Abstract
Three independent pathways of nuclear import have so far been identified in yeast, each mediated by cognate nuclear transport factors, or karyopherins. Here we have characterized a new pathway to the nucleus, mediated by Mtr10p, a protein first identified in a screen for strains defective in polyadenylated RNA export. Mtr10p is shown to be responsible for the nuclear import of the shuttling mRNA-binding protein Npl3p. A complex of Mtr10p and Npl3p was detected in cytosol, and deletion of Mtr10p was shown to lead to the mislocalization of nuclear Npl3p to the cytoplasm, correlating with a block in import. Mtr10p bound peptide repeat-containing nucleoporins and Ran, suggesting that this import pathway involves a docking step at the nuclear pore complex and is Ran dependent. This pathway of Npl3p import is distinct and does not appear to overlap with another known import pathway for an mRNA-binding protein. Thus, at least two parallel pathways function in the import of mRNA-binding proteins, suggesting the need for the coordination of these pathways.
Transport into and out of the nucleus occurs through the nuclear pore complex (NPC).1 The asymmetric distribution of macromolecules between the nucleus and cytoplasm is an essential component of cellular regulation and function. Although small molecules can diffuse through the NPC, most macromolecules are transported in an energy-dependent, regulated, and highly specific manner. The nuclear import of nuclear localization sequence (NLS)-bearing proteins has been extensively studied (for review see Corbett and Silver, 1997; Nigg, 1997). Proteins to be imported are recognized in the cytoplasm by virtue of their NLSs, which are bound by a soluble receptor protein, karyopherin α (Importin α; Kap60p in yeast; Adam and Adam, 1994; Görlich et al., 1994; Imamoto et al., 1995; Radu et al., 1995a ; Moroianu et al., 1995). Karyopherin α forms a heterodimer with another soluble protein, karyopherin β1 (Importin β; Kap95p in yeast; Chi et al., 1995; Radu et al., 1995a ; Enenkel et al., 1995; Görlich et al., 1995). This karyopherin α/β1 complex then docks the substrate at the NPC via direct interactions of karyopherin β1 and a subset of peptide repeat–containing NPC proteins (nucleoporins; Imamoto et al., 1995; Radu et al., 1995a ,b; Moroianu et al., 1995b ; Rexach and Blobel, 1995). Translocation through the NPC and subsequent disassembly of the complex is not completely understood but requires other soluble factors such as the small GTPase Ran (Melchior et al., 1993; Moore and Blobel, 1993) and its cofactor p10 (Moore and Blobel, 1994; Paschal and Gerace, 1995; Nehrbass and Blobel, 1996). As the nucleotide-bound state of Ran (GDP or GTP) is crucial for nuclear transport, proteins that regulate Ran such as the cytoplasmic RanGAP and RanBP1 and the nuclear Ran nucleotide exchange factor RCC1 are also essential components of the transport machinery (for review see Corbett and Silver, 1997).
In addition, two other pathways of nuclear import have also been described. Both are mediated by factors homologous to karyopherin β1. One is the Kap104p/karyopherin β2 (also called transportin) pathway, which imports a subset of mRNA-binding proteins (Aitchison et al., 1996; Pollard et al., 1996; Bonifaci et al., 1997; Fridell et al., 1997). The second pathway is the Kap123p pathway, via which ribosomal proteins are imported into the nucleus (Rout et al., 1997). Another karyopherin β1 homologue, Pse1p, has also been implicated in this pathway (Rout et al., 1997). In contrast to karyopherin β1, these karyopherin β1 homologues do not require karyopherin α and can interact directly with their substrates and dock at repeat-containing nucleoporins (Aitchison et al., 1996; Bonifaci et al., 1997; Rout et al., 1997). However, like the karyopherin α/β1 complex, these pathways also appear to require Ran (Bonifaci et al., 1997; Rout et al., 1997).
Sequence analysis revealed a super-family of karyopherin β homologues with members in yeast and higher eukaryotes, referred to here as β karyopherins. Several members of this family were shown to interact with Ran, and one member, human Crm1, was identified via its interaction with a repeat-containing nucleoporin (Fornerod et al., 1997a ; Görlich et al., 1997; Wolff et al., 1997). Homology to known members of the β karyopherin family suggested that these proteins may function as karyopherins, raising the possibility that nuclear transport may occur via many different pathways. Recently, two members of this family, Crm1 (in yeast and higher eukaryotes) and human CAS were shown to be karyopherins involved in nuclear export (also called exportins; Fornerod et al., 1997b ; Kutay et al., 1997b ; Stade et al., 1997). It is probable that these proteins function similarily to Kap104p and Kap123p, in that they bind directly to their substrates and to the NPC (Fornerod et al., 1997a ,b; Stade et al., 1997).
RNAs represent a major class of macromolecules exported from the nucleus. Maturation of mRNA is a prerequisite for its export into the cytoplasm. Before its export, mRNA is packaged and undergoes such modifications as splicing, polyadenylation, and capping (for review see Dreyfuss et al., 1993; Nakielny et al., 1997). All of these processes require the association of mRNA with specific proteins within the nucleus. mRNA has been shown to be complexed with many proteins, including the heterogeneous nuclear (hn) ribonucleoprotein particle family members (for review see Dreyfuss et al., 1993; Nakielny et al., 1997). These proteins are believed to function in all of the known steps of mRNA maturation and export, and some hnRNPs have been shown to accompany the mRNA into the cytoplasm (Visa et al., 1996; Kraemer and Blobel, 1997; Nakielny et al., 1997). Once in the cytoplasm, these hnRNPs rapidly return to the nucleus, where they are thought to be involved in further rounds of mRNA export (Piñol-Roma and Dreyfuss, 1992). This continuous shuttling between the nucleus and cytoplasm requires efficient import and reimport of these proteins. This import would then be a crucial step in the regulation of mRNA export.
The yeast MTR10 gene (for mRNA transport defective) was identified in a screen for mutants that accumulate polyadenylated RNA in the nucleus at 37°C, suggesting a role in mRNA export (Kadowaki et al., 1994). We show here that Mtr10p is a karyopherin that mediates a previously uncharacterized nuclear import pathway. We identify the mRNA-binding protein, Npl3p, as a cytosolic substrate for Mtr10p, and we show that deletion of Mtr10p causes mislocalization of Npl3p to the cytoplasm, probably due to a block in its import. The identification of a karyopherin and its substrate defines a new nuclear import pathway, and we show that this pathway is distinct and does not overlap with the previously described pathway for mRNA-binding protein import.
Materials and Methods
Yeast Strains and Media
All yeast strains were derived from DF5 (Finley et al., 1987); procedures for yeast manipulation were as described (Sherman et al., 1986). MTR10 was deleted by replacing the open reading frame with the HIS3-selectable marker in a diploid strain by integrative transformation (Aitchison et al., 1995). Heterozygous diploids were sporulated and tetrads dissected on YPD plates at 23°C to generate mtr10 haploid strains. The MTR10 gene was amplified by PCR from genomic DNA, cloned into pRS416 (Stratagene, La Jolla, CA), and transformed into an mtr10::HIS3/MTR10 heterozygous diploid. After sporulation and dissection this strain yielded four viable spores at 30 and 37°C. These strains were streaked on media containing 5-fluoro-orotic acid (FOA) and grown at 23 and 37°C. Diploid strains expressing the Mtr10p–protein A fusion protein (Mtr10-PrA) were constructed by integrative transformation of the coding sequence of four and a half IgG-binding repeats of protein A immediately upstream of the MTR10 stop codon as described (Aitchison et al., 1995). This resulted in chimeric Mtr10–PrA fusions under the control of the endogenous MTR10 promoter. Haploid strains were generated by sporulation and dissection.
