Abstract
The multisubunit eukaryotic exosome is an essential RNA processing and degradation machine. In its nuclear form, the exosome associates with the auxiliary factor Rrp6p, which participates in both RNA processing and degradation reactions. The crystal structure of Saccharomyces cerevisiae Rrp6p displays a conserved RNase D core with a flanking HRDC (helicase and RNase D C-terminal) domain in an unusual conformation shown to be important for the processing function of the enzyme. Complexes with AMP and UMP, the products of the RNA degradation process, reveal how the protein specifically recognizes ribonucleotides and their bases. Finally, in vivo mutational studies show the importance of the domain contacts for the processing function of Rrp6p and highlight fundamental differences between the protein and its prokaryotic RNase D counterparts.
Keywords: RNA degradation, RNA processing, x-ray crystallography, RNase D
The RNA exosome participates in a wide range of reactions, including processing and degradation of tRNA and rRNA as well as degradation of both nuclear and cytoplasmic RNA polymerase II-derived transcripts (1–6). The core eukaryotic exosome is present in both the cytoplasm and nucleus and consists of 10 proteins, with at least 7 harboring proven or predicted 3′-to-5′ exonuclease activity (one RNase II and six RNase PH type) (7–10). In the yeast nucleus, the complex is distinguished by three additional proteins, the RNase D-type enzyme Rrp6p (for ribosomal RNA processing), the DEAD-box RNA helicase Mtr4p, and the less well characterized protein Rrp47p (7, 11). Exosomes are found in both eukaryotes and archaea, and, recently, several crystal structures of archaeal subcomplexes were reported (12–14). The center of the archaeal exosome consists of three Rrp41 and three Rrp42 proteins forming an overall donut-shaped heterohexameric structure. Rrp41 and -42 are each similar to three proteins in the eukaryotic complex, which consists of six different proteins forming the “donut” (12, 14). This ring-like structure is able to bind additional proteins, forming a “cap” suggested to constrict and probably regulate the entry of RNA into the central cavity containing the phosphorolytic active sites (12).
The nuclear exosome is essential for maturation of eukaryotic ribosomal RNAs (25S, 18S, and 5.8S), which are synthesized as a single transcript (for a review, see ref. 15). Processing is initiated by endonucleolytic cleavage of the external transcribed spacers (ETSs), which are subsequently degraded by Rrp6p (16). This protein is also required for trimming of the two internal transcribed spacers 1 and 2 during maturation to produce the mature rRNAs, and a 30-nt 3′-end extended form of 5.8S rRNA appears in Δrrp6 cells (17). Similarly, many small nucleolar RNAs (snoRNAs) depend on the exosome during their maturation (18, 19), and deletion of Rrp6p in yeast also leads to accumulation of extended forms of both polycistronic snoRNAs (18) and the independently transcribed snoRNAs, such as snR33 and snR40 (20).
Rrp6p is homologous to human PM-Scl 100, a protein that is implicated in several degenerative autoimmune disorders (17, 21) and forms a part of the human nuclear exosome (7). The central part of Rrp6p is homologous to bacterial RNA-processing enzymes, such as RNase D, but the eukaryotic protein is much larger (736 vs. 375 aa) (17). Both proteins belong to the group of DEDD nucleases that includes enzymes active on both DNA and RNA and have structural homology to the exonuclease domain found in the Klenow fragment of DNA polymerase I (22). These enzymes are characterized by at least four conserved acidic residues (DEDD) required for nucleic acid degradation in the 3′-to-5′ direction via a hydrolytic reaction mechanism, where two divalent metal ions are involved in activation of a water molecule that attacks the last phosphodiester bond (23). RNase D enzymes belong to the DEDDy subgroup that is present in both bacteria and eukaryotes and contain a conserved tyrosine close to the active site. In addition, RNase D enzymes contain one or more conserved HRDC (helicase and RNase D C-terminal) domains, first identified as a putative nucleic acid-binding motif by sequence comparison within the RecQ helicase protein family (24, 25). The crystal structure of Escherichia coli RNase D revealed a compact and conserved exonuclease domain flanked by two distal HRDC domains forming a funnel-shaped ring, proposed to mediate RNA binding or specificity during stable RNA processing in bacteria (26).
