Abstract
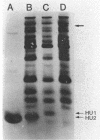
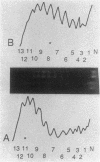
Salmonella hupA and hupB mutants were studied to determine the reasons for the high degree of conservation in HU structure in bacteria. We found one HU-1-specific effect; the F'128 plasmid was 25-fold less stable in hupB compared with hupA or wild-type cells. F' plasmids were 120-fold more unstable in hupA hupB double mutants compared with wild-type cells, and the double mutant also had a significant alteration in plasmid DNA structure. pBR322 DNA isolated from hupA hupB strains was deficient in supercoiling by 10 to 15% compared with wild-type cells, and the topoisomer distribution was significantly more heterogeneous than in wild-type or single-mutant strains. Other systems altered by HU inactivation included flagellar phase variation and phage Mu transposition. However, Mu transposition rates were only about fourfold lower in Salmonella HU double mutants. One reason that Salmonella HU double mutants may be less defective for Mu transposition than E. coli is the synthesis in double mutants of a new, small, basic heat-stable protein, which might partially compensate for the loss of HU. The results indicate that although either HU-1 or HU-2 subunit alone may accommodate the cellular need for general chromosomal organization, the selective pressure to conserve HU-1 and HU-2 structure during evolution could involve specialized roles of the individual subunits.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker T. A., Sekimizu K., Funnell B. E., Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986 Apr 11;45(1):53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Bonnefoy E., Almeida A., Rouviere-Yaniv J. Lon-dependent regulation of the DNA binding protein HU in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7691–7695. doi: 10.1073/pnas.86.20.7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles S. S., Pettijohn D. E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J Mol Biol. 1986 Jan 5;187(1):47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Botstein D., Watanabe T., Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology. 1972 Dec;50(3):883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- Craigie R., Arndt-Jovin D. J., Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K. A., Sternglanz R., Reynolds A. E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982 Nov;31(1):43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- Dürrenberger M., Bjornsti M. A., Uetz T., Hobot J. A., Kellenberger E. Intracellular location of the histonelike protein HU in Escherichia coli. J Bacteriol. 1988 Oct;170(10):4757–4768. doi: 10.1128/jb.170.10.4757-4768.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins N. P., Hillyard D. Primary structure and mapping of the hupA gene of Salmonella typhimurium. J Bacteriol. 1988 Dec;170(12):5751–5758. doi: 10.1128/jb.170.12.5751-5758.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., Faelen M., Girard D., Jaffé A., Toussaint A., Rouvière-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J Bacteriol. 1989 Jul;171(7):3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. F., Simon M. I. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986 Aug 15;46(4):531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- Marsh M., Hillyard D. R. Nucleotide sequence of the HU-1 gene of Salmonella typhimurium. Nucleic Acids Res. 1988 Jul 25;16(14B):7196–7196. doi: 10.1093/nar/16.14.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Van Keuren M. L. Gel protein stains: silver stain. Methods Enzymol. 1984;104:441–447. doi: 10.1016/s0076-6879(84)04111-2. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Wada M., Kano Y., Imamoto F., Okazaki T. DNA replication in Escherichia coli mutants that lack protein HU. J Bacteriol. 1989 Oct;171(10):5672–5679. doi: 10.1128/jb.171.10.5672-5679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983 Feb;32(2):351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- Pruss G. J. DNA topoisomerase I mutants. Increased heterogeneity in linking number and other replicon-dependent changes in DNA supercoiling. J Mol Biol. 1985 Sep 5;185(1):51–63. doi: 10.1016/0022-2836(85)90182-2. [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. Topoisomerase I mutants: the gene on pBR322 that encodes resistance to tetracycline affects plasmid DNA supercoiling. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8952–8956. doi: 10.1073/pnas.83.23.8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss G. J., Manes S. H., Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982 Nov;31(1):35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Yaniv M., Germond J. E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979 Jun;17(2):265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119(1):75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Carlson J. O., Pettijohn D. E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980 Oct;21(3):773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Storts D. R., Markovitz A. Construction and characterization of mutations in hupB, the gene encoding HU-beta (HU-1) in Escherichia coli K-12. J Bacteriol. 1988 Apr;170(4):1541–1547. doi: 10.1128/jb.170.4.1541-1547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Wada M., Kano Y., Ogawa T., Okazaki T., Imamoto F. Construction and characterization of the deletion mutant of hupA and hupB genes in Escherichia coli. J Mol Biol. 1988 Dec 5;204(3):581–591. doi: 10.1016/0022-2836(88)90357-9. [DOI] [PubMed] [Google Scholar]
- Zacharias W., Jaworski A., Larson J. E., Wells R. D. The B- to Z-DNA equilibrium in vivo is perturbed by biological processes. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7069–7073. doi: 10.1073/pnas.85.19.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]