Immunofluorescence Microscopy
After fixation in 3.7% formaldehyde for 15 min, immunofluorescence microscopy on yeast spheroplasts was done as previously described (Wente et al., 1992). Protein A tags were visualized using rabbit anti–mouse IgG (preadsorbed against formaldehyde-fixed wild-type yeast cells) followed by Cy3-conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Labs, West Grove, PA). Npl3p and Nab2p were visualized using the antibodies 1E4 and a Nab2p mouse polyclonal antiserum (Wilson et al., 1994; Aitchison et al., 1996). The SV-40 NLS-GFP was viewed directly in living cells (Shulga et al., 1996). DNA was visualized by 4′,6-diamidino-2-phenylindole (DAPI). All images were viewed under the 63× oil objective on an Axiophot microscope (Zeiss Inc., Thornwood, NY); images were collected with a video imaging system directly in Adobe Photoshop v3.
Protein Purification
Post-nuclear, post-ribosomal cytosol was prepared from the Mtr10-PrA strain as described (Aitchison et al., 1996). Using this method we can detect the majority of Mtr10-PrA in the crude cytosol fraction. Mtr10-PrA and associated proteins were immunoisolated by overnight incubation of cytosol with rabbit IgG Sepharose (Cappel, Durham, NC) as described (Aitchison et al., 1996). After washing, proteins were eluted from the Sepharose with a step gradient of MgCl2 and precipitated with methanol before analysis by SDS-PAGE. Coomassie staining bands were excised and prepared for analysis by MALDI-TOF mass spectrometry (MS), and one peptide was further analyzed by MS-MS as described (Gharahdaghi et al., 1996). Npl3p was identified by MS and confirmed by Western blotting. Procedures for Western blotting and subsequent detection by ECL were as described (Amersham, Arlington Heights, IL).
Expression Constructs and Blot Overlay Assay
An amino-terminally tagged Npl3p fusion protein was expressed under the control of the alcohol dehydrogenase (ADH) promoter from the plasmid pRS416. The coding sequence for the IgG-binding domain of protein A was cloned upstream of the ATG of the coding sequence of NPL3 in this vector and expressed in the appropriate yeast strain. Full length Mtr10p or a fragment encoding amino acids 1–500 were amplified by PCR from genomic DNA and subcloned into pGEX-5X1 (Pharmacia Fine Chemicals, Piscataway, NJ) for expression in bacteria, to create glutathione-S-transferase (GST)-fusion proteins. Expression and purification of the other bacterially expressed proteins have been described elsewhere (Kraemer et al., 1995; Rexach and Blobel, 1995). Bacterially expressed proteins were separated by SDS-PAGE and transferred to nitrocellulose. Overlay assays were performed essentially as described (Aitchison et al., 1996). Yeast cytosol from Mtr10-PrA or Kap95-PrA strains was diluted 1:5 and 1:10, respectively, and incubated on the blot overnight at 4°C. Alternatively, bacterial pellets from a 5-ml culture of induced bacteria were lysed by freeze thawing and sonication in 500 μl transport buffer (20 mM Hepes, pH 7.5, 110 mM KOAc, 2 mM MgCl2, 1 mM DTT) with 0.1% Tween-20. After dilution by 1:1 in transport buffer with 5% milk, lysates were incubated with the blot overnight. Bound proteins were detected either with rabbit IgG (Pr-A tags) or with anti-GST antibody followed by HRP-conjugated donkey anti–rabbit antibodies and ECL (GST tags).
Ran Overlay
Ran (Gsp1p) was expressed in bacteria and purified as described (Floer and Blobel, 1996). Proteins were separated by SDS-PAGE and blotted onto nitrocellulose. The blot was incubated in transport buffer with 0.5% BSA, 0.3% Tween-20 overnight, and then blocked in transport buffer with 0.1% Tween-20 and 200 μM GTP for 1 h. Gsp1p was loaded with α-[32P]GTP as described and added to the blocking solution for 1 h (Floer and Blobel, 1996). The blot was then washed 3 times in transport buffer with 0.1% Tween-20 and subjected to autoradiography.
Results
Mtr10p Shows Homology with Other Members of the β Karyopherin Family
Transport into and out of the nucleus is mediated by members of the karyopherin family. Recently a superfamily of genes with homology to karyopherin β1 or Kap95p has been noted. For reasons addressed below, we chose to focus on Mtr10p. The yeast MTR10 gene was identified in a screen for mutants that accumulate polyadenylated RNA in the nucleus at 37°C (Kadowaki et al., 1994). Although the function of MTR10 was unknown, its identification in this screen suggested that it may be involved in the maturation or transport of mRNA. Comparison of Mtr10p and Kap95p showed limited homology at the amino acid level (16% identity; Fig. 1). Comparison of Mtr10p with other members of this family suggested that it was most similar to Crm1p and Los1p (data not shown). However, the levels of homology detected with all members of the β karyopherin family were so similar that it was not possible to determine whether the differences had any functional significance. The homology was most striking in the amino terminus of members of this family (Fornerod et al., 1997b ; Görlich et al., 1997). These sequence similarities led us to determine if Mtr10p was a karyopherin and define which proteins it was responsible for transporting.
Figure 1.
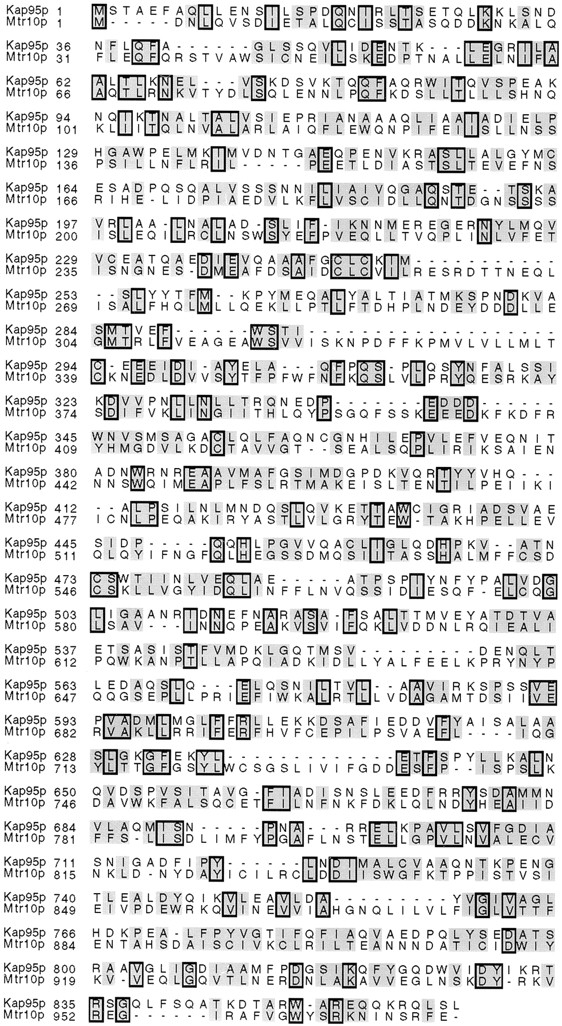
Mtr10p shares similarity with Kap95p. Comparison of amino acid sequences of Kap95p and Mtr10p. The proteins are aligned using CLUSTAL v.1.6 (DNAStar, Madison, WI). Identical amino acids are boxed; similar amino acids are shaded.