In this article, we present four crystal structures of Saccharomyces cerevisiae Rrp6p, isolated or in complexes with metal ions, AMP, or UMP. The structures show how Rrp6p interacts with metal ions, specifically recognizes ribonucleotides, and highlights the importance of the eukaryotic-specific N-terminal extension in anchoring the HRDC domain. Mutational analysis suggests that this anchoring is essential for the RNA-processing function of Rrp6p.
Results
Overall Structure of S. cerevisiae Rrp6p.
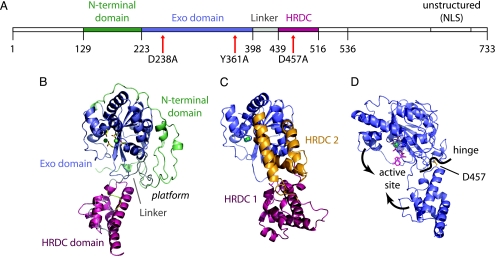
We have so far been unsuccessful in expressing full-length WT, as well as catalytically inactive, Rrp6p in E. coli. Shorter versions of WT Rrp6p also express poorly; however, by introducing a Y361A mutation into the active site, we were able to produce and crystallize a shorter form of the protein comprising residues 129–536 (molecular mass 47.7 kDa) containing part of the unique N-terminal domain, the conserved catalytic exonuclease domain (harboring the Y361A active-site mutation), and the HRDC domain (Fig. 1A and B). Single crystals diffracted to ≈2 Å and were used to determine the structure by multiple-wavelength anomalous dispersion phasing (Table 1). The refined model has a single molecule in the asymmetric unit covering residues 129–516 and 201 solvent molecules. All parts of the protein are clearly defined, with the exception of a short stretch of the linker between the exonuclease and HRDC domains (residues 425–433), which is traceable, but with relatively poor electron density. The final model has a crystallographic R-factor of 23.3% (Rfree = 29.2%).
Fig. 1.
Structure overview. (A) Rrp6p sequence overview. Colored-coded segments are part of the current structure showing the N-terminal domain (green), exonuclease domain (blue), linker (gray), and HRDC domain (red). (B) Overview of the Rrp6p structure using the same color code as A. The ions and side chains in the active site are shown as balls and sticks. (C) E. coli RNase D with the exonuclease (blue), HRDC1 (red), and HRDC2 (orange) domains shown in the same orientation as Rrp6p in B (26). (D) The contraction of Rrp6p around the active site upon binding of substrate is achieved by a rotation around a hinge point near the residue D457.
Table 1.
Data collection and structure refinement
| Space group (axes a, c), Å | P3121 (110.34, 80.26) |
| Resolution (outer shell), Å | 95–2.1 (2.18–2.10) |
| Completeness (outer shell), % | 98.5 (91.3) |
| Data redundancy (outer shell) | 4.9 (3.6) |
| Mean I/σI (outer shell) | 25.6 (2.6) |
| Rsym (outer shell), % | 5.8 (50.3) |
| Refinement R-factor (Rfree), % | 23.3 (29.2) |
| Rms on bonds, Å; angles, ° | 0.008; 1.34 |
| Luzzati error (cross-validated), Å | 0.32 (0.40) |
| Ramachandran plot statistics, % | |
| Most favorable regions | 84.3 |
| Additionally allowed regions | 14.3 |
| Generously allowed regions | 1.4 |
| Disallowed regions | 0.0 |
The overall structure has one large domain consisting of the N terminus and core exonuclease fold (residues 129–398, green and blue) and, separately, the loosely attached HRDC domain (residues 439–516, red) (Fig. 1 A and B). The core of the exonuclease domain shows the classical α/β fold seen in the Klenow fragment of E. coli DNA polymerase I (22), consisting of a six-stranded, mixed β-sheet flanked by α-helices. S. cerevisiae Rrp6p has ≈225 residues before the exonuclease domain, which we term the N-terminal extension, partly resolved in the present structure (residues 129–222, green in Fig. 1 A and B). The extension wraps around the core domain and is wedged at the exonuclease–HRDC domain interface. A convoluted linker (residues 398–438) connects the exonuclease domain to the all-helical HRDC domain and takes part in forming a platform with the N-terminal extension (Fig. 1B, marked “platform”). We hypothesize that this platform could serve as a place for protein–protein interactions in accordance with yeast two-hybrid data showing interaction between residues 1–456 and the core exosome components Rrp41p, Rrp46p, and Mtr3p (27). The HRDC domain consists of five regular helices and is topologically identical to the first HRDC domain in E. coli RNase D (rmsd, 1.4 Å); however, the orientation of the domain relative to the exonuclease domain is quite different (26) (Fig. 1 B and C). Approximately 200 residues are missing in the C terminus of the present Rrp6p structure; however, lack of sequence conservation in this part of the protein combined with functional data showing that a C-terminally truncated protein is fully capable of 5.8S rRNA processing (17) suggests it is of minor importance for the biological function of the protein.
