Abstract
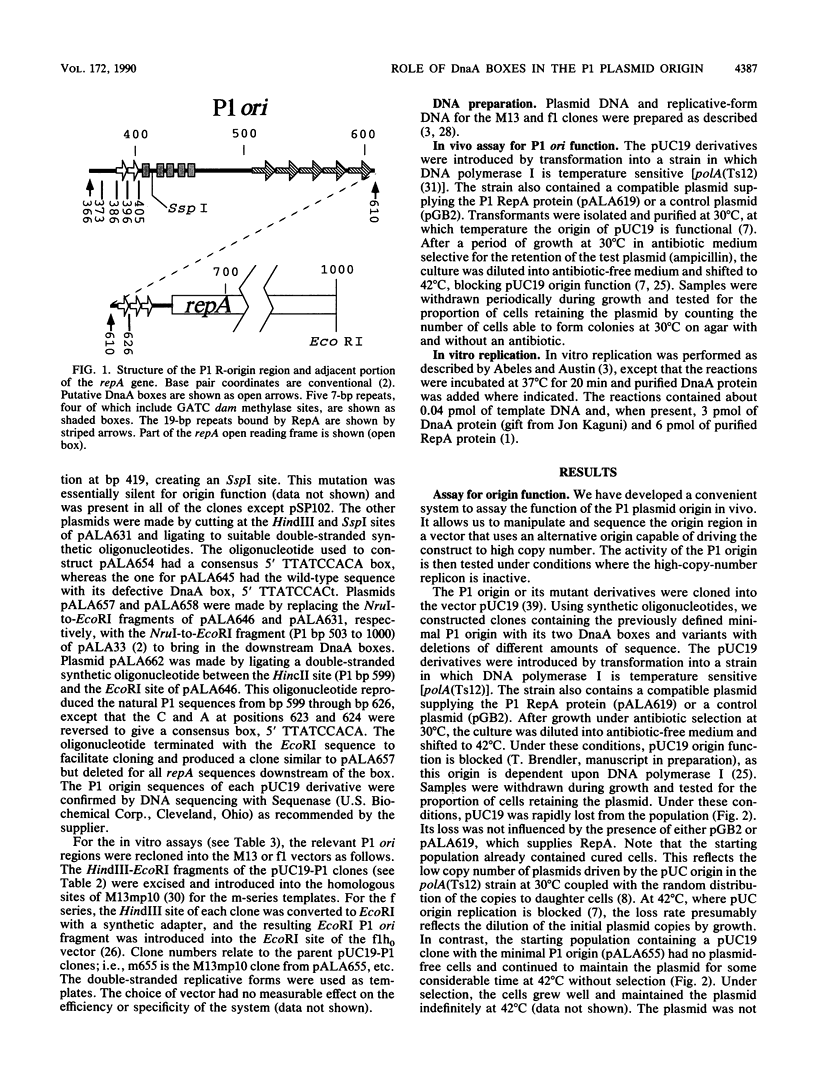
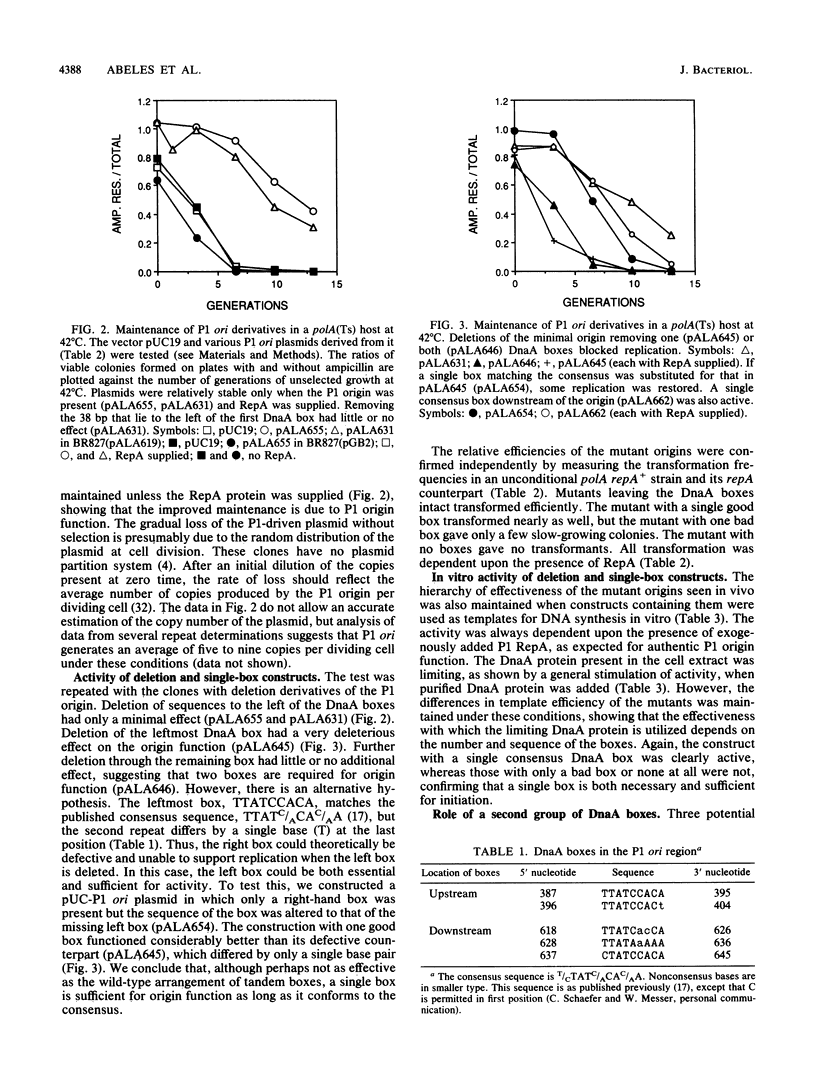
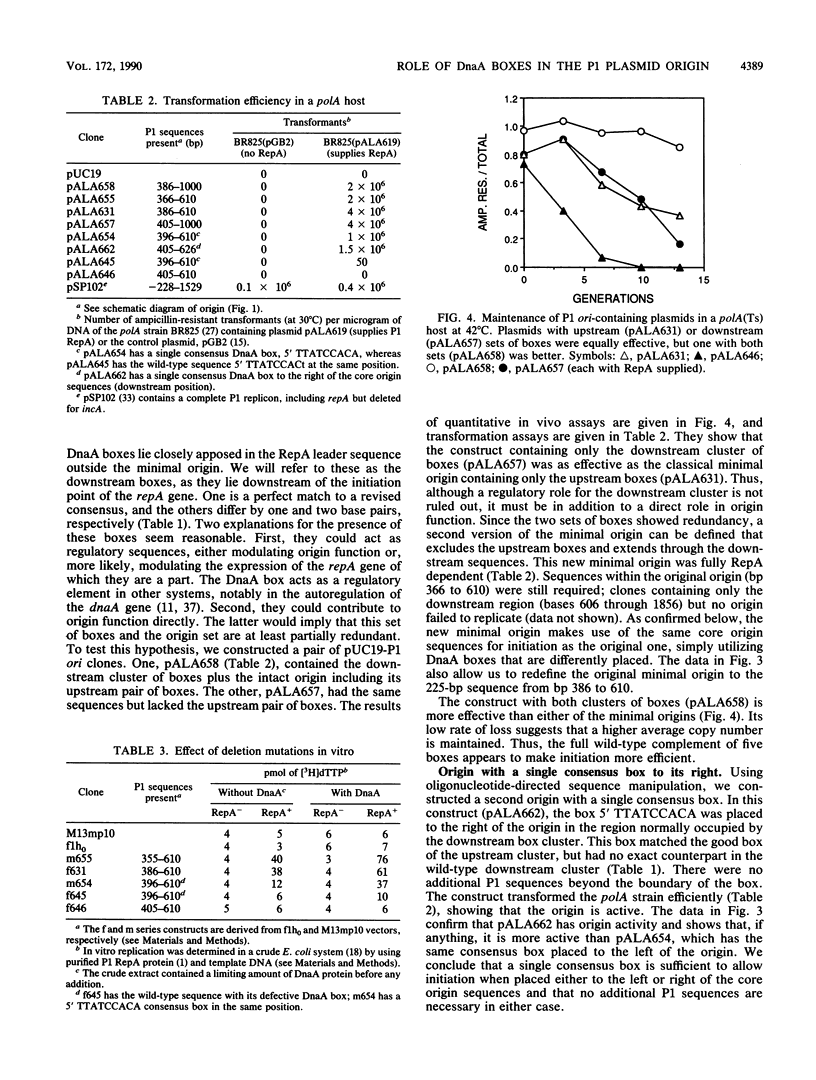
The P1 plasmid replication origin requires the host DnaA protein for function. Two DnaA-binding boxes lie in tandem within the previously defined minimal origin, constituting its left boundary. Three more boxes lie 200 base pairs to the right of these, in the leader region for the P1 repA gene. We show that either set alone is active for origin function. One of the two origin boxes is relatively inactive. Constructs with just one of the five boxes are active for specific origin function as long as the box conforms exactly to the published consensus. This single consensus box is functional when placed either to the left or right of the core origin sequences. The flexibility shown by this system suggests that the boxes play a role different from those in the host oriC origin, where the number and position of boxes are critical.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Austin S. J. P1 plasmid replication requires methylated DNA. EMBO J. 1987 Oct;6(10):3185–3189. doi: 10.1002/j.1460-2075.1987.tb02630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles A. L., Friedman S. A., Austin S. J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985 Sep 20;185(2):261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- Abeles A. L. P1 plasmid replication. Purification and DNA-binding activity of the replication protein RepA. J Biol Chem. 1986 Mar 15;261(8):3548–3555. [PubMed] [Google Scholar]
- Abeles A. L., Reaves L. D., Austin S. J. Protein-DNA interactions in regulation of P1 plasmid replication. J Bacteriol. 1989 Jan;171(1):43–52. doi: 10.1128/jb.171.1.43-52.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles A. L., Snyder K. M., Chattoraj D. K. P1 plasmid replication: replicon structure. J Mol Biol. 1984 Mar 5;173(3):307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- Alton N. K., Vapnek D. Nucleotide sequence analysis of the chloramphenicol resistance transposon Tn9. Nature. 1979 Dec 20;282(5741):864–869. doi: 10.1038/282864a0. [DOI] [PubMed] [Google Scholar]