Mtr10p Is Essential at 37°C and Is Predominantly Cytoplasmic
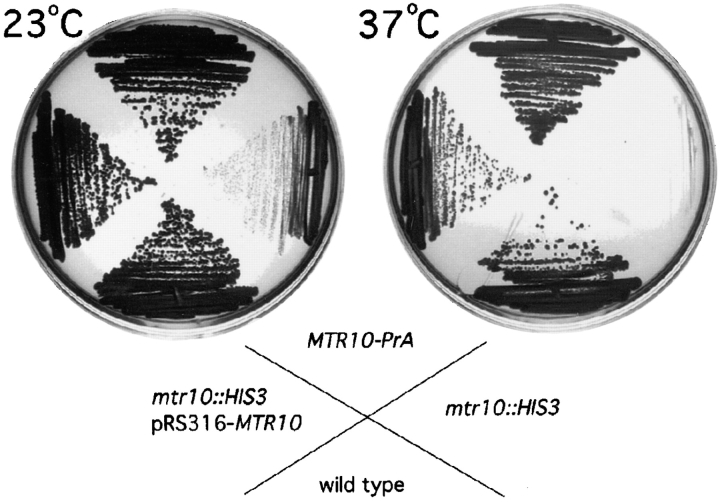
The MTR10 gene was disrupted in diploid yeast cells by replacing the coding sequence of one allele with the HIS3 gene. After sporulation and tetrad dissection, two large colonies and two very small colonies were visible after 5 to 6 d at room temperature. Only the small colonies grew on medium lacking histidine, and analysis of the genomic DNA revealed that the slow-growing colonies represented haploids with the disrupted copy of MTR10 (data not shown). Deletion of MTR10 led to extremely slow growth at 23°C and inviability at 37°C (Fig. 2). This phenotype was similar to that reported for the mtr10-1 mutant (Kadowaki et al., 1994). MTR10/mtr10::HIS3 heterozygous diploid strains were also sporulated and dissected in the presence of the MTR10 gene expressed on a low copy (CEN/URA3) plasmid. All four colonies were viable at 30 and 37°C. However, on media containing 5FOA, which selectively kills cells containing this plasmid, haploids deleted for MTR10 grew very slowly at 23°C and not at all at 37°C, presumably due to loss of the plasmid (data not shown).
Figure 2.
Deletion of MTR10 causes severe growth defects and temperature sensitivity. Wild-type, the MTR10-deleted strain (mtr10::HIS3), a strain with a protein A–tagged copy of MTR10 (Mtr10-PrA), and the MTR10-deleted strain complemented with the MTR10 gene on a plasmid (mtr10:HIS3, pRS316-MTR10) were streaked on rich medium at 23 and 37°C.
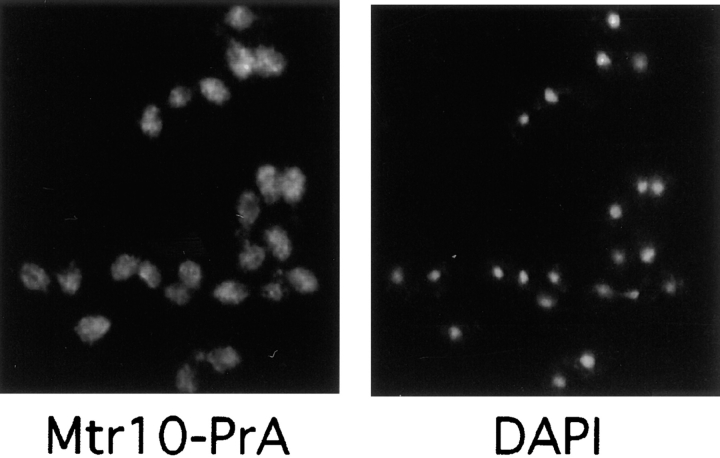
To determine the function of Mtr10p, we genomically tagged the gene with an in-frame carboxy-terminal fusion of the IgG-binding domains of protein A. Haploid strains expressing Mtr10-PrA were able to grow at 37°C (Fig. 2), demonstrating that the fusion protein was able to complement the endogenous gene. The tagged strains were used to determine the localization of Mtr10p by indirect immunofluorescence. Mtr10p was predominantly cytosolic when compared with DAPI-stained DNA, although some cells appeared to show some nuclear staining (Fig. 3). This localization was in agreement with cell fractionation data, where most of the Mtr10PrA was found in the cytosolic fraction (data not shown).
Figure 3.
Mtr10p is primarily localized to the cytoplasm. The localization of Mtr10-PrA was examined by immunofluorescent detection of the protein A tag; the coincident staining of DNA with DAPI is shown.
Mtr10p Interacts with Peptide Repeat–containing Nucleoporins
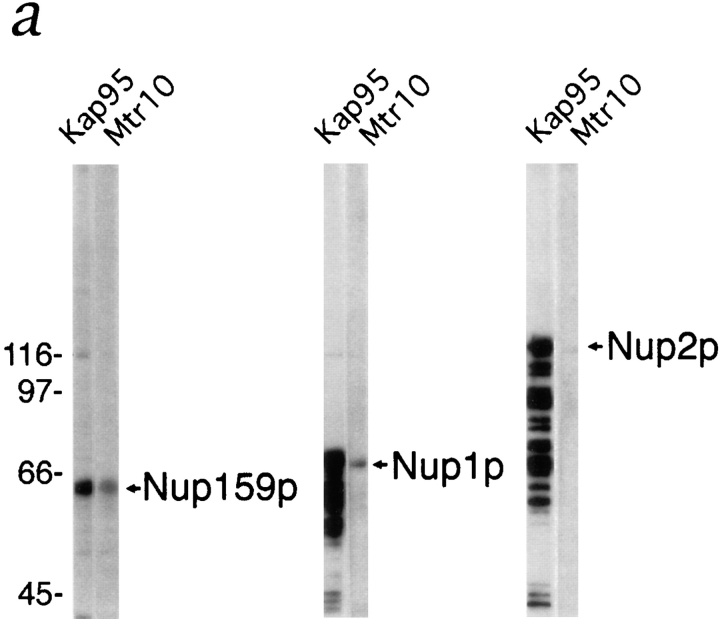
If Mtr10p is a β karyopherin it is probable that it also interacts with other components of the nuclear import machinery such as the NPC itself. Kap95p, Kap104p, and Kap123p have all been shown to interact with the NPC, and this interaction is mediated by direct interaction with several repeat-containing nucleoporins (Rexach and Blobel, 1995; Aitchison et al., 1996; Rout et al., 1997). The nucleoporin Nup2p, the repeat domain of Nup159p, and the repeat-containing domain of Nup1p were expressed in bacteria and the whole bacterial lysates were separated by SDS-PAGE and blotted onto nitrocellulose. Overlay assays were performed on nitrocellulose strips using yeast cytosol from the Mtr10-PrA strain. As a control, duplicate strips were also probed with cytosol from a Kap95-PrA strain. Mtr10p did not appear to interact with Nup2p (Fig. 4 a), demonstrating that the Mtr10-PrA did not interact nonspecifically with abundant proteins present in the whole bacterial extract. However, an interaction was detected between Mtr10p and the repeat domains of Nup159p and Nup1p (Fig. 4 a). Kap95p gave a stronger signal with all the nucleoporins tested, including Nup2p (Fig. 4 a). The ladder of bands detected with the nucleoporins Nup1p and Nup2p suggests that these proteins have undergone some proteolytic degradation and the Kap95p is able to recognize the proteolytic fragments. These results suggested that Kap95p and Mtr10p may interact with overlapping components of the NPC, however, it would appear that their nucleoporin binding specificities are not identical. It is likely that there may be significant competition for binding of the NPC proteins, as the yeast cytosol used in this assay contains other proteins such as other β karyopherins that are able to bind NPC proteins. Therefore, further studies will be necessary to determine and compare the affinities of members of the β karyopherin family for components of the NPC.