Rrp6p Requires both Mn2+ and Zn2+ to Bind Nucleotides.
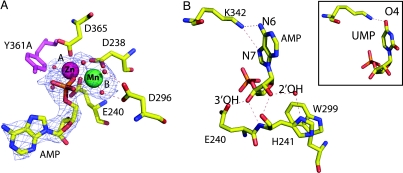
The activity of DEDD nucleases depends on two divalent metal ions in the active site (23). The ions are absent from the native Rrp6p structure, so, to visualize these, data were collected to 2.3 Å from single crystals soaked in 10 mM MnCl2 (Table 2, which is published as supporting information on the PNAS web site). The anomalous difference maps clearly showed the presence of two octahedrally coordinated Mn2+ ions at the active site. Binding of the ions is associated with a slight overall contraction of the molecule around the active site and moderate movements of several side chains within the active site. There is a relatively large distance (6.1 Å) between the ions compared with other DEDD nuclease structures (3.7–5.1 Å) (28, 29), and D238 of the DEDD motif interacts with only one of the ions. This arrangement corresponds to one of two conformations observed in E. coli RNase D (26) and illustrates that the active site of Rrp6p remains intact, despite the nearby Y361A mutation. This finding is consistent with biochemical experiments carried out on the exonuclease domain from DNA polymerase I, where mutation of the tyrosine leads to a change in the relative affinity toward single- and double-stranded DNA, without rendering the enzyme completely inactive (30).
To visualize the enzyme–product complex, crystals were soaked in MnCl2 and AMP, but, surprisingly, no nucleotide was bound under these conditions (data not shown). However, studies of the exonuclease domain from DNA polymerase I had indicated that this enzyme binds several different ions (28); so, in a subsequent soaking trial, both Mn2+ and Zn2+ were included in addition to AMP. Analysis of the 2.3-Å diffraction data revealed a single adenosine nucleotide at the active site, in a position corresponding to the posthydrolysis state resulting from cleavage of the 3′ end of RNA (Fig. 2A). Interestingly, the Rrp6p–AMP complex does not form in the presence of Zn2+ alone or both Zn2+ and the isostructural Mg2+ (data not shown), suggesting that manganese and zinc are specifically required in the active site of Rrp6p. Globally, binding of a nucleotide in the active site is associated with a closure of the active-site cleft by rotation around a hinge point defined by the exonuclease domain–HRDC domain contact (Fig. 1D). Binding of a nucleotide places a negatively charged phosphoryl oxygen between the two metal ions and drastically reduces the distance between the two metal ions (4.3 Å), and D238 is now bridged between the ions (Fig. 2A). Data collected from crystals soaked in both Mn2+ and Zn2+, but not AMP, demonstrate that this contraction is not simply due to the binding of different ions (data not shown). The identity of the ions was confirmed by analysis of the anomalous scattering and is similar to the exonuclease domain from DNA polymerase I (28). We conclude that concomitant binding of both Mn2+ and Zn2+ is required for formation of an enzyme–product complex in Rrp6p.
Fig. 2.
Binding of ions and nucleotides at the active site. (A) The active-site configuration and interaction with the phosphate group in AMP, showing charged and hydrogen-bond contacts (dashed lines). The mFo − DFc difference electron-density map is shown at 2.0σ, and the modeled position of the mutated active-site residue Y361A is shown in purple. (B) The interactions between the base and sugar of AMP and conserved residues in Rrp6p. (Inset) The interaction between K342 and UMP.
