Abstract
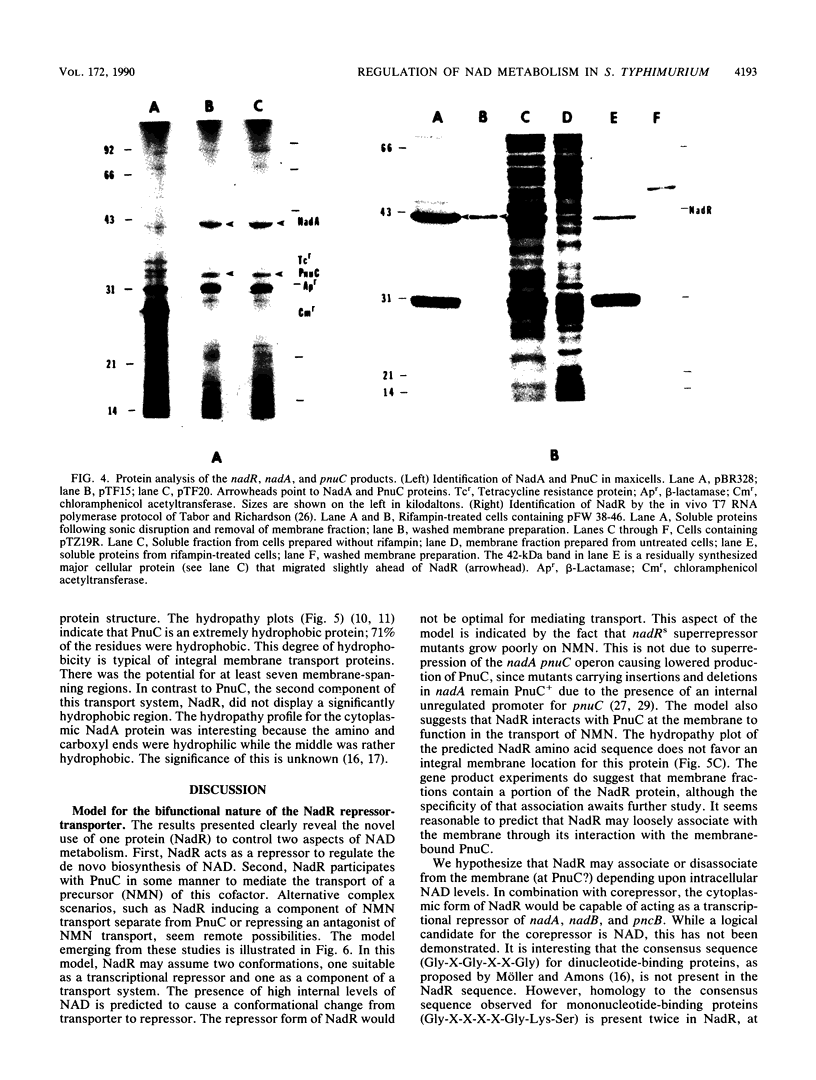
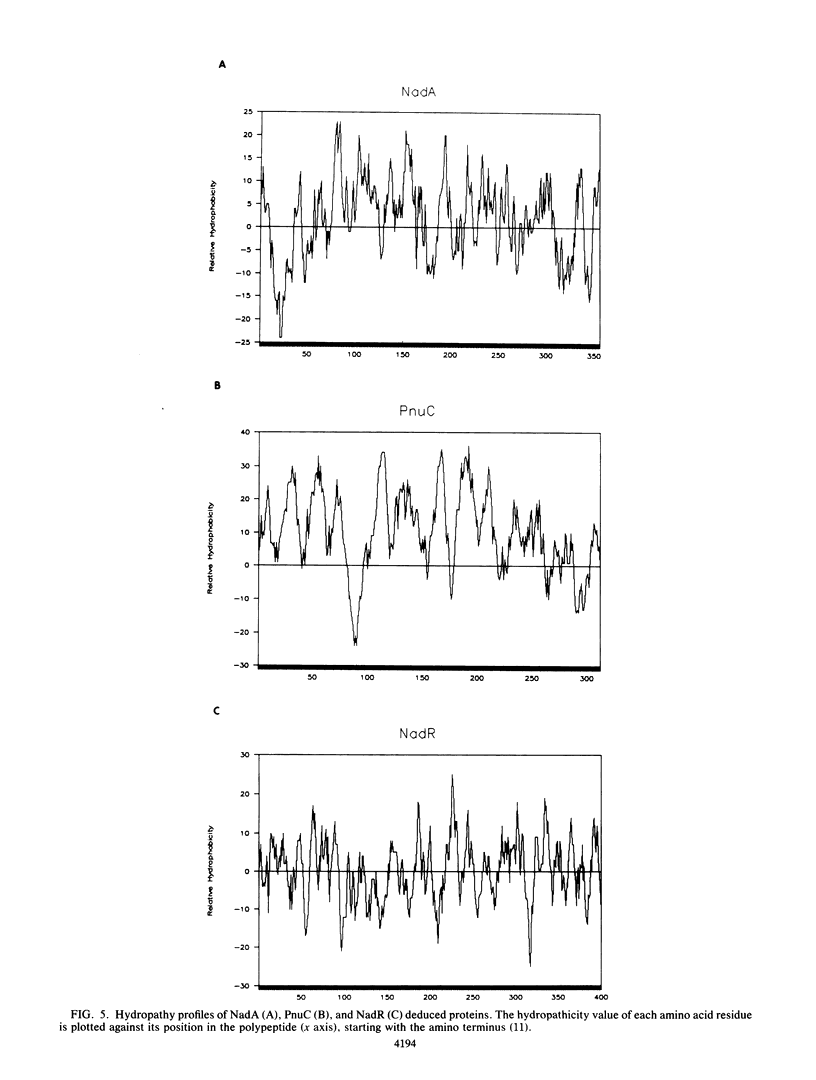
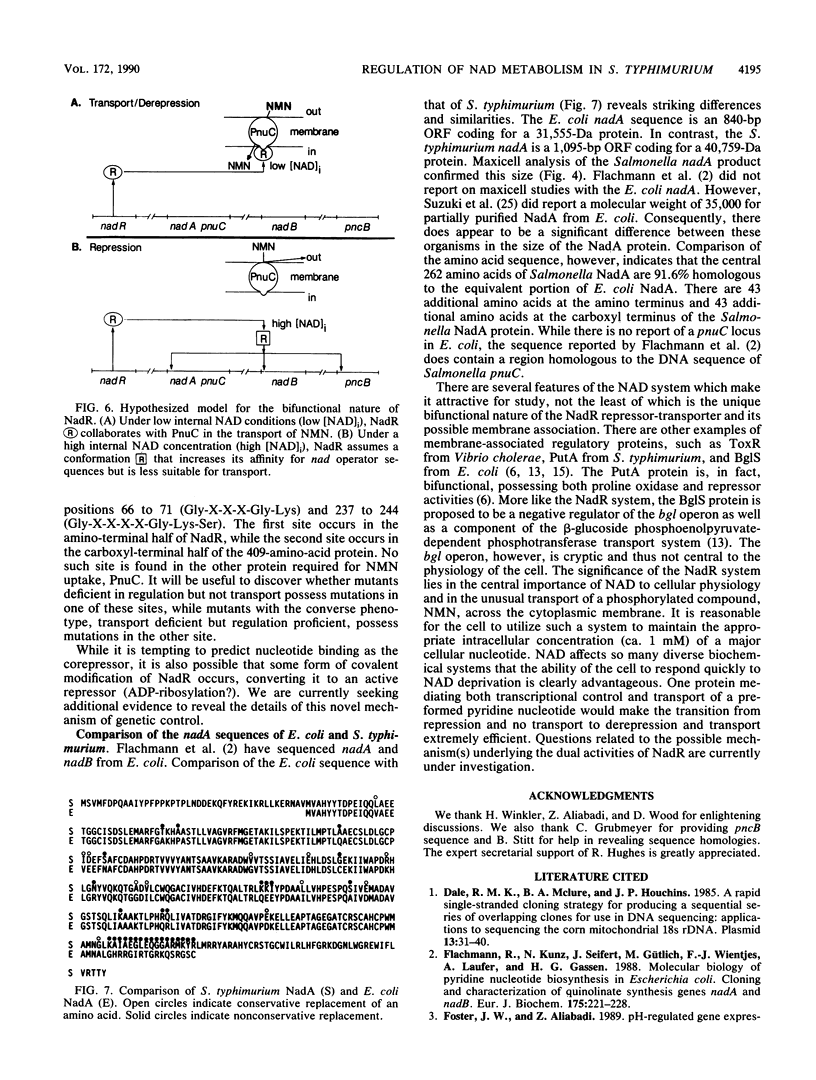
In Salmonella typhimurium, de novo synthesis of NAD is regulated through the transcriptional control of the nadA and nadB loci. Likewise, the pyridine nucleotide salvage pathway is controlled at pncB. The transcriptional expression of these three loci is coordinately regulated by the product of nadR. However, there is genetic evidence suggesting that NadR is bifunctional, serving in both regulatory and transport capacities. One class of mutations in the nadR locus imparts a transport-defective PnuA- phenotype. These mutants retain regulation properties but are unable to transport nicotinamide mononucleotide (NMN) intact across the cell membrane. Other nadR mutants lose both regulatory and transport capabilities, while a third class loses only regulatory ability. The unusual NMN transport activity requires both the PnuC and NadR proteins, with the pnuC locus residing in an operon with nadA. To prove that nadR encoded a single protein and to gain insight into a regulatory target locus, the nadR and nadA pnuC loci were cloned and sequenced. A DNA fragment which complemented both regulatory and transport mutations was found to contain a single open reading frame capable of encoding a 409-amino-acid protein (47,022 daltons), indicating that NadR is indeed bifunctional. Confirmation of the operon arrangement for nadA and pnuC was obtained through the sequence analysis of a 2.4-kilobase DNA fragment which complemented both NadA and PnuC mutant phenotypes. The nadA product, confirmed in maxicells, was a 365-amino-acid protein (40,759 daltons), while pnuC encoded a 322-amino-acid protein (36,930 daltons). The extremely hydrophobic (71%) nature of the PnuC