Figure 4.
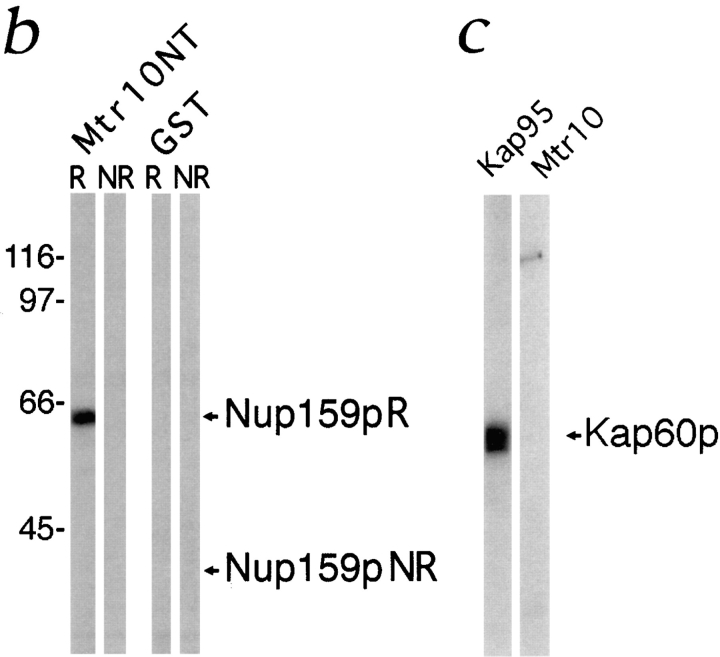
Mtr10p binds to a subset of repeat-containing nucleoporins. (a) Blot overlay assays were used to show binding of Mtr10-PrA to E. coli lysates containing repeat domain fragments of Nup159p (amino acids 441–876) and Nup1p (amino acids 432– 816) but not to bacterially expressed purified Nup2p. Blots were incubated with whole yeast cytosol from protein A–tagged strains; bound proteins were detected via the protein A moiety. Multiple bands detected with Kap95-PrA are due to its ability to bind strongly to proteolytic degradation fragments of the nucleoporin. Molecular masses in kD are shown. (b) The repeat domain (R; amino acids 441–876) and a nonrepeat-containing domain (NR; amino acids 176–440) of Nup159p were expressed in bacteria; the bacterial lysates were blotted as before. Overlay assays were carried out using a bacterial lysate containing GST-MTR10NT or unfused GST. Bound proteins were detected with a GST antiserum. Arrows indicate the position of Nup159N or Nup159NR on the blot. Molecular masses in kD are shown. (c) Kap95-PrA but not Mtr10-PrA interacts with bacterially expressed, purified Kap60p by overlay blot assay.
Using whole cytosol has the advantages of providing any cofactors necessary for the interaction. However, to determine whether the interaction between Mtr10p and the nucleoporins was direct, bacterially expressed Mtr10p was used. Full length Mtr10p (GST-Mtr10) and the amino-terminal half of the protein (GST-Mtr10NT) were expressed in bacteria as GST fusions. The GST-Mtr10NT was expressed at much higher levels, probably due to its smaller size and was therefore used for the following experiments. GST-Mtr10NT was tested by blot overlay for binding to the peptide repeat–containing domain of Nup159p (Kraemer et al., 1995). GST-Mtr10NT interacted directly with the repeat domain of Nup159p, and unfused GST-containing bacterial extracts showed no interaction with this domain (Fig. 4 b). Neither GST-Mtr10NT nor GST alone interacted with a nonrepeat-containing domain of Nup159p (Fig. 4 b). These results show that the domain of Mtr10p responsible for NPC binding resided in the amino terminus of the protein.
Kap95p, which interacts indirectly with its substrate, functions by forming a heterodimer with the NLS-binding protein Kap60p (Enenkel et al., 1995). Binding of Mtr10p to purified Kap60p was also tested by blot overlay. Unlike Kap95p, Mtr10p did not appear to interact with Kap60p (Fig. 4 c) and therefore probably interacts directly with its substrate, as has been shown for Kap104p and Kap123p (Aitchison et al., 1996; Rout et al., 1997). We also tested by blot overlay whether Mtr10p could form a heterodimer with Kap95p, however, we were unable to show an interaction (data not shown).
Mtr10p Interacts with Ran
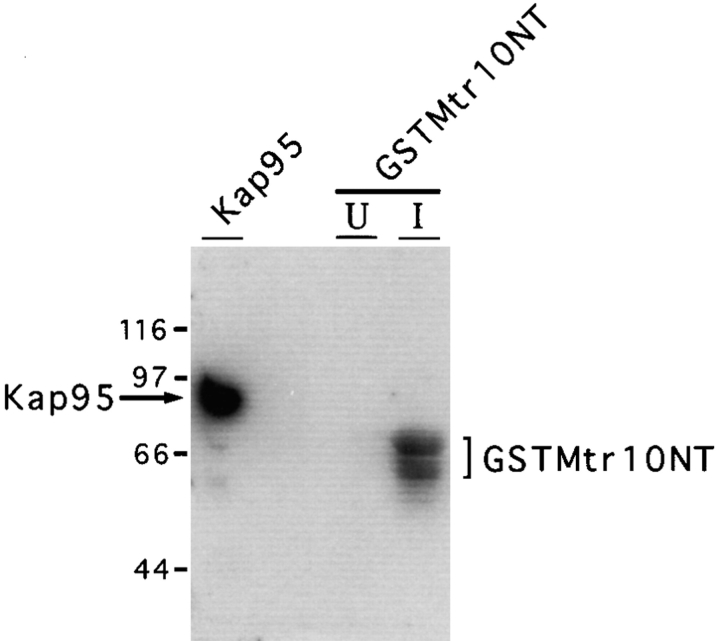
The small GTPase Ran (or Gsp1p in yeast) has been shown to be an essential cofactor for the nuclear import of NLS-bearing substrates and has been shown to directly bind Kap95p (Melchior et al., 1993; Moore and Blobel, 1993; Rexach and Blobel, 1995; Floer and Blobel, 1996). Several members of the family of β karyopherins have also been shown to bind Gsp1p (Rout et al., 1997; Görlich et al., 1997). We therefore determined whether Mtr10p was able to bind Gsp1p. Extracts prepared from bacteria expressing GST-Mtr10NT or purified Kap95p, were tested by blot overlay with purified Gsp1p, loaded with α[32P]GTP. It was not possible to test lysates expressing full length GST-Mtr10 due to the low levels of expression obtained with this fusion protein. As shown in Fig. 5, strong binding of Gsp1p-GTP was detected with lysates induced for GST-Mtr10NT expression. In contrast, no binding of Gsp1p-GTP to uninduced bacterial lysate was observed. As a positive control, the Gsp1p-GTP overlay was also performed on purified Kap95p (Fig. 5). Therefore, Mtr10p interacts with Gsp1p, and this interaction can be mediated by the amino terminus of Mtr10p.
Figure 5.