Recognition of the 3′ Ribonucleotide.
Comparison of the complex to the structure of another DEDD nuclease, human PARN bound to RNA (31), shows that the interactions of the 3′ nucleotide are likely to be conserved between the prehydrolytic and posthydrolytic states, and we can therefore use the present structure to propose mechanisms for the substrate selection by the protein. Rrp6p–AMP interactions are numerous and include specific contacts that would stabilize the correct substrate in the active site before hydrolysis (Fig. 2B). The phosphate group is sandwiched between the two metal ions, with interaction distances close to the equidistant organization in the presumed pentacoordinated transition state (28), and the sugar moiety is monitored at both the 3′ OH and 2′ OH positions to ensure specific recognition of RNA. The 3′ OH is bound tightly by two strong hydrogen bonds to E240 and the backbone of H241, and recognition of the 2′ OH is achieved by hydrogen bonds to the backbone and side chain (through a water molecule) of H241. H241 stacks against W299 and both residues are highly conserved in eukaryotic Rrp6p proteins but not bacterial RNase D or other eukaryotic RNase D enzymes, such as Pop2p or PARN (31, 32), suggesting that subtle variations in side chains near the active site play an important role in substrate specificity and recognition. Recognition of the nucleotide base is conferred by K342, which forms two specific hydrogen bonds to AMP (Fig. 2B). The lysine is highly conserved among both bacterial and eukaryotic RNase D enzymes, indicating that this could be a general recognition mechanism, although not shared in the wider DEDD family. Guanine could be recognized in a similar way, but the possibility that Rrp6p might have a preference for purines prompted us to study crystals soaked in MnCl2, ZnCl2, and UMP (Table 2). This structure shows that Rrp6p does, indeed, bind pyrimidines, but with a single hydrogen bond (Fig. 2B Inset). We conclude that there is no strong structural determinant for purine selectivity in Rrp6p apart from the subtle difference in affinity caused by the additional hydrogen bond in the AMP complex.
RNA Processing by Rrp6p Is Affected by N-Terminal Anchoring of the HRDC Domain.
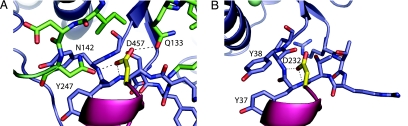
In both bacterial RNase D and Rrp6p, the position of the first HRDC domain is controlled by two points of fixation, a covalent attachment via the linker and a strong interaction between a conserved aspartic acid (D457 in Rrp6p and D232 in RNase D) in the HRDC domain and residues in the exonuclease core. The variation in orientation of the HRDC domain between the two proteins is a result of the N-terminal extension in Rrp6p, which prevents the conformation seen in RNase D (Fig. 1 B and C); however, the interaction between the conserved aspartate and the exonuclease core remains intact despite this difference. This finding suggests that there is a great deal of flexibility in these two domain enzymes and that the HRDC domain may be able to move through a range of different orientations while keeping the interdomain interactions intact.
Mutation of conserved residues in Rrp6p has indicated that the HRDC domain is critical for proper 3′-end processing of stable RNAs. Specifically, it was concluded that the D457A mutation rendered Rrp6p incapable of proper 3′-end trimming of stable RNAs (5.8S rRNA and snR40 snoRNA) while retaining the exonucleolytic activity, as judged by the removal of 5′ ETS RNA (25). Our structure shows that this mutation would disrupt the strong contacts between D457 and the residues Q133 and N142 in the N-terminal extension of the exonuclease domain, which are highly conserved in all fungal Rrp6p orthologs as well as in the human PM-Scl 100 protein (Fig. 3A; and see Fig. 5, which is published as supporting information on the PNAS web site). In bacterial RNase D enzymes that lack the N-terminal extension, interactions of the corresponding D232 in HRDC1 are to the backbone of Y37 and Y38 (Fig. 3B).
Fig. 3.
