Abstract
A major question in nuclear import concerns the identity of the nucleoporin(s) that interact with the nuclear localization sequences (NLS) receptor and its cargo as they traverse the nuclear pore. Ligand blotting and solution binding studies of isolated proteins have attempted to gain clues to the identities of these nucleoporins, but the studies have from necessity probed binding events far from an in vivo context. Here we have asked what binding events occur in the more physiological context of a Xenopus egg extract, which contains nuclear pore subcomplexes in an assembly competent state. We have then assessed our conclusions in the context of assembled nuclear pores themselves. We have used immunoprecipitation to identify physiologically relevant complexes of nucleoporins and importin subunits. In parallel, we have demonstrated that it is possible to obtain immunofluorescence localization of nucleoporins to subregions of the nuclear pore and its associated structures. By immunoprecipitation, we find the nucleoporin Nup153 and the pore-associated filament protein Tpr, previously shown to reside at distinct sites on the intranuclear side of assembled pores, are each in stable subcomplexes with importin α and β in Xenopus egg extracts. Importin subunits are not in stable complexes with nucleoporins Nup62, Nup93, Nup98, or Nup214/CAN, either in egg extracts or in extracts of assembled nuclear pores. In characterizing the Nup153 complex, we find that Nup153 can bind to a complete import complex containing importin α, β, and an NLS substrate, consistent with an involvement of this nucleoporin in a terminal step of nuclear import. Importin β binds directly to Nup153 and in vitro can do so at multiple sites in the Nup153 FXFG repeat region. Tpr, which has no FXFG repeats, binds to importin β and to importin α/β heterodimers, but only to those that do not carry an NLS substrate. That the complex of Tpr with importin β is fundamentally different from that of Nup153 is additionally demonstrated by the finding that recombinant β or β45–462 fragment freely exchanges with the endogenous importin β/Nup153 complex, but cannot displace endogenous importin β from a Tpr complex. However, the GTP analogue GMP-PNP is able to disassemble both Nup153– and Tpr–importin β complexes. Importantly, analysis of extracts of isolated nuclei indicates that Nup153– and Tpr–importin β complexes exist in assembled nuclear pores. Thus, Nup153 and Tpr are major physiological binding sites for importin β. Models for the roles of these interactions are discussed.
The import of proteins through the nuclear pore is an energy-driven process specific for proteins bearing nuclear localization sequences or NLSs1 (see Davis, 1995; Gorlich and Mattaj, 1996; Doye and Hurt, 1997; for review see Corbett and Silver, 1997). The canonical NLS is that of the SV-40 large T antigen, consisting of a single stretch of largely basic amino acids (aa; Dingwall and Laskey, 1991). A second type of NLS, more complex and found in proteins such as nucleoplasmin, is composed of two basic clusters separated by a 10-aa spacer. Still other sequences capable of conferring nuclear localization exist; these appear specific, but larger and less easily defined (Pollard et al., 1996; Michael et al., 1997).
Much progress has been made towards identifying the soluble factors required for transport of proteins through the nuclear pore. Using a digitonin-permeabilized cell assay, two proteins were found to comprise a soluble receptor that recognizes the NLS of the SV-40 T antigen and that of nucleoplasmin. Importin α (or karyopherin α) and importin β (also known as p97 or karyopherin β) bind to SV-40–type NLSs as a heterodimer and facilitate the import of an NLS-bearing protein into the nucleus (Adam and Adam, 1994; Gorlich et al., 1994, 1995a,b; Chi et al., 1995; Imamoto et al., 1995; Radu et al., 1995a ; Rexach and Blobel, 1995). Two additional proteins that do not interact directly with NLS sequences are also essential for protein import through the nuclear pore. These factors are the small GTPase Ran/TC4 and the protein NTF2 (Melchior et al., 1993; Moore and Blobel, 1993, 1994; Paschal and Gerace, 1995; Clarkson et al., 1996; Corbett and Silver, 1996; Rush et al., 1996; Wong et al., 1997).
The mechanism of import for classical NLS-bearing proteins appears to consist of an initial step where importin α recognizes and binds to the NLS of a future nuclear protein. Importin β, the second subunit of the receptor heterodimer, then mediates docking of this complex to sites on the nuclear pore. The transfer of the NLS-bearing protein into the nucleus requires GTP hydrolysis by Ran and is assisted by NTF2. Termination of import is thought to be accomplished by the binding of GTP-Ran to importin β, disrupting the importin α/β–NLS complex (for review see Gorlich, 1997 and Goldfarb, 1997). Importin β has been observed by immunoelectron microscopy to remain bound to the interior side of the pore after import, and is then believed to recycle directly to the cytoplasm (Gorlich et al., 1995b ). Immunoelectron microscopy of importin α reveals it as deeper within the nucleus after import, indicating release from the pore and slower kinetics for return of this factor to the cytoplasm. Importantly, although the docking sites for the importin α/β/NLS complex as it traverses the pore have been suggested from solution binding assays (Moroianu et al., 1995; Radu et al., 1995a,b; Rexach and Blobel, 1995; Percipalle et al., 1997), the actual sites for docking or for interaction as the receptor complex passes through the pore have not been identified in vivo or in isolated pores.