Mtr10p binds to Gsp1p. Bacterial lysates either induced (I) or uninduced (U) for GST-Mtr10NT, and bacterially expressed Kap95p were separated by SDS-PAGE and blotted. Blots were probed with Gsp1p-α[32P]GTP in an overlay assay, washed, and subjected to autoradiography. Bracket represents position of GST-Mtr10NT; additional bands are most likely degradation products of this protein. Molecular masses in kD are shown.
Mtr10p Interacts with a Cytosolic Pool of Npl3p
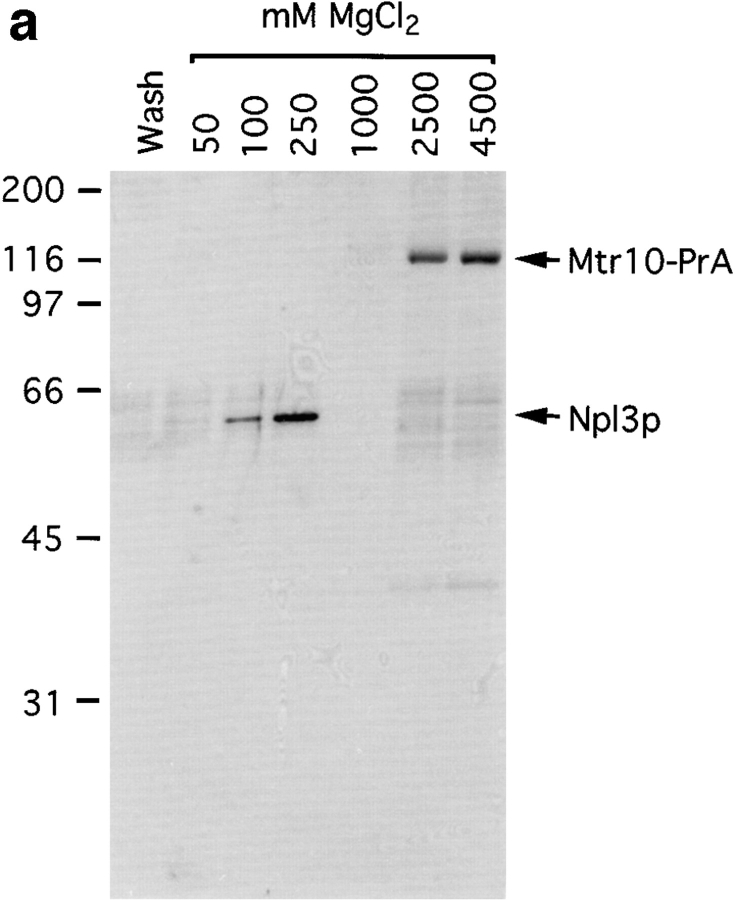
The homology with known karyopherins, together with the NPC and Ran binding, suggested that Mtr10p is a karyopherin. To determine its role in nuclear transport we attempted to identify its cognate transport substrates. Due to its abundance in the cytoplasm, Mtr10-PrA was isolated by affinity chromatography from post-nuclear, post-ribosomal cytosol prepared from the haploid Mtr10-PrA strain. Using this method it should be possible to also copurify interacting proteins from the cytosol (Aitchison et al., 1996; Rout et al., 1997). The cytosol from the protein A–tagged strain was incubated with IgG Sepharose, and interacting proteins were eluted with a step gradient ranging from 0.05 to 4.5 M MgCl2. The eluted fractions were analyzed by Coomassie blue staining of SDS-PAGE gels. Mtr10-PrA, which, by virtue of its protein A tag, binds IgG with high affinity, was eluted at high concentrations of MgCl2 (2.5–4.5 M; Fig. 6 a). A protein of ∼60-kD eluted earlier in the gradient (0.1–0.25 M; Fig. 6 a). The protein present in this band was analyzed by mass spectrometry, which suggested that it was Npl3p. The 0.25 M eluate fraction was immunoblotted with an antibody that specifically recognizes Npl3p, confirming the identity of the protein as Npl3p (Fig. 6 b). The amounts of Mtr10p and Npl3p purified by this method appeared to be roughly similar by Coomassie blue staining, suggesting that Mtr10p binds directly to cytosolic Npl3p and that Npl3p is likely a major substrate of Mtr10p.
Figure 6.
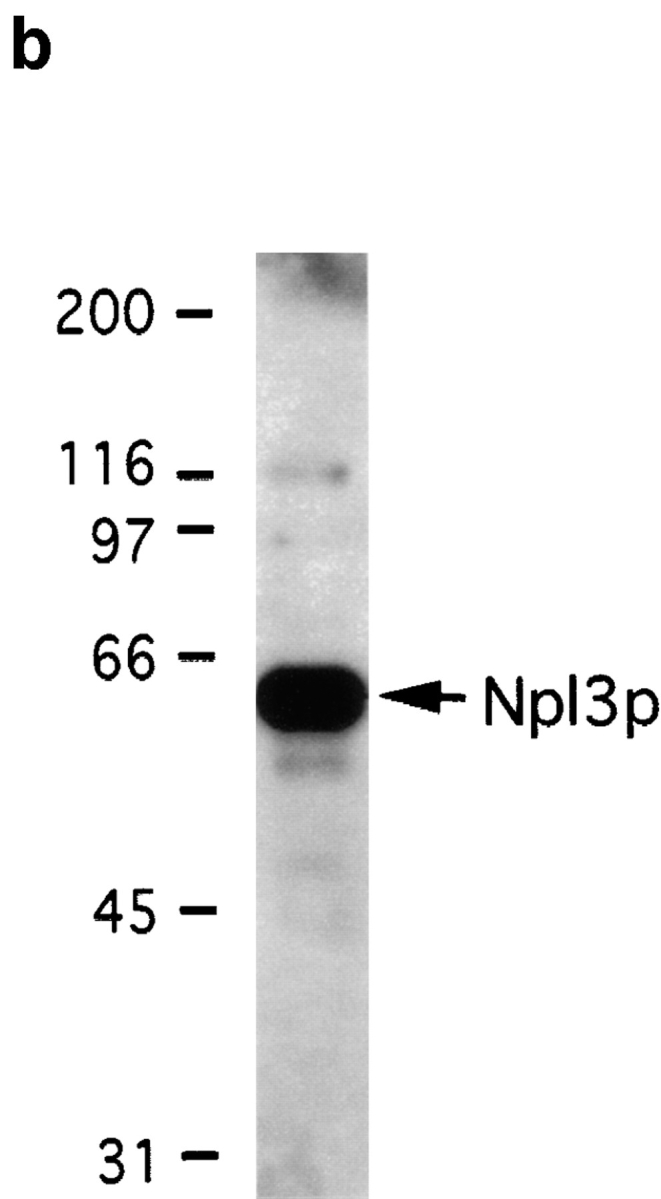
Immuno-isolation of a complex containing Mtr10-PrA and the mRNA-binding protein Npl3p. (a) Cytosol prepared from Mtr10-PrA cells was incubated with IgG sepharose; the Mtr10-PrA and complexed material were eluted in a MgCl2 step gradient. Fractions were analyzed by SDS-PAGE and Coomassie blue staining. The final wash fraction was also loaded (wash); numbers above lanes indicate molarity of MgCl2. The indicated band containing Npl3p was excised and analyzed by mass spectrometry. (b) The 250 mM eluate from a was immunoblotted with an antibody specific for Npl3p. Numbers to the left of both figures indicate molecular mass in kD.