Anchoring of the HRDC domain. (A) The exonuclease–HRDC interaction in Rrp6p. The exonuclease domain is shown as a blue cartoon with the residue 243–249 region as blue sticks, the N-terminal domain as green sticks, D457 as yellow sticks, and the HRDC-domain as a red cartoon. Dashes indicate observable hydrogen bonds. (B) The exonuclease–HRDC interaction in E. coli RNase D. The exonuclease domain is shown as a blue cartoon with the residue 33–39 region as blue sticks, D232 as yellow sticks, and HRDC1 as a red cartoon.
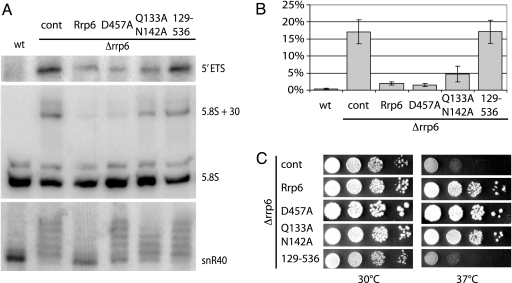
To investigate the importance of the D457–Q133/N142 interaction on Rrp6p RNA-processing efficiency, we constructed full-length D457A and Q133A+N142A mutants and analyzed their function in vivo (Fig. 4). Removal of Rrp6p results in a temperature-sensitive phenotype, which can be fully rescued by supplying a plasmid-borne allele of WT Rrp6p. Under these conditions, growth of both the D457A and Q133A+N142A mutants is indistinguishable from WT cells, whereas a truncated form of Rrp6p, corresponding to the WT version of the crystallized protein (residues 129–536), grows like an RRP6 deletion strain, presumably because it is not localized properly in the cell or lacks an interaction site (Fig. 4C). We then studied the state of 5′ ETS rRNA, snR40 snoRNA, and 5.8S rRNA in the Rrp6p mutant yeast strains by Northern blotting (Fig. 4 A and B). As expected, both the D457A and Q133A+N142A mutants are able to degrade 5′ ETS rRNA with an efficiency comparable with that of WT Rrp6p, indicating that the exonuclease activity is intact. However, probes against snR40 snoRNA revealed a clear RNA 3′-end trimming defect in the mutant strains, as extended forms of snR40 appeared. The Q133A+N142A mutant shows a reduced efficiency of 5.8S rRNA processing; however, a reduction is not seen for D457A, in somewhat of a discrepancy with previous claims (25). Taken together, these results strongly indicate that correct anchoring of the HRDC domain is required to maintain efficient processing of some substrates by Rrp6p but that the defects in RNA processing are not rate-limiting for growth.
Fig. 4.
Functional analysis of Rrp6p mutants. (A) Northern blotting analysis of RNA-processing and -degradation phenotypes using probes directed against 5′ ETS, 5.8S rRNA, or snR40 snoRNA substrates. Lane 1, WT S. cerevisiae, lanes 2–6, S. cerevisiae Δrrp6 strain complemented with empty vector control (cont), plasmid-borne full-length WT protein (Rrp6), single- (D457A) or double- (Q133A N142A) mutant constructs, or truncated protein (residues 129–536). (B) Quantitation of the level of 5.8S pre-rRNA processing. The bars show the percentage of unprocessed 5.8S + 30 pre-rRNA of total 5.8S rRNA. The results represent the average of three independent experiments with standard deviations. (C) Growth phenotypes of the S. cerevisiae Δrrp6 strains in A. Tenfold serial dilutions of cells were spotted onto agar plates grown at the indicated temperatures.
Discussion
The crystal structure of S. cerevisiae Rrp6p represents the only characterized structure of a eukaryotic exosome component. Its structural homology to the prokaryotic RNase D enzyme suggests that the exosome might have evolved from single nucleases that expanded to allow for a higher level of regulation in eukaryotes. One example of this idea is the N-terminal extension and linker of Rrp6p, which create a platform that could be used for anchoring of Rrp6p onto the core exosome. Our mutational data show that a construct corresponding to the crystallized protein is neither able to support growth nor function in nuclear RNA degradation, indicating that the missing parts are important for proper protein function (Fig. 4). Because Rrp6p lacking the C-terminal nuclear localization signals (NLS) is fully active in nuclear 5.8S rRNA processing (17), this finding indicates the N terminus as relevant for the function, and, consistently, sequence alignment shows high conservation in several regions (data not shown).