protein indicated that it was an integral membrane protein, consistent with its central role in the transport of NMN across the cytoplasmic membrane. The results presented here and in previous studies suggest a hypothetical model in which NadR interacts with PnuC at low internal NAD levels, permitting transport of NMN intact into the cell. As NAD levels increase within the cell, the affinity of NadR for the operator regions of nadA, nadB, and pncB increases, repressing the transcription of these target genes.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Flachmann R., Kunz N., Seifert J., Gütlich M., Wientjes F. J., Läufer A., Gassen H. G. Molecular biology of pyridine nucleotide biosynthesis in Escherichia coli. Cloning and characterization of quinolinate synthesis genes nadA and nadB. Eur J Biochem. 1988 Aug 1;175(2):221–228. doi: 10.1111/j.1432-1033.1988.tb14187.x. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Holley-Guthrie E. A., Warren F. Regulation of NAD metabolism in Salmonella typhimurium: genetic analysis and cloning of the nadR repressor locus. Mol Gen Genet. 1987 Jun;208(1-2):279–287. doi: 10.1007/BF00330454. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Moat A. G. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev. 1980 Mar;44(1):83–105. doi: 10.1128/mr.44.1.83-105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D. R., Myers R. S., Kent C. R., Maloy S. R. Regulation of proline utilization in Salmonella typhimurium: molecular characterization of the put operon, and DNA sequence of the put control region. Mol Gen Genet. 1988 Jul;213(1):125–133. doi: 10.1007/BF00333408. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Holley E. A., Spector M. P., Foster J. W. Regulation of NAD biosynthesis in Salmonella typhimurium: expression of nad-lac gene fusions and identification of a nad regulatory locus. J Gen Microbiol. 1985 Oct;131(10):2759–2770. doi: 10.1099/00221287-131-10-2759. [DOI] [PubMed] [Google Scholar]