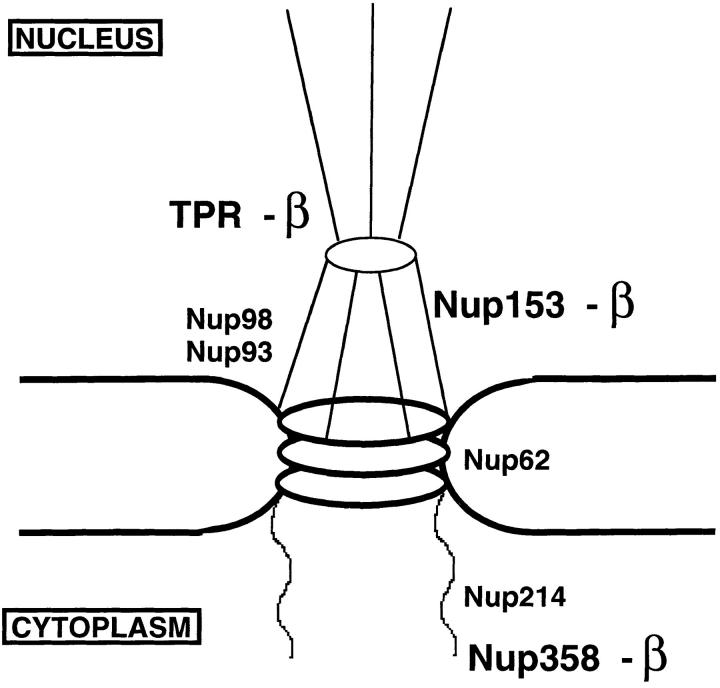
The architecture of the vertebrate nuclear pore is complex and includes many structures with which the NLS– receptor complex could potentially interact upon passage through the pore. The basic structure of the 120 million dalton pore consists of three stacked rings: a nuclear ring, a central ring of eight spokes, and a cytoplasmic ring (see Fig. 11 below; Hinshaw et al., 1992; Akey and Radermacher, 1993; for review see Pante and Aebi, 1993). Extending from the cytoplasmic ring are eight filaments that have been observed to bind NLS-complexed gold particles (Dworetzky et al., 1988; Richardson et al., 1988). The nucleoporins Nup358 and Nup214/CAN have been localized to these filaments (Kraemer et al., 1994; Fornerod, 1995; Wu et al., 1995; Yokoyama et al., 1995). A complex of nucleoporins, Nup62/58/54/45, lies near or possibly comprises the central transporter of the pore, through which import occurs (Finlay et al., 1991; Kita et al., 1993; Buss and Stewart, 1995; Guan et al., 1995). A nuclear basket composed of eight filaments connected to a small ring lies on the nucleoplasmic side of the pore and extends 500 Å into the nucleus (Ris, 1989, 1991; Goldberg and Allen, 1992; Pante and Aebi, 1993). Proteins of the basket might be involved in the terminal steps of nuclear import, the initial steps of nuclear export, or both. Several proteins have now been identified in vertebrates as, at least in part, components of the nuclear basket: Nup98, Nup93, its partner Nup205, and Nup153 (Sukegawa et al., 1993; Powers et al., 1995; Radu et al., 1995b ; Grandi et al., 1997). Indeed, when altered, both Nup98 and Nup153 lead to an inhibition of RNA export (Bastos et al., 1996; Powers et al., 1997). Lastly, Tpr, a pore-associated protein, is present on fibers that extend from the nuclear basket 2,000–3,500 Å into the nucleus (Cordes et al., 1997; Zimowska et al., 1997; see also Byrd et al., 1994). It is not known whether Tpr is the sole constituent or only one of the constituents of these fibers. Examination of the fibers at high resolution indicates that they form a regular network of branching hollow cables that lead to and from the nuclear pore (Ris, 1997).
Figure 11.
A model is shown summarizing the interactions of importin β with vertebrate nucleoporins under the in vivo–like conditions of the Xenopus egg extract and extracts of assembled nuclear pores. β indicates interaction with importin β in both the extract and in assembled pores. This occurred with nucleoporins Nup358, Nup153, and Tpr, although we were unable to assess Nup358 binding to β in rat liver nuclei due to a lack of specific anti-Nup358 antibody. A lack of β indicates no interaction of importin β with the designated nucleoporins was observed. This was found to be true for nucleoporins Nup214, Nup98, Nup93, and Nup62; Nup214 could only be assessed in Xenopus egg extracts.
To attempt to identify potential proteins of the pore that might interact with the import complex (i.e., importin α/β/ NLS-bearing protein), crude blot overlay studies were previously performed in which total nuclear envelope proteins were denatured, electrophoresed, and transferred to membrane. The membrane was then “probed” with cytosol plus NLS-HSA transport substrate (Radu et al., 1995a ). The proteins on the blot that bound NLS-HSA were identified by probing the blot with an anti-HSA antibody. In the absence of receptor-containing cytosol, the blot appeared black when probed in this manner. In the presence of cytosol, the nonspecific affinities of the probe were overcome and individual protein bands stood out and were identified as the FXFG repeat-containing nucleoporins Nup153, Nup214, and Nup358, as well as the GLFG repeat-containing nucleoporin Nup98. When a similar blot was “probed” with recombinant 35S-importin β, the same protein bands bound the probe (Moroianu et al., 1995). These ligand blot results, as well as solution binding experiments between recombinant proteins (Radu et al., 1995b ; Rexach and Blobel, 1995), were used to propose that all these proteins are in vivo binding sites for the import receptor and that nuclear import could occur by the sequential transfer from one site to the next. No indication of the involvement of the functionally important FXFG-containing nucleoporin p62 was obvious from these studies. It is not clear whether this method detects interactions that occur in vivo between each of the nucleoporins, or whether one such FXFG protein interaction occurs in vivo, but the artificial environment of the blot allows the other nucleoporins to mimic that one authentic interaction. Interaction of yeast importin β with multiple FG repeat nucleoporins on blots and by two hybrid analysis has also been observed, but in the yeast system it has been possible to assess proposed ligand blot interactions by genetic means (Iovine et al., 1995; see Iovine and Wente, 1997 and references therein). Interestingly, using carefully renatured and characterized recombinant proteins in a solution binding assay, Percipalle et al. (1997) found evidence for the binding of recombinant Nup62 to importin β. However, the importin β–binding domain of Nup62 was mapped not to the FXFG repeat-containing domain of Nup62, but to its coiled-coil domain. Although these studies indicate interactions of nucleoporins with importin β and also with importin α/β–NLS complexes in vitro, with the exception of certain of the yeast studies, they do not indicate which interactions are important or in fact even occur in vivo.
One would like to assess protein–protein interactions in a more in vivo–like context. Xenopus egg extracts, which are capable of assembling complete nuclei when DNA or chromatin is added, afford one such a context. In extract, the nuclei assemble quickly and are functional for nuclear import, DNA replication, and transcription (Lohka and Masui, 1983; Newmeyer et al., 1986; Newport and Spann, 1987; Newmeyer and Forbes, 1988; Dasso and Newport, 1990; Laskey and Leno, 1990; Cox and Laskey, 1991; Wolffe, 1993; Dasso et al., 1994; Powers et al., 1995; Ullman and Forbes, 1995). The extract contains large stores of subcomplexes of nuclear pores and other nuclear structures. Previous analysis of the Xenopus extract by immunoprecipitation and other affinity methods has revealed novel protein–protein interactions that have proved important in understanding multiple aspects of nuclear pore structure and function (Dabauvalle et al., 1990; Finlay and Forbes, 1990; Finlay, 1990, 1991; Macaulay et al., 1995; Saitoh et al., 1996). In addition, coimmunoprecipitation from mammalian nuclear envelope extracts has revealed exciting and unexpected partners for known nucleoporins, such as the recently discovered export receptor, exportin 1/CRM (Fornerod et al., 1997a,b), making this a valuable approach to understanding in vivo interactions within the nuclear pore.