Although the overall fold of the exonuclease core is conserved between kingdoms, there are significant structural differences in the close vicinity of the active site that can account for subtle variations in substrate specificity between the enzymes. The present Rrp6p structure suggests that H241 is uniquely required for RNA specificity through interaction with the ribose 2′ OH group and that this contact could be modulated through the tethering of the region harboring H241 to the HRDC domain. Thereby, even small rearrangements of the two domains relative to each other, for example, through protein–protein interactions in the platform area, could directly affect substrate selection and the catalytic function of the enzyme. Recognition of the base is conferred by the side chain of K342, and, although the protein can bind both purines and pyrimidines, the additional hydrogen bond to purines could improve affinity toward GA-rich sequences.
The HRDC domain has been implicated in nucleic acid binding, but it is not immediately clear from the structure how this binding would take place. However, the structural conservation of the domain between kingdoms shows the importance of its fold. Apart from directly affecting the active site, as mentioned above, binding of nucleic acids to the HRDC domain could serve to present a complex substrate in the correct orientation at the active site. In an analogy with earlier results, our in vivo mutational analysis of Rrp6p shows that interruption of the exonuclease–HRDC contact (as predicted in the D457A and Q133A/N142A mutants) does not affect housekeeping functions, such as the removal of 5′ ETS RNA, indicating that correct positioning of the HRDC domain is not required for removal of unstructured RNAs. However, for RNAs that must be precisely trimmed, e.g., snR40, we observe substrate-specific defects in the two mutant strains. The processing of 5.8S rRNA is not affected by the D457A mutation, whereas the Q133A+N142A mutant shows a partial phenotype, indicating that the regulation conferred by the HRDC domain is substrate-dependent. Several snoRNAs, including snR40, accumulate as oligoadenylated forms upon deletion of Rrp6p as a result of the activity of the nuclear polyadenylation complex, TRAMP, an activator of nuclear exosome degradation (4–6, 20, 33, 34). It is possible that there exists a kinetic competition between Rrp6p and TRAMP for the free 3′ end of snoRNAs and that this equilibrium is shifted toward polyadenylation upon crippling of Rrp6p. We imagine that disruption of the exonuclease–HRDC domain contact either reduces the affinity of Rrp6p for its substrate or prevents proper orientation of certain substrates in the active site. Therefore, trimming of substrates (such as snR40) that are targeted by the TRAMP-mediated polyadenylation apparatus could be more sensitive to mutation in Rrp6p than, e.g., 5.8S rRNA.
What is the basis for the requirement for an additional single nuclease in the nuclear exosome? The reason is presumably related to differences in specificity or accessibility. Rrp6p is the only RNase D-type exonuclease associated with the exosome and, together with the core subunit Rrp44p, the only hydrolytic nuclease. The presence of different types of nucleases (hydrolytic and phophorolytic) in the exosome may reflect that some RNA substrates are more efficiently degraded by using one of the motifs and that both are required to process all substrates. Another reason for retaining single nucleases that are not firmly associated with the exosome core can be sterical hindrance at the active site of the core. The structure of the archaeal exosome showed that only three of the six core exonucleases contain functional active sites (12, 14). The path to the active sites goes through a relatively narrow constriction at one end of the core, a distance of some 50–60 Å away, so the exosome will never be able to trim stable RNAs closer than ≈10 nucleotides from the last structured element in the 3′ end (12). This finding explains the need for isolated nucleases but also implies that the 3′ end of the RNA must leave the core exosome before final trimming can occur. A deeper mechanistic understanding of this process awaits further functional and structural characterization, but it is clear that modulation of the conformation of the HRDC domain in Rrp6p through protein–protein interactions at the platform could play a part in regulating such a response.
Materials and Methods
Protein Expression and Purification.