- Austin S. J., Mural R. J., Chattoraj D. K., Abeles A. L. Trans- and cis-acting elements for the replication of P1 miniplasmids. J Mol Biol. 1985 May 25;183(2):195–202. doi: 10.1016/0022-2836(85)90212-8. [DOI] [PubMed] [Google Scholar]
- Austin S., Friedman S., Ludtke D. Partition functions of unit-copy plasmids can stabilize the maintenance of plasmid pBR322 at low copy number. J Bacteriol. 1986 Nov;168(2):1010–1013. doi: 10.1128/jb.168.2.1010-1013.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Braun R. E., O'Day K., Wright A. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell. 1985 Jan;40(1):159–169. doi: 10.1016/0092-8674(85)90319-8. [DOI] [PubMed] [Google Scholar]
- Chattoraj D. K., Snyder K. M., Abeles A. L. P1 plasmid replication: multiple functions of RepA protein at the origin. Proc Natl Acad Sci U S A. 1985 May;82(9):2588–2592. doi: 10.1073/pnas.82.9.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj D., Cordes K., Abeles A. Plasmid P1 replication: negative control by repeated DNA sequences. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6456–6460. doi: 10.1073/pnas.81.20.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney R. H., Scott J. R., Vapnek D. Integration of the plasmid prophages P1 and P7 into the chromosome of Escherichia coli. J Mol Biol. 1979 May 15;130(2):161–173. doi: 10.1016/0022-2836(79)90424-8. [DOI] [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Funnell B. E., Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984 Oct;38(3):889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- Fuller R. S., Kaguni J. M., Kornberg A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7370–7374. doi: 10.1073/pnas.78.12.