Abstract
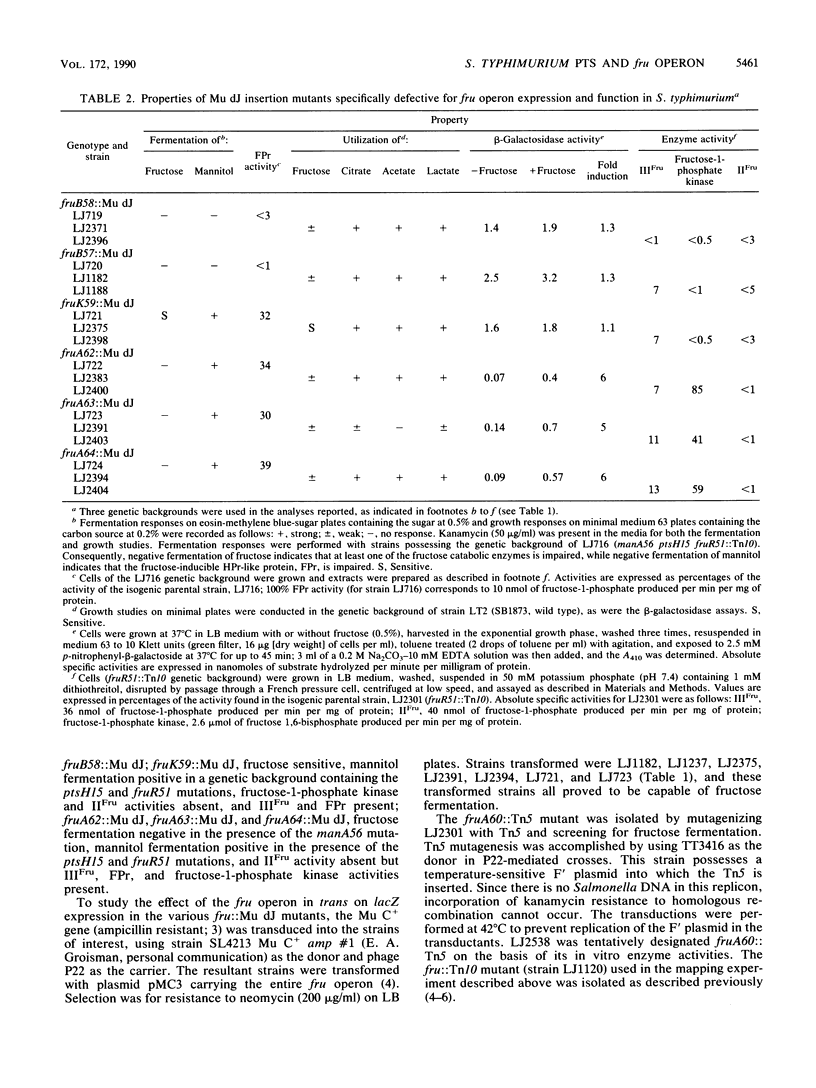
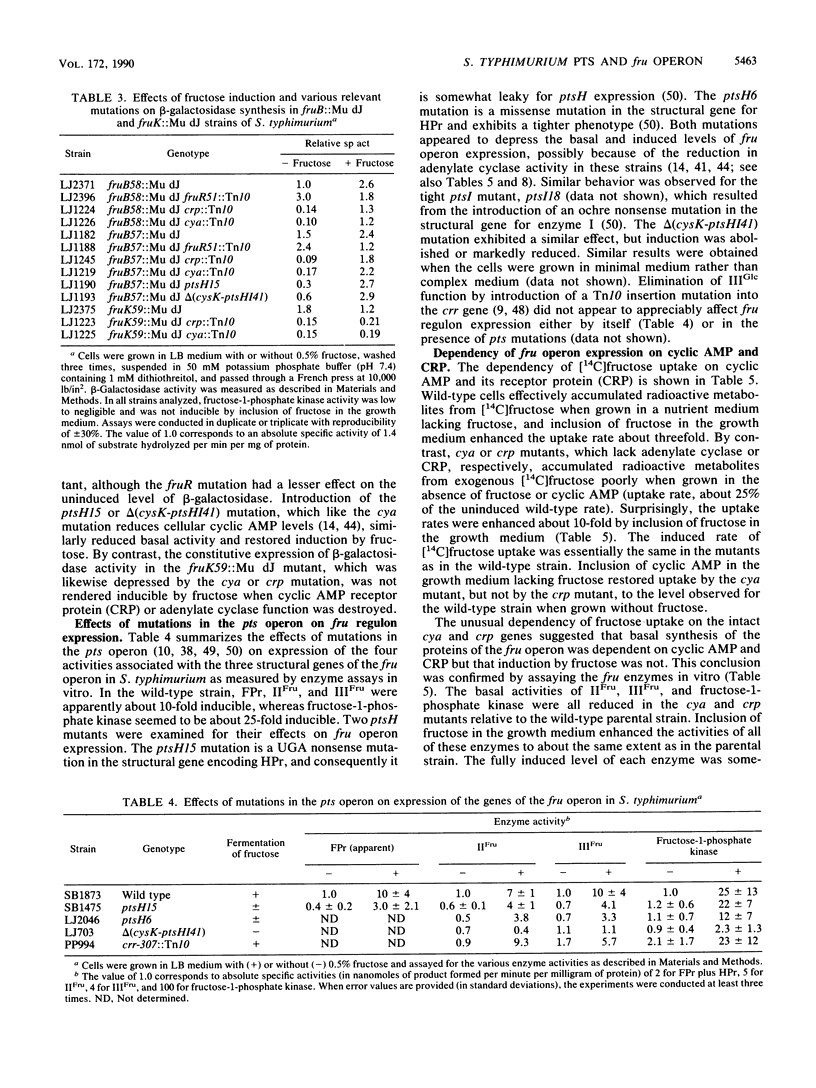
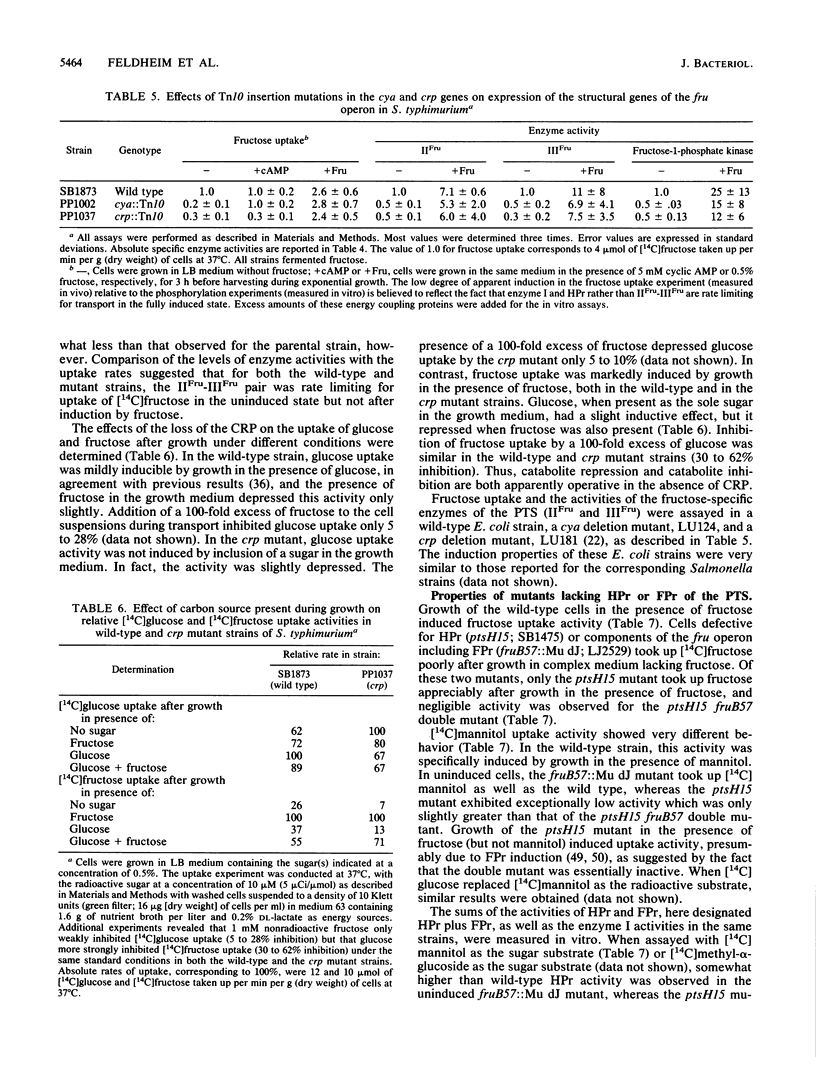
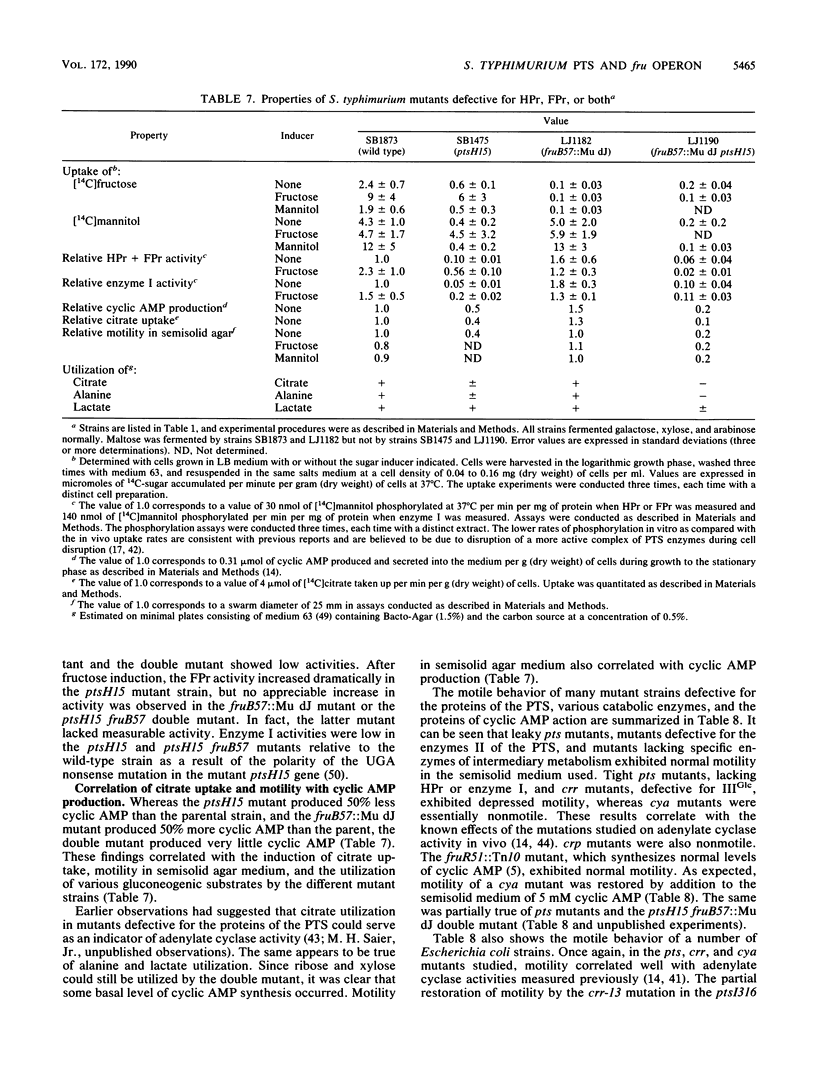
Mutants of Salmonella typhimurium defective in the proteins of the fructose operon [fruB(MH)KA], the fructose repressor (fruR), the energy-coupling enzymes of the phosphoenolpyruvate:sugar phosphotransferase system (PTS) (ptsH and ptsI), and the proteins of cyclic AMP action (cya and crp) were analyzed for their effects on cellular physiological processes and expression of the fructose operon. The fru operon consists of three structural genes: fruB(MH), which encodes the enzyme IIIFru-modulator-FPr tridomain fusion protein of the PTS; fruK, which encodes fructose-1-phosphate kinase; and fruA, which encodes enzyme IIFru of the PTS. Among the mutants analyzed were Tn10 insertion mutants and lacZ transcriptional fusion mutants. It was found that whereas a fruR::Tn10 insertion mutant, several fruB(MH)::Mu dJ and fruK::Mu dJ fusion mutants, and several ptsHI deletion mutants expressed the fru operon and beta-galactosidase at high constitutive levels, ptsH point mutants and fruA::Mu dJ fusion mutants retained inducibility. Inclusion of the wild-type fru operon in trans did not restore fructose-inducible beta-galactosidase expression in the fru::Mu dJ fusion mutants. cya and crp mutants exhibited reduced basal activities of all fru regulon enzymes, but inducibility was not impaired. Surprisingly, fruB::Mu dJ crp or cya double mutants showed over 10-fold inducibility of the depressed beta-galactosidase activity upon addition of fructose, even though this activity in the fruB::Mu dJ fusion mutants that contained the wild-type cya and crp alleles was only slightly inducible. By contrast, beta-galactosidase activity in a fruK::Mu dJ fusion mutant, which was similarly depressed by introduction of a crp or cya mutation, remained constitutive. Other experiments indicated that sugar uptake via the PTS can utilize either FPr-P or HPr-P as the phosphoryl donor, but that FPr is preferred for fructose uptake whereas HPr is preferred for uptake of the other sugars. Double mutants lacking both proteins were negative for the utilization of all sugar substrates of the PTS, were negative for the utilization of several gluconeogenic carbon sources, exhibited greatly reduced adenylate cyclase activity, and were largely nonmotile. These phenotypic properties are more extreme than those observed for tight ptsH and ptsI mutants, including mutants deleted for these genes. A biochemical explanation for this fact is proposed.