subtilis. J Bacteriol. 1973 Sep;115(3):1063–1070. doi: 10.1128/jb.115.3.1063-1070.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., LeCoq D., Spence J., Ferrari E., Hoch J. A. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A. 1985 May;82(9):2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Matsumura P., Simon M. I. Protein phosphorylation is involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7609–7613. doi: 10.1073/pnas.84.21.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A. Genetics of bacterial sporulation. Adv Genet. 1976;18:69–98. doi: 10.1016/s0065-2660(08)60437-x. [DOI] [PubMed] [Google Scholar]
- Hoch J. A., Trach K., Kawamura F., Saito H. Identification of the transcriptional suppressor sof-1 as an alteration in the spo0A protein. J Bacteriol. 1985 Feb;161(2):552–555. doi: 10.1128/jb.161.2.552-555.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kawamura F., Saito H. Isolation and mapping of a new suppressor mutation of an early sporulation gene spoOF mutation in Bacillus subtilis. Mol Gen Genet. 1983;192(3):330–334. doi: 10.1007/BF00392171. [DOI] [PubMed] [Google Scholar]
- Lupas A., Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989 Oct 15;264(29):17337–17342. [PubMed] [Google Scholar]
- Monod M., Mohan S., Dubnau D. Cloning and analysis of ermG, a new macrolide-lincosamide-streptogramin B resistance element from Bacillus sphaericus. J Bacteriol. 1987 Jan;169(1):340–350. doi: 10.1128/jb.169.1.340-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon B. T., Ronson C. W., Ausubel F. M. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7850–7854. doi: 10.1073/pnas.83.20.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi K., Kandala J., Freese E. Evidence that Bacillus subtilis sporulation induced by the stringent response is caused by the decrease in GTP or GDP. J Bacteriol. 1982 Aug;151(2):1062–1065. doi: 10.1128/jb.151.2.1062-1065.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Cole S. P., Burbulys D., Trach K., Hoch J. A. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989 Nov;171(11):6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Spiegelman G. B., Hoch J. A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988 Nov;2(6):689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976 Dec;40(4):908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D. A., Gillece-Castro B. L., Stock A. M., Burlingame A. L., Koshland D. E., Jr Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989 Dec 25;264(36):21770–21778. [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji K., Hiratsuka S., Kawamura F., Kobayashi Y. New suppressor mutation sur0B of spo0B and spo0F mutations in Bacillus subtilis. J Gen Microbiol. 1988 Dec;134(12):3249–3257. doi: 10.1099/00221287-134-12-3249. [DOI] [PubMed] [Google Scholar]
- Stock A. M., Mottonen J. M., Stock J. B., Schutt C. E. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989 Feb 23;337(6209):745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch M. A., Perego M., Burbulys D., Hoch J. A. The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol Microbiol. 1989 Sep;3(9):1203–1209. doi: 10.1111/j.1365-2958.1989.tb00270.x. [DOI] [PubMed] [Google Scholar]
- Strauch M. A., Spiegelman G. B., Perego M., Johnson W. C., Burbulys D., Hoch J. A. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989 May;8(5):1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach K. A., Chapman J. W., Piggot P. J., Hoch J. A. Deduced product of the stage 0 sporulation gene spo0F shares homology with the Spo0A, OmpR, and SfrA proteins. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7260–7264. doi: 10.1073/pnas.82.21.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trach K., Hoch J. A. The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J Bacteriol. 1989 Mar;171(3):1362–1371. doi: 10.1128/jb.171.3.1362-1371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Gerbaud G., Lambert T., Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985 Dec 16;4(13A):3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg R. J., Mahler I., Tipper D. J., Halvorson H. O. Cloning the Bacillus subtilis 168 aroC gene encoding dehydroquinase. Gene. 1984 Dec;32(1-2):57–66. doi: 10.1016/0378-1119(84)90032-5. [DOI] [PubMed] [Google Scholar]