- Kinney D. M., Foster J. W. Identification of a cis-acting regulatory region in the pncB locus of Salmonella typhimurium. Mol Gen Genet. 1985;199(3):512–517. doi: 10.1007/BF00330767. [DOI] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Liu G., Foster J., Manlapaz-Ramos P., Olivera B. M. Nucleoside salvage pathway for NAD biosynthesis in Salmonella typhimurium. J Bacteriol. 1982 Dec;152(3):1111–1116. doi: 10.1128/jb.152.3.1111-1116.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan S., Reynolds A. E., Wright A. Positive and negative regulation of the bgl operon in Escherichia coli. J Bacteriol. 1987 Jun;169(6):2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. L., Taylor R. K., Mekalanos J. J. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987 Jan 30;48(2):271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- Möller W., Amons R. Phosphate-binding sequences in nucleotide-binding proteins. FEBS Lett. 1985 Jul 1;186(1):1–7. doi: 10.1016/0014-5793(85)81326-0. [DOI] [PubMed] [Google Scholar]
- Nasu S., Wicks F. D., Gholson R. K. L-Aspartate oxidase, a newly discovered enzyme of Escherichia coli, is the B protein of quinolinate synthetase. J Biol Chem. 1982 Jan 25;257(2):626–632. [PubMed] [Google Scholar]
- Nasu S., Wicks F. D., Gholson R. K. The mammalian enzyme which replaces B protein of E. coli quinolinate synthetase is D-aspartate oxidase. Biochim Biophys Acta. 1982 Jun 4;704(2):240–252. doi: 10.1016/0167-4838(82)90152-2. [DOI] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Brown N. L., Air G. M., Barrell B. G., Coulson A. R., Hutchison C. A., 3rd, Sanger F. DNA sequence at the C termini of the overlapping genes A and B in bacteriophage phi X174. Nature. 1977 Feb 24;265(5596):702–705. doi: 10.1038/265702a0. [DOI] [PubMed] [Google Scholar]