Npl3p Is Not Imported into the Nucleus in the Absence of Mtr10p
As Npl3p was a candidate substrate for Mtr10p, we determined in vivo, by immunofluorescence microscopy, whether deletion of Mtr10p affected the localization of Npl3p. Although Npl3p shuttles between the nucleus and cytoplasm, using an anti-Npl3p monoclonal antibody, it appeared primarily nuclear and colocalized with DAPI-stained DNA (Fig. 7 a). However, at 23°C, in strains that were deleted for mtr10, Npl3p was mislocalized to the cytoplasm in all cells (Fig. 7 b). These results suggested that nuclear import of Npl3p did not occur in the absence of Mtr10p. After a shift to 37°C for 3 h, the mislocalization appeared, if anything, more dramatic (Fig. 7 b).
Figure 7.
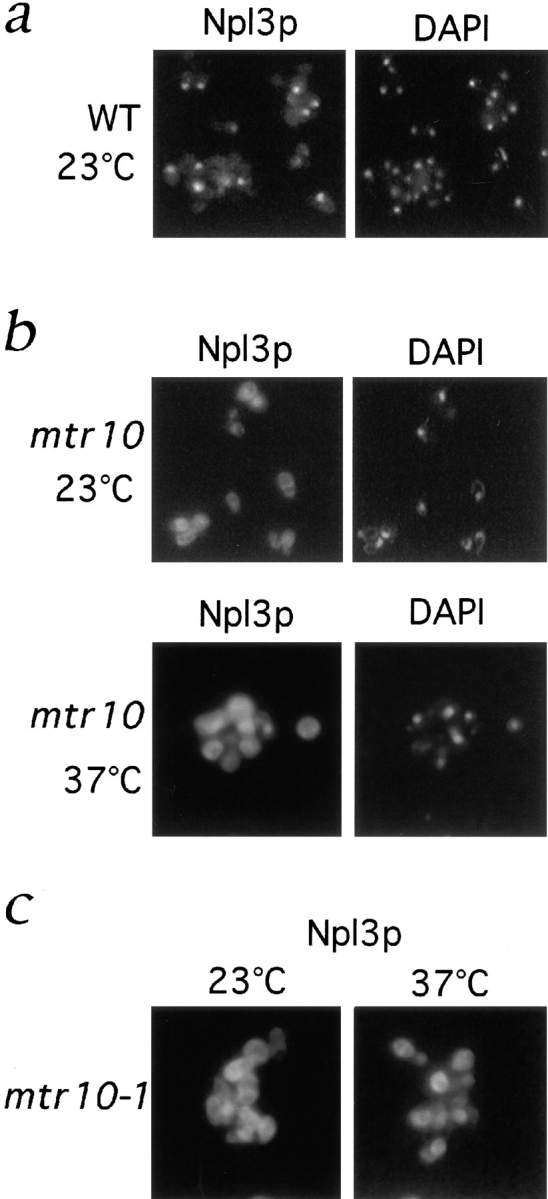
Npl3p is mislocalized to the cytoplasm in the absence of Mtr10p. (a) Immunofluorescent localization of Npl3p in wild-type (WT) cells colocalizes with the nucleus as visualized by DAPI-stained DNA. (b) Localization of Npl3p in the mtr10 deletion strain grown at 23 and 37°C shows that Npl3p is mislocalized to the cytoplasm when compared with the coincident DAPI staining. (c) Localization of Npl3p in the mtr10-1 mutant strain grown at 23 and 37°C shows that Npl3p is mislocalized to the cytoplasm when compared with the coincident DAPI staining.
The mtr10-1 mutant is temperature sensitive for growth and mRNA export defects (Kadowaki et al., 1994). Growth at 37°C leads to a nuclear accumulation of polyadenylated RNA followed by cell death. mtr10-1 mutant cells were grown at 23°C and shifted to 37°C for 3 h. Npl3p was mislocalized to the cytoplasm in cells grown at both 23 and 37°C (Fig. 7 c). As Npl3p directly binds mRNA and mtr10 mutants also have a defect in mRNA export, it was possible that Mtr10p may be responsible for retention of Npl3p in some intranuclear site and that we were just observing passive diffusion out of the nucleus. Therefore, we protein A tagged Npl3p, which increased its calculated molecular mass to >70 kD. This size would be predicted to be well above the cut-off point for passive diffusion through the NPC. The protein A–tagged Npl3p appeared exclusively nuclear in wild-type cells, however in the mtr10 deletion strain it was observed in the cytoplasm (data not shown). This suggests that the mislocalization is due to a specific import defect.
Mtr10p Imports Npl3p by a Distinct and Nonoverlapping Import Pathway
To determine how general the effect on nuclear import was, we looked at the localization of an NLS-green fluorescent protein (GFP) reporter, presumably imported by the Kap60p/Kap95p complex. Wild-type, mtr10 deletion cells, and mtr10-1 cells were transformed with this reporter, and the GFP was observed directly in living cells. In wild-type, mtr10 deletion cells, and mtr10-1 cells (data not shown) the NLS reporter was exclusively nuclear (Fig. 8 a). This suggests that the Npl3p/Mtr10p import pathway is independent of the NLS/Kap60p/Kap95p pathway.
Figure 8.
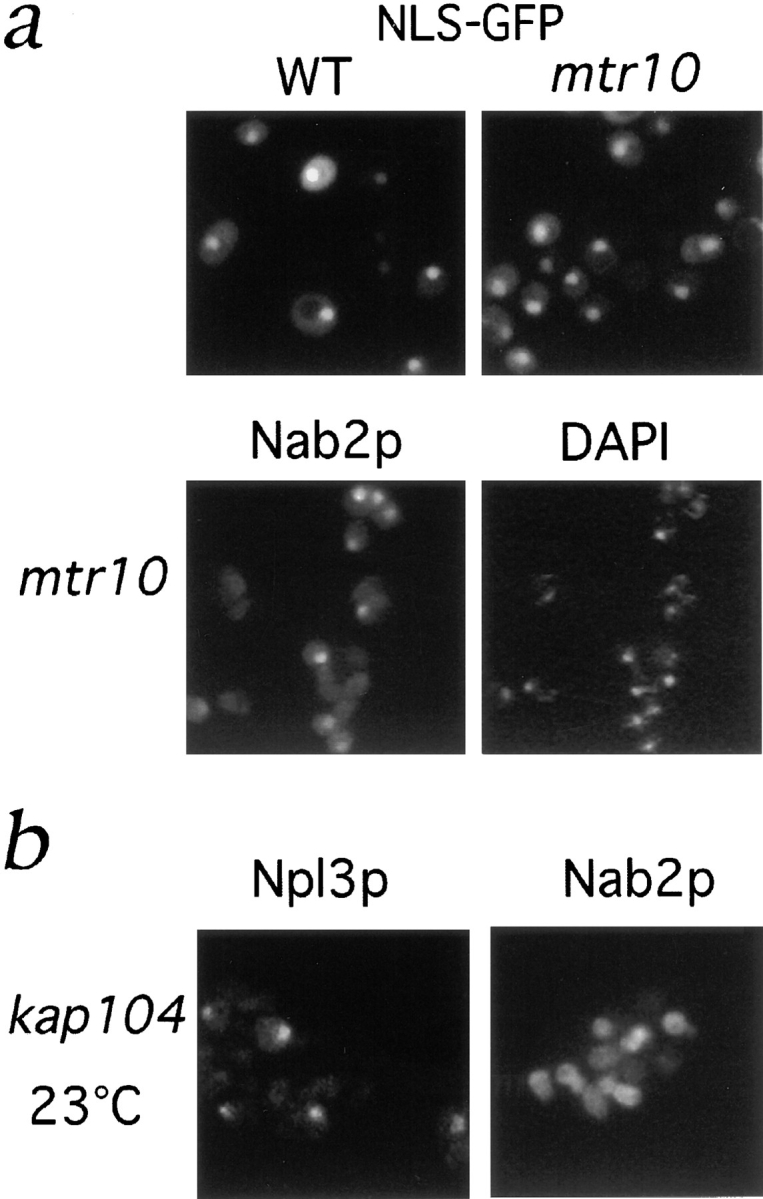
The Npl3p import pathway does not overlap with other import pathways. (a) Localization of an SV-40 NLS-GFP reporter was examined in living WT and mtr10 deletion cells and appeared nuclear in both cases. (b) Immunofluorescent localization of Nab2p in the mtr10 deletion strain shows correct nuclear localization. (c) Localization of Npl3p and Nab2p in a kap104:: HIS3 deletion strain at 23°C shows only mislocalization of Nab2p.