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Hansen E. B., Yarmolinsky M. B. Host participation in plasmid maintenance: dependence upon dnaA of replicons derived from P1 and F. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4423–4427. doi: 10.1073/pnas.83.12.4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Kamio Y., Terawaki Y. Essential DNA sequence for the replication of Rts1. J Bacteriol. 1987 Mar;169(3):1153–1160. doi: 10.1128/jb.169.3.1153-1160.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Terawaki Y. Replication properties of mini-Rts1 derivatives deleted for DnaA boxes in the replication origin. Plasmid. 1989 May;21(3):242–246. doi: 10.1016/0147-619x(89)90048-6. [DOI] [PubMed] [Google Scholar]
- Kamio Y., Tabuchi A., Itoh Y., Katagiri H., Terawaki Y. Complete nucleotide sequence of mini-Rts1 and its copy mutant. J Bacteriol. 1984 Apr;158(1):307–312. doi: 10.1128/jb.158.1.307-312.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970 Dec 24;41(6):1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- Lahue R. S., Su S. S., Modrich P. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1482–1486. doi: 10.1073/pnas.84.6.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke D. N., Eichorn B. G., Austin S. J. Plasmid-partition functions of the P7 prophage. J Mol Biol. 1989 Oct 5;209(3):393–406. doi: 10.1016/0022-2836(89)90005-3. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Monk M., Kinross J. Conditional lethality of recA and recB derivatives of a strain of Escherichia coli K-12 with a temperature-sensitive deoxyribonucleic acid polymerase I. J Bacteriol. 1972 Mar;109(3):971–978. doi: 10.1128/jb.109.3.971-978.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Austin S. J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- Pal S. K., Mason R. J., Chattoraj D. K. P1 plasmid replication. Role of initiator titration in copy number control. J Mol Biol. 1986 Nov 20;192(2):275–285. doi: 10.1016/0022-2836(86)90364-5. [DOI] [PubMed] [Google Scholar]
