Abstract
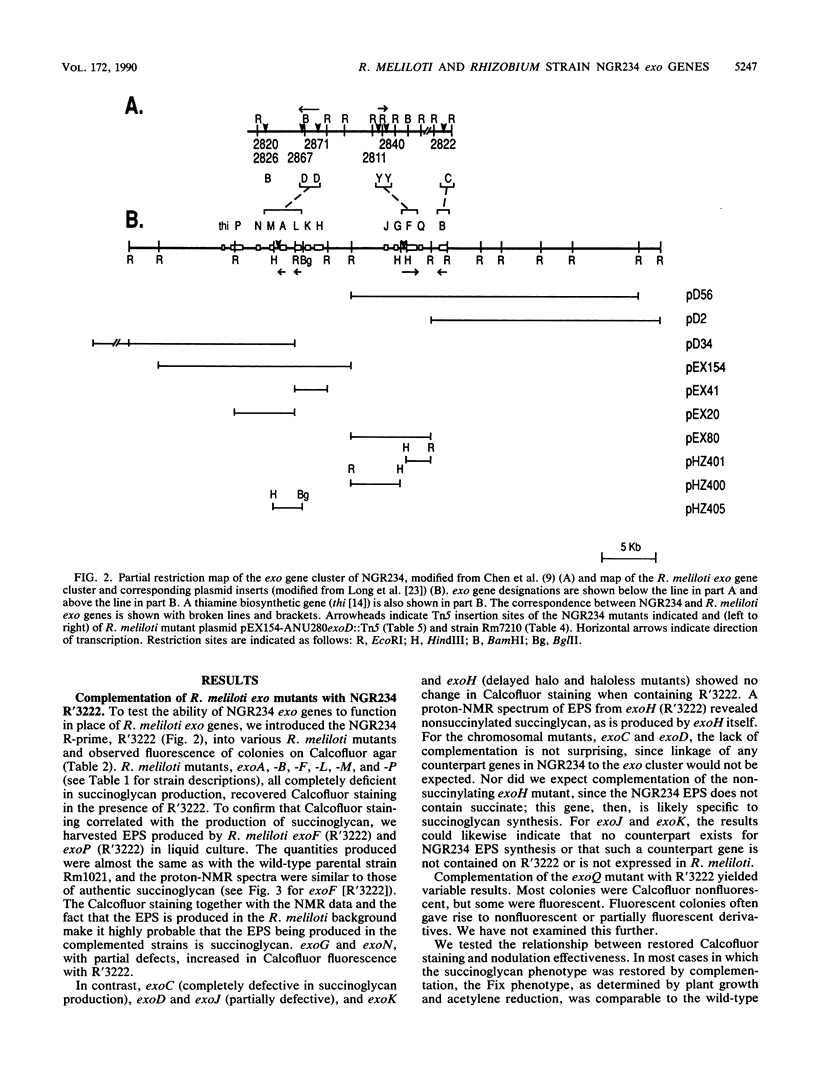
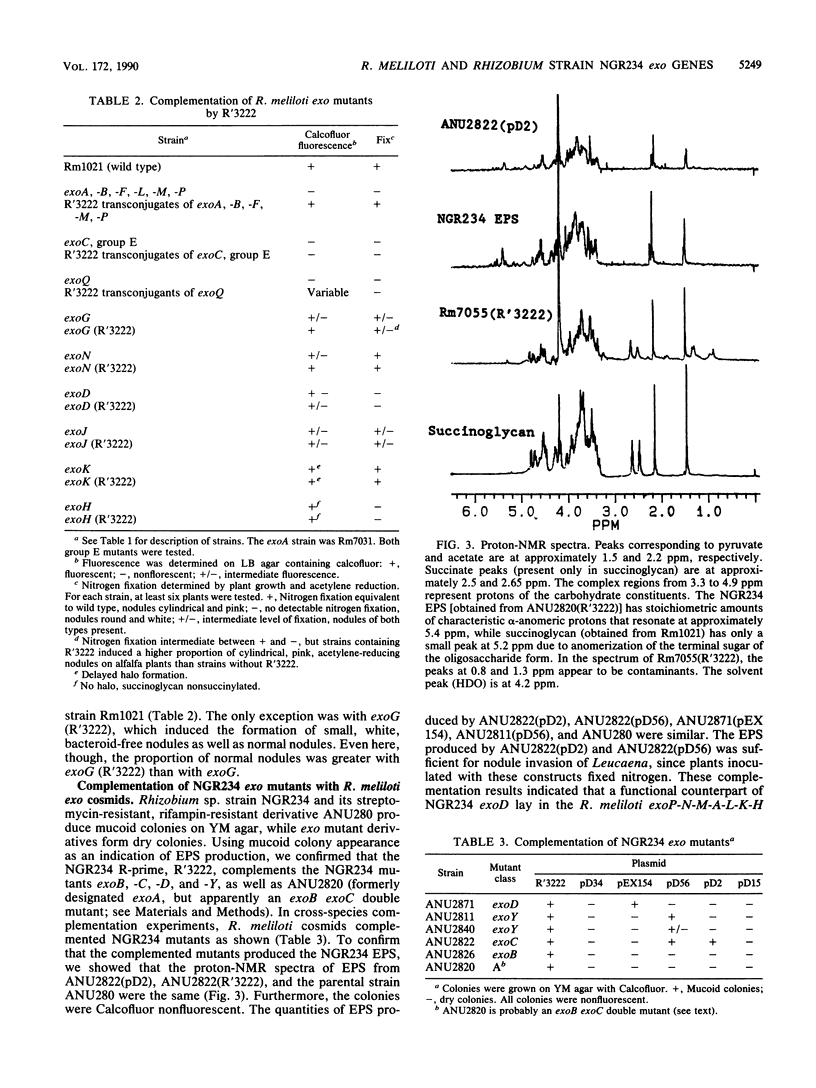
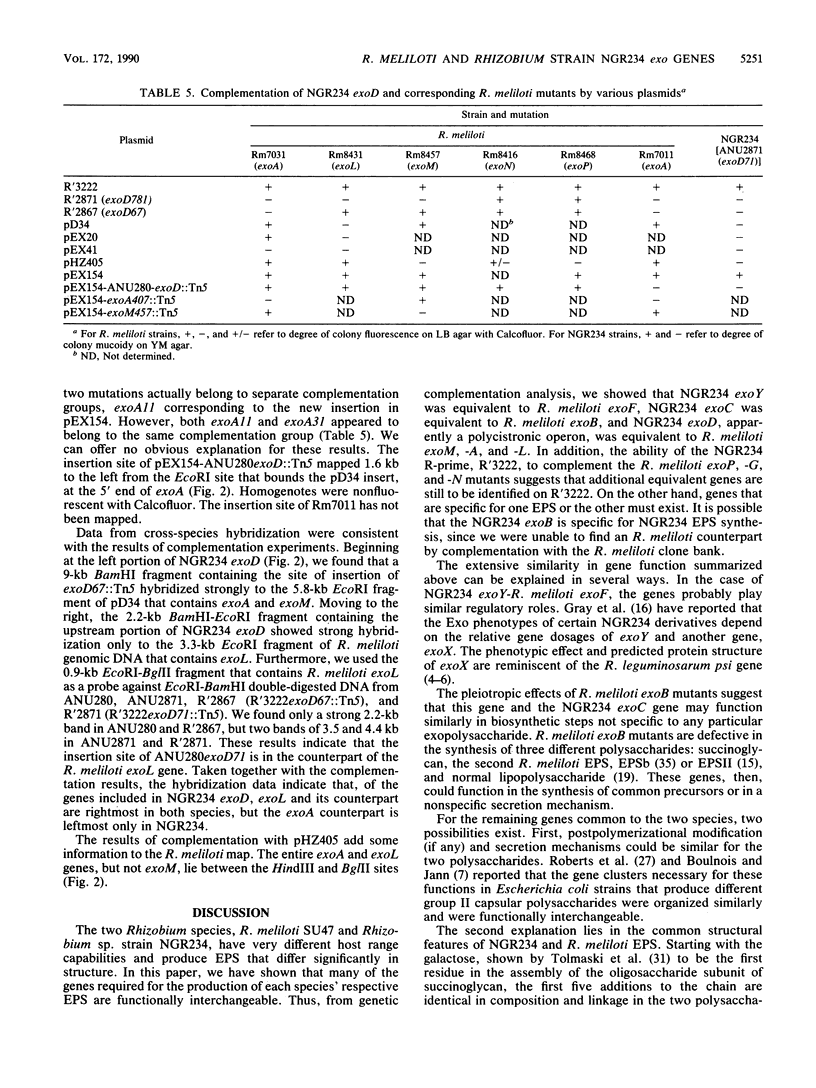
Rhizobium meliloti SU47 and Rhizobium sp. strain NGR234 produce distinct exopolysaccharides that have some similarities in structure. R. meliloti has a narrow host range, whereas Rhizobium strain NGR234 has a very broad host range. In cross-species complementation and hybridization experiments, we found that several of the genes required for the production of the two polysaccharides were functionally interchangeable and similar in evolutionary origin. NGR234 exoC and exoY corresponded to R. meliloti exoB and exoF, respectively. NGR234 exoD was found to be an operon that included genes equivalent to exoM, exoA, and exoL in R. meliloti. Complementation of R. meliloti exoP, -N, and -G by NGR234 R'3222 indicated that additional equivalent genes remain to be found on the R-prime. We were not able to complement NGR234 exoB with R. meliloti DNA. In addition to functional and evolutionary equivalence of individual genes, the general organization of the exo regions was similar between the two species. It is likely that the same ancestral genes were used in the evolution of both exopolysaccharide biosynthetic pathways and probably of pathways in other species as well.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borthakur D., Barker R. F., Latchford J. W., Rossen L., Johnston A. W. Analysis of pss genes of Rhizobium leguminosarum required for exopolysaccharide synthesis and nodulation of peas: their primary structure and their interaction with psi and other nodulation genes. Mol Gen Genet. 1988 Jul;213(1):155–162. doi: 10.1007/BF00333413. [DOI] [PubMed] [Google Scholar]