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amster-Choder O., Houman F., Wright A. Protein phosphorylation regulates transcription of the beta-glucoside utilization operon in E. coli. Cell. 1989 Sep 8;58(5):847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- Aymerich S., Steinmetz M. Cloning and preliminary characterization of the sacS locus from Bacillus subtilis which controls the regulation of the exoenzyme levansucrase. Mol Gen Genet. 1987 Jun;208(1-2):114–120. doi: 10.1007/BF00330431. [DOI] [PubMed] [Google Scholar]
- Chin A. M., Feucht B. U., Saier M. H., Jr Evidence for regulation of gluconeogenesis by the fructose phosphotransferase system in Salmonella typhimurium. J Bacteriol. 1987 Feb;169(2):897–899. doi: 10.1128/jb.169.2.897-899.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A. M., Sutrina S., Feldheim D. A., Saier M. H., Jr Genetic expression of enzyme I activity of the phosphoenolpyruvate:sugar phosphotransferase system in ptsHI deletion strains of Salmonella typhimurium. J Bacteriol. 1987 Feb;169(2):894–896. doi: 10.1128/jb.169.2.894-896.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaro J. C., Roseman S. Deletion mapping of the genes coding for HPr and enzyme I of the phosphoenolpyruvate: sugar phosphotransferase system in Salmonella typhimurium. J Bacteriol. 1972 Oct;112(1):17–29. doi: 10.1128/jb.112.1.17-29.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G. A., Drews G., Saier M. H., Jr Properties of a Tn5 insertion mutant defective in the structural gene (fruA) of the fructose-specific phosphotransferase system of Rhodobacter capsulatus and cloning of the fru regulon. J Bacteriol. 1988 Apr;170(4):1698–1703. doi: 10.1128/jb.170.4.1698-1703.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Reuse H., Danchin A. The ptsH, ptsI, and crr genes of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system: a complex operon with several modes of transcription. J Bacteriol. 1988 Sep;170(9):3827–3837. doi: 10.1128/jb.170.9.3827-3837.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dills S. S., Apperson A., Schmidt M. R., Saier M. H., Jr Carbohydrate transport in bacteria. Microbiol Rev. 1980 Sep;44(3):385–418. doi: 10.1128/mr.44.3.385-418.