Here we report the use of soluble Xenopus egg extracts to probe for molecular interactions between known nucleoporins and the importin α/β-NLS receptor. We find that the nuclear pore is disassembled into subcomplexes, a subset of which contain as a major component the NLS receptor. Specifically, the nucleoporin Nup153, a protein on the nuclear basket, and the pore-associated nuclear filament protein Tpr are in stable complexes with the NLS receptor, importin α/β. We also find that Nup358, a protein of the cytoplasmic filaments of the pore, is in a complex with importin α/β in the extract. Interestingly, other FXFG or GLFG repeat-containing pore proteins, specifically Nup62, Nup98, and Nup214/CAN, do not bind importin α/β in these more physiological conditions, nor does nucleoporin Nup93. The Nup153 and Tpr complexes were examined in detail and were both found to bind to the NLS receptor through the importin β subunit and to be disrupted by GMP-PNP. However, the two complexes are inherently different in their ability to bind to NLS substrate, in the domains used for interaction, and in their stability. We further find that the Nup153– and the Tpr–importin β complexes are present in extracts of assembled nuclear pores, strongly suggesting that Nup153 and Tpr are in vivo binding sites for the import factor, importin β.
Materials and Methods
Immunoscreening
To screen for Nup153 cDNA clones, an oligo dT–primed ZAP (Stratagene, La Jolla, CA) cDNA library made from Xenopus blastocyst RNA was plated at ∼25,000 plaque-forming units per 150-mm plate and screened using standard immunoscreening techniques. Filters were incubated first with the anti-XFXFG repeat monoclonal antibody, mAb 414 (ascites fluid, no. MMS-120R-500; BAbCO, Richmond, CA), diluted 1:3,000 with 5% milk in PBS, 0.2% Tween, and subsequently with goat anti–mouse conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA; diluted 1:5,000 in same buffer). Filters were then soaked with chemiluminescence reagent (Dupont-NEN, Boston, MA) and exposed to film for 10 min. Positive plaques (6 from 100,000 screened) were picked from plates and placed in SM buffer (10 mM NaCl, 12 mM MgSO4, 50 mM Tris, pH 7.5, 2% gelatin). Secondary and tertiary screens were performed in order to obtain pure phage clones. Inserts were excised from the phage as described in Stratagene's “zapping” protocol. These inserts were initially sequenced using T7 and T3 primers. Two clones showed significant homology to the rat and human Nup153 genes. One clone with an insert of 4.2 kb was analyzed further and extends from the equivalent of amino acid 377 in human Nup153 into the 3′ untranslated region (Fig. 1).
Figure 1.

Alignment of the Xenopus Nup153 partial cDNA with human Nup153. An alignment was performed using the human Nup153 aa sequence (aa 350–1475 are shown) and the aa sequence deduced from the partial Xenopus Nup153 cDNA. Identical aa are boxed while homologous aa are shaded in gray. The Xenopus Nup153 partial amino acid sequence is 38% identical to human Nup153, contains 38 FXFG-related repeats (16 FXFG), and 5 zinc finger repeats. The Zn fingers in Xenopus Nup153 are underlined. The Xenopus Nup153 sequence data is available from GenBank/EMBL/DDBJ under accession number AF045567.
To isolate an extended Xenopus Tpr clone, the same Xenopus cDNA library was screened with a DNA fragment of a Xenopus integrin variant clone, α5tr, known to contain a fusion with a partial cDNA of Tpr in its untranslated region (Joos et al., 1995). To prepare the probe for library screening, the integrin clone (Cordes et al., 1997) was digested with EcoRI and the fragment representing the partial TPR cDNA was isolated. This fragment was labeled using a random priming kit (Stratagene) and used to screen the Xenopus cDNA library using standard procedures. A clone of 5.0 kb was isolated from ∼200,000 plaques screened and matched a shorter partial Xenopus Tpr clone, which was published during the course of this study (Cordes et al., 1997). Sequence analysis confirmed it to be a partial cDNA of Xenopus TPR corresponding to the equivalent segment from amino acid 642 to the COOH terminus of human Tpr.
Constructs and Expression of Protein Fragments
To obtain subclones of Nup153 for sequencing and coupled transcription/ translation reactions, the partial cDNA of Nup153 (Fig. 1; plasmid 5c) was digested with different combinations of restriction enzymes and fragments were subcloned into pET28 vectors. The amino acid numbers given in the following descriptions of the resulting subclones correspond to those shown in Fig. 1 for the Xenopus clone; the different constructs are depicted in Fig. 9 A. A 1.5-kb fragment corresponding to aa 334–828 was cloned into pET28c (Construct 1; plasmid 5c-1.5). A fragment corresponding to aa 334–618 was put into pET28c (Construct 2; plasmid 5c-0.9). A 1.0-kb fragment (aa 53–334) was put into pET28b (Construct 3; plasmid 5c-1.0). A subclone of the COOH-terminal region of the partial cDNA 5c (aa 618–1219) was cloned into pET28c (Construct 5; plasmid 5c-3′). A subfragment of this was generated corresponding to aa 618-828 (Construct 8; plasmid 5c-3′ no. 3). PCR was used to generate other fragments of 5c representing aa 828–1219 (Construct 6; plasmid 5c-3′ no. 1) and 1110–1210 (Construct 7; plasmid 5c-3′ no. 2), which were then cloned into pET28 vectors. A separate Xenopus Nup153 partial cDNA (clone no. 1c) was used to generate a subclone in pET28c containing aa 618–1109 (Construct 4; plasmid 1c-1.5).
Figure 9.