- Borthakur D., Johnston A. W. Sequence of psi, a gene on the symbiotic plasmid of Rhizobium phaseoli which inhibits exopolysaccharide synthesis and nodulation and demonstration that its transcription is inhibited by psr, another gene on the symbiotic plasmid. Mol Gen Genet. 1987 Apr;207(1):149–154. doi: 10.1007/BF00331502. [DOI] [PubMed] [Google Scholar]
- Boulnois G. J., Jann K. Bacterial polysaccharide capsule synthesis, export and evolution of structural diversity. Mol Microbiol. 1989 Dec;3(12):1819–1823. doi: 10.1111/j.1365-2958.1989.tb00168.x. [DOI] [PubMed] [Google Scholar]
- Diebold R., Noel K. D. Rhizobium leguminosarum exopolysaccharide mutants: biochemical and genetic analyses and symbiotic behavior on three hosts. J Bacteriol. 1989 Sep;171(9):4821–4830. doi: 10.1128/jb.171.9.4821-4830.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudman W. F., Lacey M. J. Identification of pyruvated monosaccharides in polysaccharides by gas-liquid chromatography mass spectrometry. Carbohydr Res. 1986 Jan 1;145(2):175–191. doi: 10.1016/s0008-6215(00)90428-2. [DOI] [PubMed] [Google Scholar]
- Finan T. M. Genetic and physical analyses of group E exo- mutants of Rhizobium meliloti. J Bacteriol. 1988 Jan;170(1):474–477. doi: 10.1128/jb.170.1.474-477.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Kunkel B., De Vos G. F., Signer E. R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986 Jul;167(1):66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Walker G. C. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989 Feb 24;56(4):661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- Gray J. X., Djordjevic M. A., Rolfe B. G. Two genes that regulate exopolysaccharide production in Rhizobium sp. strain NGR234: DNA sequences and resultant phenotypes. J Bacteriol. 