- Spector M. P., Hill J. M., Holley E. A., Foster J. W. Genetic characterization of pyridine nucleotide uptake mutants of Salmonella typhimurium. J Gen Microbiol. 1985 Jun;131(6):1313–1322. doi: 10.1099/00221287-131-6-1313. [DOI] [PubMed] [Google Scholar]
- Stirling D. A., Hulton C. S., Waddell L., Park S. F., Stewart G. S., Booth I. R., Higgins C. F. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol Microbiol. 1989 Aug;3(8):1025–1038. doi: 10.1111/j.1365-2958.1989.tb00253.x. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Carlson J., Griffith G., Gholson R. K. Studies on the de novo biosynthesis of NAD in Escherichia coli. V. Properties of the quinolinic acid synthetase system. Biochim Biophys Acta. 1973 Apr 28;304(2):309–315. doi: 10.1016/0304-4165(73)90249-3. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirgari S., Spector M. P., Foster J. W. Genetics of NAD metabolism in Salmonella typhimurium and cloning of the nadA and pnuC loci. J Bacteriol. 1986 Sep;167(3):1086–1088. doi: 10.1128/jb.167.3.1086-1088.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Olivera B. M., Roth J. R. Genetic characterization of the pnuC gene, which encodes a component of the nicotinamide mononucleotide transport system in Salmonella typhimurium. J Bacteriol. 1989 Aug;171(8):4402–4409. doi: 10.1128/jb.171.8.4402-4409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Olivera B. M., Roth J. R. Identification of a repressor gene involved in the regulation of NAD de novo biosynthesis in Salmonella typhimurium. J Bacteriol. 1988 Jan;170(1):117–125. doi: 10.1128/jb.170.1.117-125.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]