The S. cerevisiae RRP6 gene encoding residues 129–536 was amplified by PCR from a plasmid containing the active-site mutant Y361A and inserted into the pET30 Ek/LIC vector (Novagen) for expression in E. coli BL21 (DE3) Rosetta. Cells were grown at 37°C in 2× TY media until A600 = 1.0, followed by 3 h of expression at 30°C after induction with 1 mM isopropyl-β-d-thiogalactopyranoside. Lysate was prepared by sonication, cleared by centrifugation, and purified on a 5-ml Ni2+-chelating agarose column (Amersham Pharmacia Biosciences). Recombinant tobacco etch virus (Tev) protease was used to cleave off the fusion tag, which was removed by a second Ni2+ agarose run. The protein was further purified on a Mono Q 4.6/100 PE anion-exchange column (Amersham Pharmacia Biosciences), followed by precipitation overnight in 66% (wt/vol) (NH4)2SO4 and final separation on a Superdex 200 10/300 GL (Amersham Pharmacia Biosciences) equilibrated in 20 mM Tris·Cl, pH 8.0, 100 mM KCl, and 5 mM β-mercaptoethanol (BME). Peak fractions were concentrated by using a Centricon YM-30 filtration device (Amicon) to 10 mg/ml before crystallization.
Crystallization and Structure Determination.
Single crystals appeared overnight in 12–14% PEG 20.000 and 100 mM Mes or Hepes at pH 6–7.5 and grew to full size in 2–4 weeks. Heavy-atom derivatives (10 mM HgCl2, 25 mM trimethyl lead acetate, 10 mM K2PtCl4, 500 mM KI, or 1 mM YbCl3 in 20% PEG 20.000, 17% ethylene glycol, and 100 mM Mes or Hepes, pH 6–7.5) were prepared by soaking the crystals overnight, followed by snap-freezing in liquid nitrogen. Diffraction data sets for both native and derivative crystals were collected at the BW7A and X13 beam lines at EMBL DESY (Hamburg, Germany). For Pt, a three-wavelength multiple-wavelength anomalous dispersion data set was collected at the LIII-absorption edge (11.56 keV). For I, two wavelengths corresponding to high and low anomalous signal (1.5218 and 1.072 Å, respectively) were collected, and, for the remaining derivatives, single-wavelength data sets were collected at wavelengths that maximized anomalous contribution. All data were processed with DENZO/SCALEPACK (35). Four Pt positions were located by SHELXC/D/E (36) and used for phasing in SHARP (37) and expanded by use of residual maps to include contribution from 7 Pt, 11 I, 2 Hg, 2 Pb, and 2 Yb atoms (see Table 3, which is published as supporting information on the PNAS web site). Phasing was followed by solvent flattening and automated model building in ARP/wARP (38). The 80%-complete model was manually corrected and expanded in the program O (39) and refined by using the standard procedure for ML-restrained refinement in the program CNS by iterative rebuilding in σA-weighted 2Fo − Fc difference maps (40). The quality of the final model was checked by using the program PROCHECK (41) (Table 1). For the complexes with ions and nucleotides, native crystals of Rrp6p were soaked for 10 min in 5 mM MnCl2, 0.5 mM ZnCl2, and 5 mM AMP or UMP, as appropriate, in stable buffer, frozen as described above, and used for data collection at 2.1–3.0 Å at a synchrotron or home source (Table 2). The positions of the two divalent metal ions in the active site were identified by using the anomalous signal, and all complex structures were refined in the program CNS like the native.
Growth Assays and RNA Analysis.
For yeast growth assays, an S. cerevisiae rrp6Δ strain was transformed with RRP6 WT or mutant plasmids and grown at 25°C overnight, diluted to A600 = 0.3, spotted in 10-fold dilutions onto YPD plates, and incubated at 30°C or 37°C for 2 days (for plasmid constructs, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). For Northern blot analysis, total RNA was extracted from exponentially growing yeast cells by using hot acid phenol extraction. Three micrograms of RNA was run on a 10% denaturing polyacrylamide gel, electroblotted onto a Hybond-N+ filter (Amersham Pharmacia Biosciences), and fixed by UV cross-linking. Prehybridization and hybridization steps were done in ULTRAhyb-Oligo Hybridization Buffer (Ambion). Northern blot probes OBS275 (snR40) (25), o5.8S (5.8S rRNA) (17), and 2′ (5′ETS) (42) (see Table 4, which is published as supporting information on the PNAS web site) were 5′-end-labeled with 32P, and probe signals were detected by PhosphorImaging.