1990 Jan;172(1):193–203. doi: 10.1128/jb.172.1.193-203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H., Tully R. E., Keister D. L. Exopolysaccharide-Deficient Mutants of Rhizobium fredii HH303 Which Are Symbiotically Effective. Appl Environ Microbiol. 1989 Jul;55(7):1852–1854. doi: 10.1128/aem.55.7.1852-1854.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J. A., Lee C. C. Characterization of polysaccharides of Rhizobium meliloti exo mutants that form ineffective nodules. J Bacteriol. 1988 Aug;170(8):3327–3332. doi: 10.1128/jb.170.8.3327-3332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J. A., Reed J. W., Hanks J. F., Hirsch A. M., Walker G. C. Rhizobium meliloti mutants that fail to succinylate their calcofluor-binding exopolysaccharide are defective in nodule invasion. Cell. 1987 Nov 20;51(4):579–587. doi: 10.1016/0092-8674(87)90127-9. [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Signer E. R., Walker G. C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levery S. B., Hakomori S. Microscale methylation analysis of glycolipids using capillary gas chromatography-chemical ionization mass fragmentography with selected ion monitoring. Methods Enzymol. 1987;138:13–25. doi: 10.1016/0076-6879(87)38004-8. [DOI] [PubMed] [Google Scholar]
- Long S., Reed J. W., Himawan J., Walker G. C. Genetic analysis of a cluster of genes required for synthesis of the calcofluor-binding exopolysaccharide of Rhizobium meliloti. J Bacteriol. 1988 Sep;170(9):4239–4248. doi: 10.1128/jb.170.9.4239-4248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison N. A., Cen Y. H., Chen H. C., Plazinski J., Ridge R., Rolfe B. G. Mobilization of a Sym plasmid from a fast-growing cowpea Rhizobium strain. J Bacteriol. 1984 Oct;160(1):483–487. doi: 10.1128/jb.160.1.483-487.