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feucht B. U., Saier M. H., Jr Fine control of adenylate cyclase by the phosphoenolpyruvate:sugar phosphotransferase systems in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1980 Feb;141(2):603–610. doi: 10.1128/jb.141.2.603-610.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet A., Arnaud M., Klier A., Rapoport G. Genetics of the phosphotransferase system of Bacillus subtilis. FEMS Microbiol Rev. 1989 Jun;5(1-2):175–182. doi: 10.1016/0168-6445(89)90022-3. [DOI] [PubMed] [Google Scholar]
- Fraenkel D. G. The phosphoenolpyruvate-initiated pathway of fructose metabolism in Escherichia coli. J Biol Chem. 1968 Dec 25;243(24):6458–6463. [PubMed] [Google Scholar]
- Gachelin G. A new assay of the phosphotransferase system in Escherichia coli. Biochem Biophys Res Commun. 1969 Feb 21;34(4):382–387. doi: 10.1016/0006-291x(69)90392-1. [DOI] [PubMed] [Google Scholar]
- Geerse R. H., Izzo F., Postma P. W. The PEP: fructose phosphotransferase system in Salmonella typhimurium: FPr combines enzyme IIIFru and pseudo-HPr activities. Mol Gen Genet. 1989 Apr;216(2-3):517–525. doi: 10.1007/BF00334399. [DOI] [PubMed] [Google Scholar]
- Geerse R. H., Ruig C. R., Schuitema A. R., Postma P. W. Relationship between pseudo-HPr and the PEP: fructose phosphotransferase system in Salmonella typhimurium and Escherichia coli. Mol Gen Genet. 1986 Jun;203(3):435–444. doi: 10.1007/BF00422068. [DOI] [PubMed] [Google Scholar]
- Geerse R. H., van der Pluijm J., Postma P. W. The repressor of the PEP:fructose phosphotransferase system is required for the transcription of the pps gene of Escherichia coli. Mol Gen Genet. 1989 Aug;218(2):348–352. doi: 10.1007/BF00331288. [DOI] [PubMed] [Google Scholar]
- Gershanovitch V. N., Bolshakova T. N., Molchanova M. L., Umyarov A. M., Dobrynina OYu, Grigorenko YuA, Erlagaeva R. S. Fructose-specific phosphoenolpyruvate dependent phosphotransferase system of Escherichia coli: its alterations and adenylate cyclase activity. FEMS Microbiol Rev. 1989 Jun;5(1-2):125–133. doi: 10.1111/j.1574-6968.1989.tb14108.x. [DOI] [PubMed] [Google Scholar]