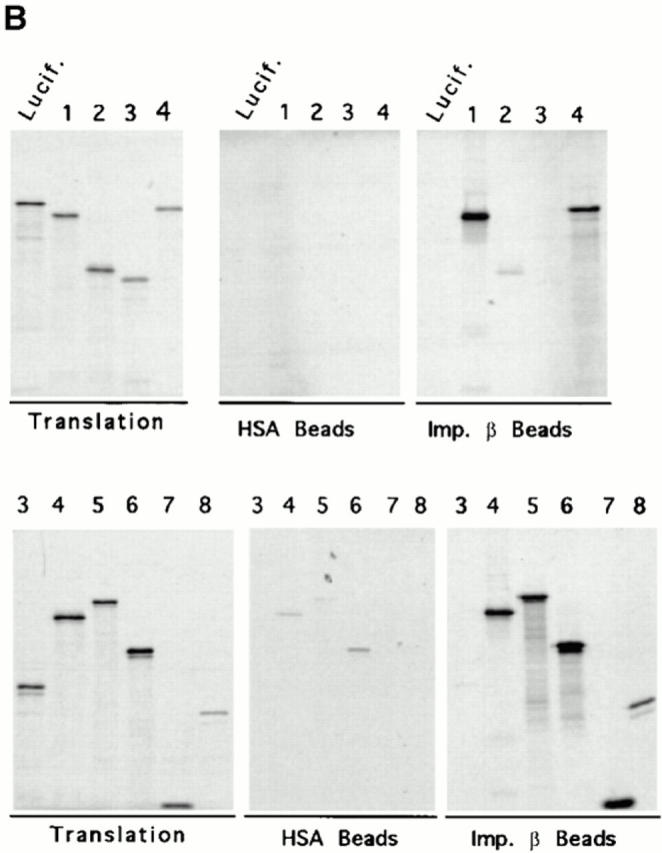
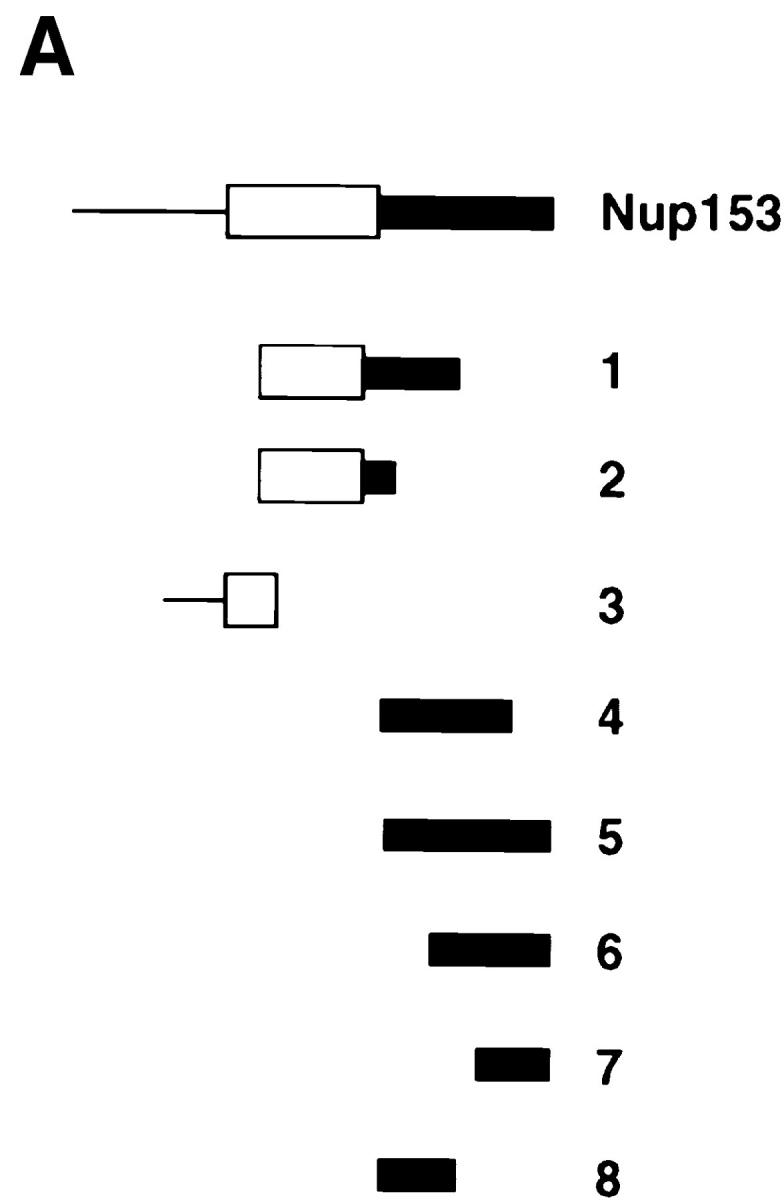
Mapping of the region of Nup153 that binds importin β in vitro. (A) Xenopus Nup153 contains a unique NH2-terminal domain (thin line), a zinc finger domain (box), and a COOH-terminal domain containing multiple FXFG repeats (thick line). Fragments were subcloned and expressed corresponding to aa 334–828 (Construct 1), aa 334–618 (Construct 2), aa 53–334 (Construct 3), and aa 618–1109 (Construct 4), aa 618–1219 (Construct 5), aa 828–1219 (Construct 6), aa 1110–1219 (Construct 7), and aa 618–828 (Construct 8) of the Xenopus Nup153 partial cDNA in Fig. 1. The cloned DNAs encoding the Nup153 fragments, shown in A, were transcribed and translated in [35S]methionine, along with a luciferase cDNA as a control. (B) An aliquot (2.5%) of the Nup153 radiolabeled protein fragments were analyzed by SDS-PAGE and exposed to film (Translation panel, Constructs 1–8); lane numbers are identical to the construct numbers. The individual radiolabeled Nup153 fragments were added to importin β–Sepharose beads or HSA–Sepharose beads and incubated for 2 h. The bound proteins were then separated using SDS-PAGE and the gel exposed to film. No binding of the Nup153 fragments to HSA-beads (HSA beads, top and bottom; Constructs 1–8) was observed. Constructs containing portions of the FXFG region of Nup153 bound strongly to importin β beads (Imp β beads, top and bottom; Constructs 1, 4, 5, 6, 7, and 8), while Constructs 2 and 3 did not (Imp β beads, Constructs 2 and 3).
The partial cDNA of Xenopus TPR was digested with PstI and a fragment 1.5 kb in length (among other bands) was subcloned into pRSETB (pSTU126). Sequence analysis confirmed this subclone to be a fragment of Xenopus TPR that corresponds to the aa 1668–2203 of human TPR.
To obtain protein fragments of Tpr and Nup153 for the purpose of raising antisera, the partial Xenopus Tpr clone, pSTU126, and the Xenopus Nup153 Construct 1 (5c-1.5; aa 334–828 of Fig. 1; antiserum 361) and Construct 3 (5c-1.0; aa 53–334 of Fig. 1; antisera 380 and 381) plasmids were transformed into the BL21/DE3 strain of Escherichia coli. Overnight cultures of the transformed plasmids were grown and diluted 1 to 10 in LB with kanamycin (25 μg/ml). After incubation of the cultures at 37°C for 45 min, the cultures were induced with 1 mM IPTG for 3 h. The bacteria were lysed in 0.5 M NaCl, 5 mM imidazole, 20 mM Tris, pH 8.0, by sonication and the soluble induced protein was purified using Ni-NTA resin (QIAGEN Inc., Chatsworth, CA). Purified Nup153 and Tpr protein expressed fragments were then used to immunize rabbits.