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip-Hollingsworth S., Hollingsworth R. I., Dazzo F. B., Djordjevic M. A., Rolfe B. G. The effect of interspecies transfer of Rhizobium host-specific nodulation genes on acidic polysaccharide structure and in situ binding by host lectin. J Biol Chem. 1989 Apr 5;264(10):5710–5714. [PubMed] [Google Scholar]
- Roberts I. S., Mountford R., Hodge R., Jann K. B., Boulnois G. J. Common organization of gene clusters for production of different capsular polysaccharides (K antigens) in Escherichia coli. J Bacteriol. 1988 Mar;170(3):1305–1310. doi: 10.1128/jb.170.3.1305-1310.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser T. J., Helinski D. R. Regions of broad-host-range plasmid RK2 involved in replication and stable maintenance in nine species of gram-negative bacteria. J Bacteriol. 1985 Oct;164(1):446–455. doi: 10.1128/jb.164.1.446-455.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland I. W. Biosynthesis and composition of gram-negative bacterial extracellular and wall polysaccharides. Annu Rev Microbiol. 1985;39:243–270. doi: 10.1146/annurev.mi.39.100185.001331. [DOI] [PubMed] [Google Scholar]
- Tolmasky M. E., Staneloni R. J., Leloir L. F. Lipid-bound saccharides in Rhizobium meliloti. J Biol Chem. 1982 Jun 25;257(12):6751–6757. [PubMed] [Google Scholar]
- Uttaro A. D., Cangelosi G. A., Geremia R. A., Nester E. W., Ugalde R. A. Biochemical characterization of avirulent exoC mutants of Agrobacterium tumefaciens. J Bacteriol. 1990 Mar;172(3):1640–1646. doi: 10.1128/jb.172.3.1640-1646.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan H. J., Levery S. B., Lee C. C., Leigh J. A. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc Natl Acad Sci U S A. 1989 May;86(9):3055–3059. doi: 10.1073/pnas.86.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]