- Groisman E. A., Casadaban M. J. Cloning of genes from members of the family Enterobacteriaceae with mini-Mu bacteriophage containing plasmid replicons. J Bacteriol. 1987 Feb;169(2):687–693. doi: 10.1128/jb.169.2.687-693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman P. E. Some improved methods in P22 transduction. Genetics. 1974 Apr;76(4):625–631. doi: 10.1093/genetics/76.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin C. H., Betsholtz C., Claesson-Welsh L., Westermark B. Subversion of growth regulatory pathways in malignant transformation. Biochim Biophys Acta. 1987 Nov 25;907(3):219–244. doi: 10.1016/0304-419x(87)90007-2. [DOI] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988 May;119(1):9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACOB F., MONOD J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961 Jun;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Kofoid E. C., Parkinson J. S. Transmitter and receiver modules in bacterial signaling proteins. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4981–4985. doi: 10.1073/pnas.85.14.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg H. L. Fructose transport by Escherichia coli. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1236):505–513. doi: 10.1098/rstb.1990.0028. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Mahadevan S., Wright A. A bacterial gene involved in transcription antitermination: regulation at a rho-independent terminator in the bgl operon of E. coli. Cell. 1987 Jul 31;50(3):485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- O'Neill M. C., Chiafari F. Escherichia coli promoters. II. A spacing class-dependent promoter search protocol. J Biol Chem. 1989 Apr 5;264(10):5531–5534. [PubMed] [Google Scholar]
- O'Neill M. C. Escherichia coli promoters. I. Consensus as it relates to spacing class, specificity, repeat substructure, and three-dimensional organization. J Biol Chem. 1989 Apr 5;264(10):5522–5530. [PubMed] [Google Scholar]
- Peri K. G., Waygood E. B. Sequence of cloned enzyme IIN-acetylglucosamine of the phosphoenolpyruvate:N-acetylglucosamine phosphotransferase system of Escherichia coli. Biochemistry. 1988 Aug 9;27(16):6054–6061. doi: 10.1021/bi00416a034. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. A. Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol Microbiol. 1989 Apr;3(4):505–515. doi: 10.1111/j.1365-2958.1989.tb00197.x. [DOI] [PubMed] [Google Scholar]
- Prior T. I., Kornberg H. L. Nucleotide sequence of fruA, the gene specifying enzyme IIfru of the phosphoenolpyruvate-dependent sugar phosphotransferase system in Escherichia coli K12. J Gen Microbiol. 1988 Oct;134(10):2757–2768. doi: 10.1099/00221287-134-10-2757. [DOI] [PubMed] [Google Scholar]