In a number of experiments, purified human importin α and β and importin β45–462 were used. These were produced as previously described (Gorlich et al., 1994; Kutay et al., 1997).
Affinity Purification of Antibodies
Affinity-purified antibodies were produced by coupling the respective antigen to which they were raised to CNBr–Sepharose (Pharmacia Biotech, Inc., Piscataway, NJ). Serum, diluted with an equal volume of 1M NaCl, 0.4% Triton X-100 (vol/vol), 50 mM Tris, pH 8.0, was applied to the antigen column. The column was then washed with 5 vol of 0.5 M NaCl, 0.2% Triton X-100, 50 mM Tris, pH 8.0, and 5 vol of PBS to remove nonspecifically bound proteins. Specific antibody was eluted from the column with 20 vol of 100 mM glycine, pH 2.5. The eluted antibody was mixed with 1/10th volume 1 M Tris, pH 8.0. The antibody was then concentrated using a spin concentrator (10K MWCO; Millipore Corp., Bedford, MA) and the antibody was then buffer exchanged with 10 vol of PBS (repeated three times; final buffer 1× PBS, 0.1 mM glycine, 0.1 mM Tris). Preimmune antiserum was protein A purified using standard procedures (Harlow and Lane, 1988).
Immunoblot Analysis
Protein samples were mixed with 2× sample buffer, boiled for 3 min, and loaded on a SDS–polyacrylamide gel (Sambrook et al., 1989). After separation, the separated proteins were electrophoretically transferred to polyvinylidene difluoride (PVDF) membrane (Millipore Corp., Waters Chromatography, Bedford, MA). The membrane was blocked with 5% nonfat dry milk in PBS, 0.2% Tween for 30 min at room temperature. To probe for the presence of a protein, the primary antibody was diluted in PBS, 0.2% Tween and incubated for 1 h at room temperature. Antibodies were used at the following dilutions: anti-Xenopus Nup153 (rabbit 380; aa 55–334) 1:2000; anti-Xenopus Nup153 (rabbit 381; aa 55–334) 1:500; anti- Xenopus Tpr, 1:20,000; anti-Xenopus Nup98, 1:1500 (Powers et al., 1995); anti-human Nup93, 1:10,000 (Grandi et al., 1997); mAb 414, 1:2,000 (BabCO, Richmond, CA); anti–importin β (Gorlich et al., 1995a ) 1:40,000; anti-Xenopus Nup214, 1:1000 (Macaulay et al., 1995); anti–importin α, 1:40,000 (Gorlich et al., 1994); and anti-rat Nup98, 1:500. The antisera to importin α and β, as well as the clones encoding the importins used below, were the kind gift of Dr. Dirk Görlich. After incubation with the appropriate primary antibodies, membranes were then washed and incubated with goat anti–mouse or goat anti–rabbit secondaries conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories) for 60 min. For visualization of the reactive bands, blots were soaked with chemiluminescence reagent (Dupont-NEN) and exposed to film (Dupont-NEN).
Immunofluorescence Microscopy
To characterize the anti-Nup153 and anti-Tpr antisera raised here, Xenopus XL177 (Warshawsky and Miller, 1995) cells were grown on coverslips and rinsed with PBS, 1 mM MgCl2. Cells were fixed with 4.0% (vol/vol) formaldehyde (16% ultrapure, EM grade; Polysciences Inc., Warington, PA) in PBS, 1 mM MgCl2, for 5 min at room temperature. Coverslips were washed twice with PBS, then three times with PBS, 0.1% (vol/vol) Triton X-100 to permeabilize the cells. Coverslips were then incubated for 10 min with Buffer IF1 (PBS, 0.1% [vol/vol] Triton X-100, 5% [vol/vol] FCS). Anti-Tpr and anti-Nup153 (380) affinity-purified antibodies were coupled directly to rhodamine or fluorescein, using the isothiocyanate derivative of each (Calbiochem-Novabiochem Corp., La Jolla, CA), as per manufacturer's protocol. Coverslips containing fixed, permeabilized XL177 cells were incubated with 1.8 μg of anti-Nup153 (380) antibody conjugated to fluorescein and 0.67 μg of anti-Tpr antibody conjugated to rhodamine in 50 μl of buffer IF1 for 1 h at room temperature in the dark. When mAb414 antibody was used, an Oregon Green-labeled goat anti–mouse secondary antibody (Molecular Probes, Inc., Eugene, OR) was used to visualize it. The coverslips were then washed three times with PBS, 5% (vol/ vol) FCS for 5 min each. The coverslips were mounted over a drop of 90% (vol/vol) glycerol, 10% PBS, containing 1 μg/ml Hoechst 33258 fluorescent DNA dye, and 1 mg/ml antifading agent p-phenylenediamine, followed by sealing of the edges of the coverslip with clear nail polish. Samples were observed with a confocal microscope (Nikon PCM2000) using a 60× objective, set at 6× Zoom and slow scan.
For comparison of anti-Nup214 staining with anti-Tpr staining, XL177 cells were fixed and blocked as described above. Anti-Nup214 antibody, diluted 1:15 in PBS, 0.1% (vol/vol) Triton X-100, 5% (vol/vol) FCS, was incubated with the coverslips at 4°C overnight. Coverslips were washed with buffer IF2 (PBS, 5% [vol/vol] FCS) three times for 5 min each, then incubated for 1 h in the dark with goat anti–rabbit antibodies conjugated to rhodamine (Jackson ImmunoResearch Laboratories), diluted 1:200 in buffer IF2. After this, coverslips were blocked in the dark for 20 min with 0.2 mg/ml rabbit IgG in buffer IF2, and washed with IF2 three more times (5 min each in the dark). FITC-conjugated anti-Tpr antibody (0.16 μg), diluted into 50 μl of buffer IF2, was incubated with the coverslips for 1 h in the dark. The coverslips were washed two times (10 min each) with the same buffer, then mounted as above, and viewed at 60×, 6× Zoom with a Nikon confocal microscope (model PCM2000). The images were captured on computer and sections were magnified using Adobe Photoshop 4.0.