A nuclear import pathway mediated by Kap104p in yeast has been shown to be responsible for the import of another shuttling mRNA-binding protein, Nab2p (Aitchison et al., 1996). Using a Nab2p polyclonal antiserum, it was possible to show that, in contrast to Npl3p, Nab2p appeared to be correctly localized to the nucleus in the mtr10 deletion strain (Fig. 8 b) and the mtr10-1 strain (data not shown). Conversely, in strains lacking Kap104p, Nab2p but not Npl3p was mislocalized to the cytoplasm (Fig. 8 c; Aitchison et al., 1996). This shows that the deletion of Mtr10p also does not affect the Kap104p import pathway, demonstrating that the Mtr10p/Npl3p import pathway is a new pathway of nuclear import. It also shows that at least two mRNA-binding proteins, Npl3p and Nab2p, are transported into the nucleus by distinct and nonoverlapping pathways.
Discussion
Here we have identified a new pathway of nuclear protein import. Mtr10p is shown to be β karyopherin and Npl3p its primary substrate. Thus this represents a new import pathway for mRNA-binding proteins. Npl3p was isolated as a substrate by virtue of its direct interaction with Mtr10p in cytosol. In the absence of Mtr10p, we can show that Npl3p is excluded from the nucleus. This suggests that Mtr10p binds Npl3p in the cytoplasm and transports it to the nucleus.
In common with other import pathways, the Mtr10p-mediated import pathway of Npl3p probably involves a docking step at repeat-containing nucleoporins (Rexach and Blobel, 1995; Aitchison et al., 1996; Bonifaci et al., 1997; Rout et al., 1997). In support of this we observed direct binding of Mtr10p to repeat-containing nucleoporins. Mtr10p also bound Gsp1p, the yeast Ran; thus the Mtr10p import pathway, like other nuclear import pathways, probably requires Ran to function. Interestingly, Npl3p has been used as a reporter for many nuclear import studies. The nuclear localization of Npl3p has previously been shown to be affected by mutants of Gsp1p, as well as mutants of Rna1p, the Gsp1p GTPase activating protein, and Rcc1p, the Gsp1p nucleotide exchange factor (Corbett et al., 1995; Koepp et al., 1996; Wong et al., 1997). This provides further evidence that the import pathway of Npl3p is Ran dependent. We have mapped the Ran and NPC-binding site to the amino-terminal half of Mtr10p. This correlates with the position of these binding determinants in karyopherin β1 and Pse1p (Moroianu et al., 1996; Görlich et al., 1997; Kutay et al., 1997a ). The similarity between β karyopherin family members is most apparent in the amino-terminal portion of the proteins, and it is likely that this domain mediates their common functions. It is possible that more divergent domains of these proteins will be shown to mediate their substrate specificity. We have shown that the Mtr10p import pathway is independent of Kap60p and Kap95p. Corroboration of this is provided by previously published experiments, where kap60 and kap95 mutant strains showed correct localization of Npl3p but mislocalized NLS reporters (Loeb et al., 1995; Koepp et al., 1996).
Npl3p appears to be the major import substrate of Mtr10p, but additional substrates for this karyopherin may yet be found. The fact that only Npl3p was isolated with Mtr10p from the cytosol, in approximately stoichiometric amounts, strongly suggests that these proteins directly interact. It is not clear whether Mtr10p directly participates in the export of proteins or mRNA. At present we do not know if any karyopherins function in both import and export, and this represents an important future question in the field of nuclear transport. By purifying Mtr10p from cytosol, it is probable that only import substrates would be isolated. This is consistent with studies that show the NLS–karyopherin complex is stable in the presence of Ran-GDP but dissociated by Ran-GTP (Rexach and Blobel, 1995; Floer and Blobel, 1996; Görlich et al., 1996). Conversely, export substrates are believed to bind their karyopherins only in the presence of Ran-GTP, and Ran in the cytoplasm is thought to be predominantly in the GDP-bound form (Fornerod et al., 1997b ; Kutay et al., 1997b ).
Both Mtr10p and Npl3p have been identified in screens for mutants that accumulate polyadenylated RNA in the nucleus, leading to the suggestion that they are directly involved in mRNA export (Kadowaki et al., 1994; Singleton et al., 1995). However, screens for nuclear accumulation of polyadenylated RNA have identified a wide range of mutants (for review see Schneiter et al., 1995). Mutations have been isolated in genes encoding for a heat shock protein, an RNAase, Gsp1p and its exchange factor and GTPase activating protein, at least nine nucleoporins, Npl3p, a subunit of RNA pol I, and a lipid biosynthetic enzyme (Schneiter et al., 1995, 1996). Thus this phenotype may not reflect a direct role in mRNA export and only the identification of specific export substrates will define Mtr10p as a karyopherin involved in nuclear export. That Mtr10p functions as an import factor for the mRNA-binding protein, Npl3p, has been shown here. Therefore, it is also possible that the mRNA export phenotype observed in mtr10-1 mutant results from the mislocalization of Npl3p.
The mRNA-binding protein Npl3p was identified in several screens, such as those for mutants defective in nuclear protein localization and mutants that accumulated polyadenylated RNA in the nucleus (Bossie et al., 1992; Kadowaki et al., 1994; Singleton et al., 1995). Npl3p has also been identified by direct crosslinking to RNA and by its similarity to the glycine- and arginine-rich domain of the nucleolar protein, Nop1p (Russell and Tollervey, 1992; Wilson et al., 1994). Deletion of NPL3 leads to inviability or very severe growth defects, depending on the yeast strain investigated. Npl3p shuttles in and out of the nucleus and has structural similarities to other RNA-binding proteins (Russell and Tollervey, 1992; Flach et al., 1994; Henry et al., 1996; Lee et al., 1996). These similarities include two central RNA recognition motifs flanked by a carboxy-terminal glycine-rich domain that contains a series of RGGY/F repeats (Wilson et al., 1994). The precise function of Npl3p is not known; however, it has striking homology to the SR type splicing factors, and many mutants of Npl3p lead to accumulation of polyadenylated RNA within the nucleus (Birney et al., 1993; Lee et al., 1996). It has been proposed that Npl3p forms a complex with mRNA and participates as a carrier for RNA export to the cytoplasm (Lee et al., 1996). Many temperature-sensitive mutants of Npl3p have been characterized (Lee et al., 1996). The majority of mutations analyzed were mapped to the RNA recognition motif domain of Npl3p (Lee et al., 1996). These mutations led to the mislocalization of Npl3p to the cytoplasm at the permissive temperature and accumulation of polyadenylated RNA in the nucleus at the nonpermissive temperature. However, Npl3p appeared to be correctly localized to the nucleus at the nonpermissive temperature (Lee et al., 1996). This led to the suggestion that as both mRNA and Npl3p are blocked in the nucleus at the nonpermissive temperature, the export of Npl3p and mRNA are tightly linked (Lee et al., 1996). It is possible that the mutations in the RRM domain affect the interaction of Npl3p with mRNA leading to a defect in the export of mRNA.