- Rephaeli A. W., Saier M. H., Jr Regulation of genes coding for enzyme constituents of the bacterial phosphotransferase system. J Bacteriol. 1980 Feb;141(2):658–663. doi: 10.1128/jb.141.2.658-663.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznikoff W. S., Siegele D. A., Cowing D. W., Gross C. A. The regulation of transcription initiation in bacteria. Annu Rev Genet. 1985;19:355–387. doi: 10.1146/annurev.ge.19.120185.002035. [DOI] [PubMed] [Google Scholar]
- Saffen D. W., Presper K. A., Doering T. L., Roseman S. Sugar transport by the bacterial phosphotransferase system. Molecular cloning and structural analysis of the Escherichia coli ptsH, ptsI, and crr genes. J Biol Chem. 1987 Nov 25;262(33):16241–16253. [PubMed] [Google Scholar]
- Saier M. H., Jr, Cox D. F., Feucht B. U., Novotny M. J. Evidence for the functional association of enzyme I and HPr of the phosphoenolpyruvate-sugar phosphotransferase system with the membrane in sealed vesicles of Escherichia coli. J Cell Biochem. 1982;18(2):231–238. doi: 10.1002/jcb.1982.240180210. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Daniels G. A., Boerner P., Lin J. Neutral amino acid transport systems in animal cells: potential targets of oncogene action and regulators of cellular growth. J Membr Biol. 1988 Aug;104(1):1–20. doi: 10.1007/BF01871898. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Feucht B. U. Coordinate regulation of adenylate cyclase and carbohydrate permeases by the phosphoenolpyruvate:sugar phosphotransferase system in Salmonella typhimurium. J Biol Chem. 1975 Sep 10;250(17):7078–7080. [PubMed] [Google Scholar]
- Saier M. H., Jr, Feucht B. U., Roseman S. Phosphoenolpyruvate-dependent fructose phosphorylation in photosynthetic bacteria. J Biol Chem. 1971 Dec 25;246(24):7819–7821. [PubMed] [Google Scholar]
- Saier M. H., Jr, Grenier F. C., Lee C. A., Waygood E. B. Evidence for the evolutionary relatedness of the proteins of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. J Cell Biochem. 1985;27(1):43–56. doi: 10.1002/jcb.240270106. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr Involvement of the bacterial phosphotransferase system in diverse mechanisms of transcriptional regulation. Res Microbiol. 1989 Jul-Aug;140(6):349–352. doi: 10.1016/0923-2508(89)90010-7. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Microbiol Rev. 1989 Mar;53(1):109–120. doi: 10.1128/mr.53.1.109-120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Jr, Roseman S. Sugar transport. The crr mutation: its effect on repression of enzyme synthesis. J Biol Chem. 1976 Nov 10;251(21):6598–6605. [PubMed] [Google Scholar]