- Prentki P., Chandler M., Caro L. Replication of prophage P1 during the cell cycle of Escherichia coli. Mol Gen Genet. 1977 Mar 28;152(1):71–76. doi: 10.1007/BF00264942. [DOI] [PubMed] [Google Scholar]
- Swack J. A., Pal S. K., Mason R. J., Abeles A. L., Chattoraj D. K. P1 plasmid replication: measurement of initiator protein concentration in vivo. J Bacteriol. 1987 Aug;169(8):3737–3742. doi: 10.1128/jb.169.8.3737-3742.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X. B., Womble D. D., Rownd R. H. DnaA protein is not essential for replication of IncFII plasmid NR1. J Bacteriol. 1989 Oct;171(10):5290–5295. doi: 10.1128/jb.171.10.5290-5295.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. P., Kaguni J. M. Transcriptional repression of the dnaA gene of Escherichia coli by dnaA protein. Mol Gen Genet. 1987 Oct;209(3):518–525. doi: 10.1007/BF00331158. [DOI] [PubMed] [Google Scholar]
- Wickner S. H., Chattoraj D. K. Replication of mini-P1 plasmid DNA in vitro requires two initiation proteins, encoded by the repA gene of phage P1 and the dnaA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3668–3672. doi: 10.1073/pnas.84.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yung B. Y., Crooke E., Kornberg A. Fate of the DnaA initiator protein in replication at the origin of the Escherichia coli chromosome in vitro. J Biol Chem. 1990 Jan 25;265(3):1282–1285. [PubMed] [Google Scholar]
- Yung B. Y., Kornberg A. The dnaA initiator protein binds separate domains in the replication origin of Escherichia coli. J Biol Chem. 1989 Apr 15;264(11):6146–6150. [PubMed] [Google Scholar]
- Zyskind J. W., Cleary J. M., Brusilow W. S., Harding N. E., Smith D. W. Chromosomal replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1164–1168. doi: 10.1073/pnas.80.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]