Immunoprecipitation from Xenopus Egg Extracts
To search for novel protein–protein interactions, the soluble fraction of a Xenopus egg extract was diluted 25–100-fold in either RL buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin, 0.1% [vol/vol] NP-40) or ELBS buffer (50 mM KCl, 2.5 mM MgCl2, 10 mM Hepes, pH 7.5, 1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin). 10 μl of protein A–Sepharose (Pharmacia Fast Flow; Pharmacia Biotech, Inc.) was added to this mixture, along with the appropriate antibody. Antibodies were used in the following amounts for immunoprecipitation: 2 μg anti-Nup153 (380; 12 μg if coupled to protein A–Sepharose), 1.6 μg anti-Nup153 (361), 10 μg anti-Tpr (342; coupled to protein A–Sepharose), 100 ng anti-rat Nup62 (Finlay et al., 1991), 4 μg anti-Xenopus Nup98 (Powers et al., 1995), 2 μg anti-rat Nup98, 1–3 μg anti-Nup93 (Grandi et al., 1997), 5 μl of mAb 414 (ascites fluid; BAbCO ascites MMS-120R-500), and 12 μg control rabbit IgG (coupled to protein A–Sepharose; Calbiochem-Novabiochem Corp.). Anti-Nup153 preimmune antiserum (for rabbits 380 and 361) were protein A purified as in Harlow and Lane (1988). Preimmune antisera were used at microgram amounts equivalent to the affinity-purified anti-nucleoporin antibodies. After adding the antibodies to the extract, immunoprecipitations were rotated at 4°C for 2 h, centrifuged at 2,600 g for 20 s, and then washed with RL buffer or ELBS buffer for 5 min (repeated four times). Immunoprecipitated pellets in RL buffer were further washed once with RW buffer (150 mM NaCl, 10 mM Tris, pH 6.8, 1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ ml leupeptin). The immunoprecipitated proteins were eluted with standard gel sample buffer (see above) or, for immunoprecipitations done with protein A–Sepharose-coupled antibody, with 100 mM glycine, pH 2.5. Glycine eluates were neutralized with 1/10th volume 1 M Tris, pH 8.0, in sample buffer. Antibodies were coupled to protein A–Sepharose as described in Harlow and Lane (1988). The immunoprecipitate and supernatant samples were analyzed either by immunoblotting or by silver staining.
For immunoprecipitations from egg extract in the presence of GMP-PNP or AMP-PNP, Xenopus egg extract was first mixed with 1/10th volume of the appropriate nucleotide analogue (100 mM in 100 mM Hepes, pH 8.0) and incubated at room temperature for 10 min. The extract was further diluted fivefold with ELBS buffer also containing the analogue at 10 mM (without PMSF). After an additional 10 min at room temperature, the extract was diluted 20-fold with ice-cold ELBS or RL buffer. The diluted extract was then spun at 17,000 g for 10 min (4°C) to remove any particulate aggregates. This soluble fraction was then used for immunoprecipitation in the manner described above.
Immunoprecipitations in the presence of the recombinant importin β45–462 fragment were done as follows. For immunoprecipitations in egg extract, 2 μg of recombinant β45–462 fragment and 5 μl of egg extract were diluted to 500 μl with RL buffer or ELBS. Immunoprecipitations were then carried out as described above.
For experiments where immunoprecipitated Xenopus Nup153 or Tpr was stripped of any bound coimmunoprecipitated protein and then assayed for subsequent binding of recombinant importin β, the following procedure was done. Nup153 and Tpr were immunoprecipitated from Xenopus egg extract that had been pretreated with GMP-PNP in buffer RL (as described above; GMP-PNP treatment was to promote dissociation of endogenous bound importin β). The immunoprecipitated Nup153 or Tpr were washed two times (5 min each) with buffer RL, then once with buffer RL containing additional NaCl (0.5 M final) to further remove any other bound proteins. The Nup153 and Tpr immunoprecipitates were washed twice more with RL buffer alone, resuspended into 1 ml RL, and then each was divided into two tubes. One aliquot of the immunoprecipitate was incubated with 350 ng of recombinant importin β in buffer RL while the other was incubated with buffer RL alone for 90 min at 4°C. After this, the immunoprecipitates were washed three times with buffer RL (5 min each), and additionally with buffer RW for 5 min. Bound protein was eluted from the antibodies with 100 mM glycine, pH 2.5, as described earlier, and analyzed on gels by immunoblotting.
NLS-HSA Affinity Column
To construct a NLS-HSA affinity column, SV-40 large T antigen NLS peptide (CTPPKKKRKV; Newmeyer et al., 1986) was coupled to HSA in multiple copies, as described previously (Newmeyer et al., 1986). Rabbit antiserum to HSA (Sigma Chemical Co., St. Louis, MO) was affinity purified and coupled to protein A–Sepharose (Harlow and Lane, 1988). 5 μl of egg extract, diluted into 495 ml ELBS, was mixed with anti-HSA antibody (6 μg) coupled to protein A–Sepharose, and HSA-NLS (1.9 μg), or HSA alone (2.4 μg). This mixture was rotated at 4°C for 2 h. The anti-HSA antibody bound the NLS-HSA to the protein A–Sepharose beads, creating an NLS affinity column (Gorlich, 1995a). The NLS-HSA/anti-HSA/protein A beads were pelleted from the extract and washed four times with ELBS for 5 min each. Elution of the bound protein was accomplished with 100 mM glycine, pH 2.5. In some experiments, GMP-PNP or AMP-PNP (0.5 mM) or importin β45–462 fragment (2 μg/500 μl diluted extract) were added at the outset and the experiments performed as described above. The presence of individual proteins in the eluate from the NLS-column was determined by immunoblot analysis with anti-pore and anti-importin antibodies. Estimation of the percentage of bound proteins was accomplished by comparing the amount of the eluted protein to the amount of that protein present in a dilution series of egg extract similarly immunoblotted.
Immunoprecipitation from Rat Liver Nuclei
To determine whether individual nucleoporins are in complex with importin β in extracts of the assembled nuclear pores of rat nuclei, rat liver nuclei were isolated as described previously (Newmeyer et al., 1986; Newport and Spann, 1987). Rat liver nuclei (150 μl; 5 × 105 nuclei/μl; ∼9 × 107 nuclei) were incubated with 1.0 ml of PBS-ALP-TX buffer (PBS, 2.0% vol/vol Triton X-100, 1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin) for 5 min on ice to extract nuclear proteins, including proteins of the nuclear pore. This mixture was then vortexed for 1 min (setting no. 5, Vortex Genie 2, Fisher) and then centrifuged at 17,000 g for 10 min (4°C). An aliquot of the supernatant (180 μl; extracted from ∼1.5 × 107 nuclei) was mixed with 320 μl of PBS-ALP buffer (PBS, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mM PMSF) and used for immunoprecipitation by addition of affinity-purified anti-Nup153, Tpr, Nup93, or Nup98 antiserum in the amounts described above for Xenopus egg extract (final Triton X-100 concentration was 0.7% vol/vol). The immunoprecipitation reactions were rotated for 2 h at 4°C, and then centrifuged and washed four times (5 min each) with PBS-ALP buffer plus 0.1% Triton X-100 (vol/vol). For elution, 100 mM glycine, pH 2.5, was used as described above. The immunoprecipitates were mixed with sample buffer, boiled, electrophoresed on an 8% SDS–polyacrylamide gel, transferred, and analyzed by immunoblotting.