- Saier M. H., Jr, Simoni R. D., Roseman S. The physiological behavior of enzyme I and heat-stable protein mutants of a bacterial phosphotransferase system. J Biol Chem. 1970 Nov 10;245(21):5870–5873. [PubMed] [Google Scholar]
- Saier M. H., Jr, Yamada M., Erni B., Suda K., Lengeler J., Ebner R., Argos P., Rak B., Schnetz K., Lee C. A. Sugar permeases of the bacterial phosphoenolpyruvate-dependent phosphotransferase system: sequence comparisons. FASEB J. 1988 Mar 1;2(3):199–208. doi: 10.1096/fasebj.2.3.2832233. [DOI] [PubMed] [Google Scholar]
- Schnetz K., Rak B. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988 Oct;7(10):3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz K., Toloczyki C., Rak B. Beta-glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987 Jun;169(6):2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni R. D., Roseman S., Saier M. H., Jr Sugar transport. Properties of mutant bacteria defective in proteins of the phosphoenolpyruvate: sugar phosphotransferase system. J Biol Chem. 1976 Nov 10;251(21):6584–6597. [PubMed] [Google Scholar]
- Sutrina S. L., Chin A. M., Esch F., Saier M. H., Jr Purification and characterization of the fructose-inducible HPr-like protein, FPr, and the fructose-specific enzyme III of the phosphoenolpyruvate: sugar phosphotransferase system of Salmonella typhimurium. J Biol Chem. 1988 Apr 15;263(11):5061–5069. [PubMed] [Google Scholar]
- Wu L. F., Tomich J. M., Saier M. H., Jr Structure and evolution of a multidomain multiphosphoryl transfer protein. Nucleotide sequence of the fruB(HI) gene in Rhodobacter capsulatus and comparisons with homologous genes from other organisms. J Mol Biol. 1990 Jun 20;213(4):687–703. doi: 10.1016/S0022-2836(05)80256-6. [DOI] [PubMed] [Google Scholar]
- Yamada M., Saier M. H., Jr Positive and negative regulators for glucitol (gut) operon expression in Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):569–583. doi: 10.1016/0022-2836(88)90193-3. [DOI] [PubMed] [Google Scholar]
- den Blaauwen J. L., Postma P. W. Regulation of cyclic AMP synthesis by enzyme IIIGlc of the phosphoenolpyruvate:sugar phosphotransferase system in crp strains of Salmonella typhimurium. J Bacteriol. 1985 Oct;164(1):477–478. doi: 10.1128/jb.164.1.477-478.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]