Supplementary Material
Acknowledgments
We thank Cyril Saguez, Louise V. Laursen, Gregers Rom Andersen, and Morten Kjeldgaard for reagents, help, and discussion of the manuscript and MAXLAB, DESY, and BESSY for synchrotron beam time. This work was supported by the Danish National Research Foundation (Grundforshningsfonden), DANSYNC, the Human Frontier Science Program (HFSP), the Novo Nordisk Foundation, and the Lundbeck Foundation.
Abbreviations
- ETS
external transcribed spacer
- snoRNA
small nucleolar RNA.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: Atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2HBJ (native), 2HBK (Mn-bound), 2HBL (AMP-bound), and 2HBM (UMP-bound)].
References
- 1.Anderson J. S., Parker R. P. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bousquet-Antonelli C., Presutti C., Tollervey D. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 3.Hilleren P., McCarthy T., Rosbash M., Parker R., Jensen T. H. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 4.Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G., Anderson J. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuai L., Fang F., Butler J. S., Sherman F. Proc. Natl. Acad. Sci. USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyers F., Rougemaille M., Badis G., Rousselle J. C., Dufour M. E., Boulay J., Regnault B., Devaux F., Namane A., Seraphin B., et al. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 7.Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell P., Tollervey D. Nat. Struct. Biol. 2000;7:843–846. doi: 10.1038/82817. [DOI] [PubMed] [Google Scholar]
- 10.van Hoof A., Parker R. Cell. 1999;99:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell P., Petfalski E., Houalla R., Podtelejnikov A., Mann M., Tollervey D. Mol. Cell. Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buttner K., Wenig K., Hopfner K. P. Mol. Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Lorentzen E., Conti E. Mol. Cell. 2005;20:473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Lorentzen E., Walter P., Fribourg S., Evguenieva-Hackenberg E., Klug G., Conti E. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 15.Morrissey J. P., Tollervey D. Trends Biochem. Sci. 1995;20:78–82. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- 16.Butler J. S. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- 17.Briggs M. W., Burkard K. T., Butler J. S. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 18.Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fatica A., Morlando M., Bozzoni I. EMBO J. 2000;19:6218–6229. doi: 10.1093/emboj/19.22.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Hoof A., Lennertz P., Parker R. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reichlin M., Maddison P. J., Targoff I., Bunch T., Arnett F., Sharp G., Treadwell E., Tan E. M. J. Clin. Immunol. 1984;4:40–44. doi: 10.1007/BF00915286. [DOI] [PubMed] [Google Scholar]
- 22.Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 23.Steitz T. A., Steitz J. A. Proc. Natl. Acad. Sci. USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morozov V., Mushegian A. R., Koonin E. V., Bork P. Trends Biochem. Sci. 1997;22:417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 25.Phillips S., Butler J. S. RNA. 2003;9:1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuo Y., Wang Y., Malhotra A. Structure (London) 2005;13:973–984. doi: 10.1016/j.str.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Lehner B., Sanderson C. M. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brautigam C. A., Steitz T. A. J. Mol. Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- 29.Hamdan S., Carr P. D., Brown S. E., Ollis D. L., Dixon N. E. Structure (London) 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 30.Derbyshire V., Grindley N. D., Joyce C. M. EMBO J. 1991;10:17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu M., Reuter M., Lilie H., Liu Y., Wahle E., Song H. EMBO J. 2005;24:4082–4093. doi: 10.1038/sj.emboj.7600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thore S., Mauxion F., Seraphin B., Suck D. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 34.Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski Z., Minor W. In: Methods in Enzymology. Carter C. W., Sweet R. M., editors. Vol. 276. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 36.Uson I., Sheldrick G. M. Curr. Opin. Struct. Biol. 1999;9:643–648. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 37.de La Fortelle E., Bricogne G. In: Methods in Enzymology. Carter C. W., Sweet R. M., editors. Vol. 276. New York: Academic; 1997. pp. 472–494. [DOI] [PubMed] [Google Scholar]
- 38.Perrakis A., Morris R., Lamzin V. S. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 39.Jones T. A., Cowan S., Zou J.-Y., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Brunger A. T., Adams P. D., Clore G. M., Delano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J.-S., Kuszewski J., Nilges N., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 42.de la Cruz J., Kressler D., Tollervey D., Linder P. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.