Recombinant Nup153/Importin β Binding Assay
For the purpose of mapping the important regions of Nup153 and importin β involved in formation of a complex, recombinant proteins were produced. Once expressed, a full-length recombinant importin β affinity column was produced: 700 μg of recombinant importin β or 700 μg of HSA were coupled to 100 μl of CNBr Sepharose 4B beads (Pharmacia Biotechnology, Inc.). The beads were blocked for 2 h with 0.2 M ethanolamine, pH 8.0, on ice, then washed with 5 vol of 0.5 M NaCl, 100 mM Tris, pH 8.0, and then 5 vol of 0.5 M NaCl, 100 mM sodium acetate, pH 4.6 (repeated three times).
Radiolabeled fragments of Nup153 protein were produced by in vitro transcription and translation of the Nup153 subclones 5c-1.5, 5c-1.0, 5c-0.9, and 1c-1.5 (see Fig. 9 A) using the Promega TNT system and [35S]methionine (10-μl reactions). Control radiolabeled luciferase was also produced from the vector in this system. After 90 min of translation, the reactions were spun at 17,000 g for 10 min (4°C). Seven μl of the supernatant was diluted into 1.0 ml RL buffer containing 8 mg/ml BSA. Half of this (500 μl) was then added to 5 μl of HSA–Sepharose beads or 5 μl of the importin β–Sepharose beads. The reactions were rotated for 2 h at 4°C. The beads were then washed three times for 5 min with RL buffer and once with RW buffer for 5 min. Bound proteins were eluted by the addition of 20 μl of sample buffer plus boiling (3 min), of which 15 μl was analyzed by SDS-PAGE. 0.25 μl of the initial translation reactions were also analyzed in parallel by SDS-PAGE. Radioactivity was quantitated using a phosphorimaging system (Molecular Dynamics, Inc., Sunnyvale, CA).
Results
Production of Xenopus Nup153 and Tpr Antibody Reagents
Five nucleoporins have now been identified that reside on the nucleoplasmic side of the pore. These include the nucleoporins Nup98, Nup153, Tpr, and, most recently, Nup93 and its partner Nup205 (Sukegawa and Blobel, 1993; Byrd et al., 1994; Powers et al., 1995; Radu et al., 1995b ; Bastos et al., 1996; Cordes et al., 1997; Grandi et al., 1997; Zimowska et al., 1997). In examining the nuclear pore, we wished to probe for potential complexes between these proteins and novel molecular partners. Antisera to Xenopus Nup93 and Nup98, as well as antibodies to Nup62 and Nup214, were in hand (Finlay et al., 1991; Macaulay et al., 1995; Powers et al., 1995; Grandi et al., 1997). For the present study, antibodies to Xenopus Nup153 and Tpr were also needed.
To obtain anti-Nup153 and anti-Tpr antisera, we cloned partial Xenopus cDNAs of these genes (see Materials and Methods). To identify a Xenopus Nup153 clone, a cDNA library derived from Xenopus blastocysts was immunoscreened with monoclonal antiserum mAb414. Four reactive clones, when sequenced, overlapped with one another and showed strong homology with rat Nup153. The longest cDNA clone of Xenopus Nup153 gene extends from the equivalent of amino acid 377 in human Nup153 into the 3′ untranslated region (Fig. 1). This partial cDNA codes for a protein with the same organization as human Nup153. Specifically, it contains the three regions typical of human Nup153: (a) a unique region, (b) preceding a region of zinc fingers, followed by (c) a region of FG repeats; these domains have 41, 40, and 35% identity, respectively, to the domains of human Nup153. Of the FG repeats, 21 of 23 are conserved between Xenopus and human. Overall, the cDNA, which we designate Xenopus Nup153, is 38% identical and 68% similar to its human Nup153 counterpart. Although few Xenopus nucleoporin genes have been cloned, for comparison, Xenopus p62 is 36% identical in its FG domain and 88% identical in its unique domain to the respective domains of human p62. Of note with respect to Xenopus Nup153 is the finding that it encodes five zinc finger repeats, whereas human and rat Nup153 encode four. The Xenopus Nup153 sequence contains certain inserted amino acid regions (Fig. 1) suggests that the full-length xNup153 will be slightly larger in size than the rat and human homologues; indeed, the xNup153 protein runs slightly higher than the rat Nup153 protein (data not shown; see Fig. 10 A below).
Figure 10.