We show here that in the mtr10-1 mutant, Npl3p appears to be constitutively mislocalized to the cytoplasm at all temperatures. However, RNA export defects have only been reported in mtr10-1 at the nonpermissive temperature (Kadowaki et al., 1994; Singleton et al., 1995). Similarily, in another mutant strain npl3-27, the mutant Npl3p protein is also mislocalized at all temperatures, although this strain does not have an mRNA export defect (Lee et al., 1996). This mutant has been mapped to the extreme carboxy terminus of Npl3p, the domain also reponsible for conferring nuclear localization on this protein (Flach et al., 1994). It remains to be determined whether the carboxy terminus of Npl3p contains the recognition sequence for Mtr10p and whether the npl3-27 mutant protein is mislocalized because it cannot interact with Mtr10p. These results also suggest that the RNA export defect observed in the mtr10-1 strain may be independent of the localization of Npl3p. However, further experiments are needed to determine that a small amount of Npl3p, not detected by immunofluorecent localization, has not entered the nucleus by another pathway.
The mRNA-binding protein Nab2p, like Npl3p, can be readily UV crosslinked to RNA (Anderson et al., 1993). The precise function of Nab2p is not known; however, it too is believed to participate in mRNA export (Anderson et al., 1993). The import pathway for Nab2p (and Hrp1p) has been identified and is mediated by the β karyopherin Kap104p (Aitchison et al., 1996). This protein is homologous to karyopherin β2, which has been shown to import hnRNP A1 (Pollard et al., 1996; Bonifaci et al., 1997; Cullen et al., 1997). The specific NLS-type sequence recognized by Kap104p has not been determined. However, due to their different substrate specificities, it is probable that the cognate NLS-type sequence for Mtr10p would be different from that recognized by Kap104p. We show here for the first time that two pathways of import for mRNA-binding proteins are distinct and nonoverlapping. A novel NLS-type sequence has also been mapped for the hnRNP K protein in Xenopus (Michael et al., 1997). This sequence appears to confer import by a pathway independent of the karyopherin α/β1 complex or karyopherin β2, suggesting it is a substrate for what may be a new pathway (Michael et al., 1997). However, the corresponding karyopherin has not yet been identified. Some hnRNPs also appear to contain classical NLS sequences, although it has not been shown whether they are transported via karyopherin α/β1, representing an additional pathway of hnRNP import (Michael et al., 1997). Thus, it is possible that several distinct pathways will also function in the import of mRNA-binding proteins in higher eukaryotes.
Eukaryotic cells have to continuously export mRNA. To facilitate the processing and export of this mRNA, it is necessary to have efficient and rapid import and reimport of mRNA-binding proteins. We propose that Mtr10p imports Npl3p into the nucleus; Npl3p there participates in mRNA maturation and export. Npl3p would then be exported into the cytoplasm with the mRNA, where, rebinding by Mtr10p would lead to a further round of import of Npl3p. It has yet to be determined whether Mtr10p is involved in the export of mRNA directly or via Npl3p. Little is known about the regulation and coordination of mRNA maturation and export, but it is believed that the association with mRNA-binding proteins is essential for these processes. The existence of parallel pathways to import these proteins into the nucleus provides a possible mechanism for differential regulation of their functions.
In summary we have identified a new pathway of nuclear protein import. It will be important to define all of the pathways of nuclear protein import and export. Only a complete understanding of all these pathways of nuclear transport and their substrate specificities can show us how the spatial regulation of the localization of macromolecules between cellular compartments is achieved.
Acknowledgments
We thank Farzin Gharahdaghi of the Rockefeller University Protein/ DNA Technology Center for the MS analysis, Mike Rout for Kap95-PrA cytosol, John Aitchison for the kap104 strain and the Nab2p antiserum, Maurice Swanson for the 1E4 antibody, Alan Tartakoff for the mtr10-1 strain, and Monique Floer, Ulf Nehrbass, and Michael Rexach for bacterially expressed proteins and anti-GST antibody. We thank members of the Blobel lab for helpful discussion.
Abbreviations used in this paper
- ADH
alcohol dehydrogenase
- GFP
green fluorescent protein
- GST
glutathione-S-transferase
- hn
heterogeneous nuclear
- NPC
nuclear pore complex
Footnotes
J.S. Rosenblum was supported by a postdoctoral fellowship from the National Institutes of Health.
Address all correspondence to Günter Blobel, Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, New York, NY 10021. Tel.: (212) 327-8181. Fax: (212) 327-7880. E-mail: blobel @rockvax.rockefeller.edu
References
- Adam EJH, Adam S. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–555. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Anderson JT, Wilson SM, Datar KV, Swanson MS. NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin β2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie MA, DeHoratius C, Barcelo G, Silver P. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Adam SA. Sequence and characterization of cytoplasmic nuclear import factor p 97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Koepp DM, Schlenstedt G, Lee MS, Hopper AK, Silver PA. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins DA, Silver PA. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Blobel G. The nuclear transport factor karyopherin β binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur Mol Biol Organ) J. 1997a;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997b;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-β. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F, Kirchner M, Fernandez J, Mische SM. Peptide-mass profiles of polyvinylidene difluoride-bound proteins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in the presence of nonionic detergents. Anal Biochem. 1996;233:94–99. doi: 10.1006/abio.1996.0012. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills A, Hartmann E, Laskey RA. Distinct functions for the two importin subunits is nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M, Borland CZ, Bossie M, Silver PA. Potential RNA binding proteins in Saccharomyces cerevisiaeidentified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-tageting complex in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1995b;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiaemRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Wong DH, Corbett AH, Silver PA. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer DM, Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci USA. 1997;94:9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO (Eur Mol Biol Organ) J. 1997a;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997b;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Loeb JD, Schlenstedt G, Pellman D, Kornitzer D, Silver PA, Fink GR. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO (Eur Mol Biol Organ) J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin α and together with karyopherin β docks import substrate at the nuclear pore complex. Proc Natl Acad Sci USA. 1995a;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2bind nuclear localization signal and β interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995b;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. Nuclear protein import: Ran-GTP dissociates the karyopherin α/β heterodimer by displacing α from an overlapping binding site on β. Proc Natl Acad Sci USA. 1996;93:7059–7062. doi: 10.1073/pnas.93.14.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Fischer U, Michael WM, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995a;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995b;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Russell ID, Tollervey D. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J Cell Biol. 1992;119:737–747. doi: 10.1083/jcb.119.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Kadowaki T, Tartakoff AM. mRNA transport in yeast: time to reinvestigate the functions of the nucleolus. Mol Biol Cell. 1995;6:357–370. doi: 10.1091/mbc.6.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter RT, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Chen S, Hitomi M, Kumagai C, Tartakoff AM. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J Cell Sci. 1995;108:265–272. doi: 10.1242/jcs.108.1.265. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Bjorkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through nuclear pores and into polysomes. Cell. 1996;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Datar KV, Paddy MR, Swedlow JR, Swanson M. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. . J Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff B, Sanglier J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- Wong DH, Corbett AH, Kent HM, Stewart M, Silver PA. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]