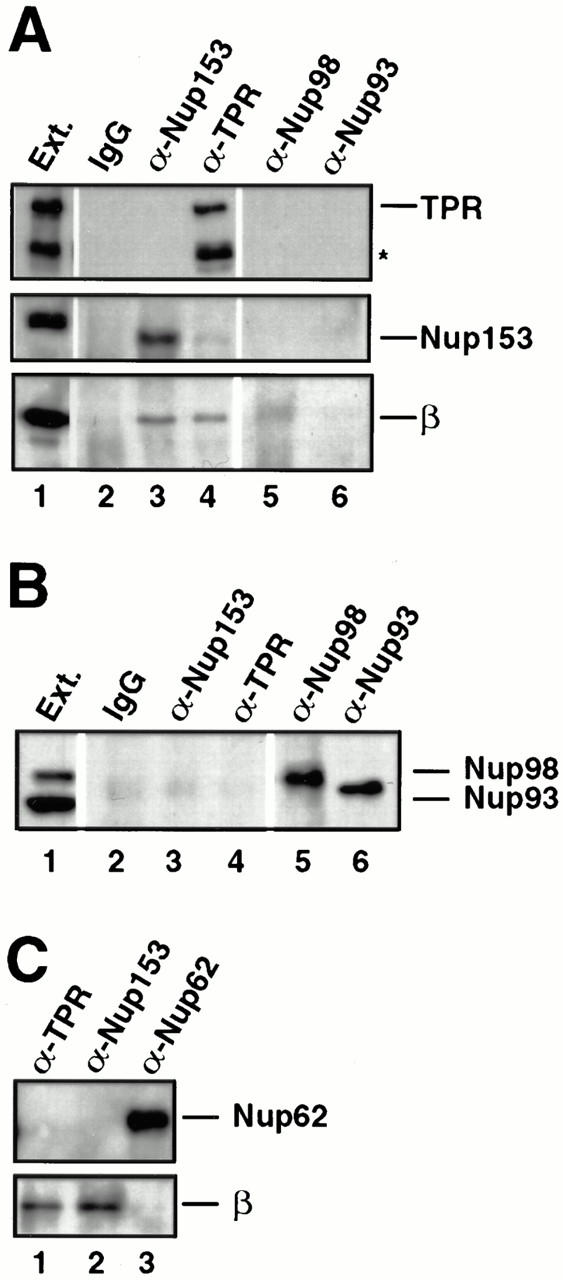
Nup153– and Tpr–importin β complexes are present in extracts of fully assembled nuclear pores. (A and B) Isolated rat liver nuclei were extracted with PBS Triton to release nuclear pore proteins. The nuclear extract was diluted and immunoprecipitated with anti-Nup153 (380), anti-Tpr, or rabbit IgG antibodies coupled to protein A–Sepharose, or with anti–rat Nup98 and anti-Nup93 antibodies uncoupled to Sepharose. The immunoprecipitates were split in half, electrophoresed on two separate gels, and immunoblotted. One blot (A) was probed with anti-Tpr (top strip; lanes 1–6), mAb 414 (middle strip), and anti–importin β antibodies (bottom strip). A second blot (B) was probed with a mix of anti–rat Nup98 and anti-Nup93 antibodies to demonstrate that Nup98 and Nup93 did immunoprecipitate (B, lanes 5 and 6). An aliquot of egg extract (0.05 μl) was run for size comparison (lane 1). Nup153 immunoprecipitated in a complex with importin β from rat liver nuclei (A, lane 3), as did Tpr (A, lane 4). Neither Nup98 nor Nup93 immunoprecipitated with importin β from rat liver nuclear extracts (A, lanes 5 and 6). (C) Identical immunoprecipitations were performed from rat liver nuclear extracts with anti-Tpr, anti-Nup153, or anti–rat Nup62 antisera. The blot was cut in two and probed with mAb 414 to detect Nup62 (top) and anti–importin β (bottom). Importin β coimmunoprecipitated with Nup153 (lane 2) and Tpr (lane 1), but not with Nup62 (lane 3).
To produce antibodies to Xenopus Nup153, the partial cDNA was subcloned and expressed in bacteria. Rabbit polyclonal antisera were produced to two protein fragments corresponding to aa 53–334 (antisera 380 and 381) and aa 334–828 (antiserum 361) of the Xenopus Nup153 partial sequence in Fig. 1. Affinity-purified antiserum 380 was used for immunoprecipitation from the soluble fraction of a Xenopus egg extract. When an immunoblot of the pellet was probed with Nup153 antiserum (380; Fig. 2 A, lane 6) or by mAb 414 which is known to react with Nup153 (Fig. 2 A, lane 4), the same single ∼180-kD band was detected by both. The Xenopus ∼180-kD reactive band was similarly detected on an immunoblot by antisera 381 and 361 (data not shown). Rat Nup153 is also known to migrate by SDS-PAGE at ∼180 kD (Sukegawa et al., 1993). When antiserum 380 was used to immunodeplete an extract of Xenopus eggs and then a blot of the depleted extract was probed with mAb414, all of the ∼180-kD reactive band size was specifically missing (data not shown), indicating the antiserum recognizes authentic Xenopus Nup153. When immunofluorescence was performed with Nup153 antiserum (380), a punctate nuclear rim stain was observed on Xenopus XL177 cultured cells (FITC; Fig. 3, f–i). Taken together, these data confirmed that the antisera were suitable for use in characterizing Xenopus Nup153.
Figure 2.
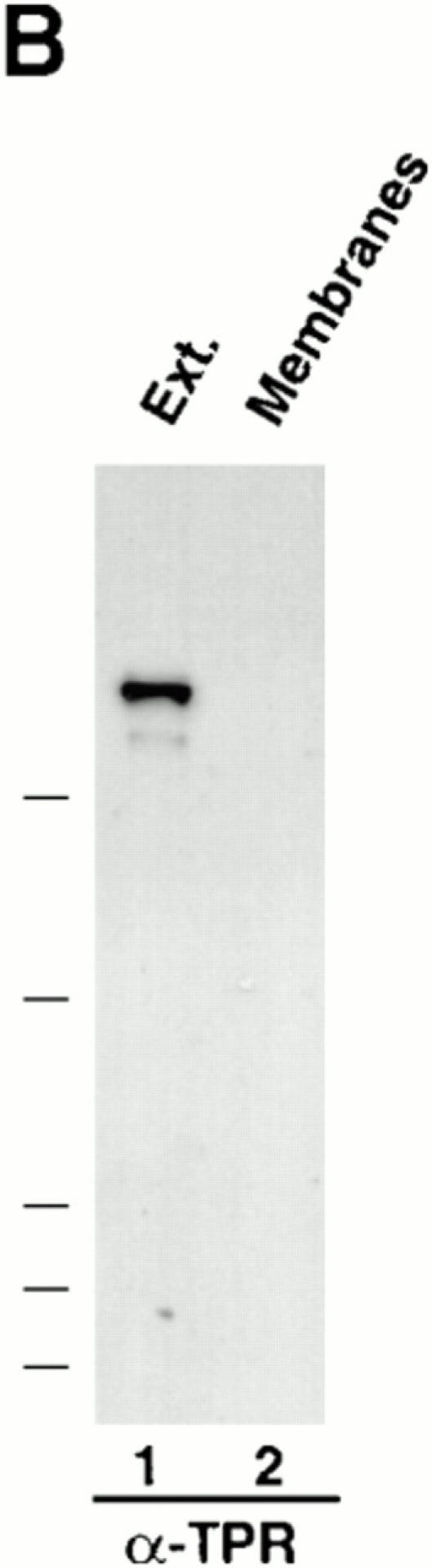
Antibody probes to Xenopus Nup153 and Tpr. (A) Immunoprecipitations were conducted with anti-Nup153 (380) antibody, mAb414, and rabbit IgG from Xenopus egg extract diluted 1:250 with buffer RL, as described in Materials and Methods. Pellets from the immunoprecipitations, along with an equivalent amount of diluted Xenopus egg extract (Ext.) for comparison, were then electrophoresed and immunoblotted independently with mAb 414 (lanes 1–4) and anti-Nup153 (380) antibody (lanes 5 and 6). The antibody used for immunoprecipitation is shown at the top of the figure, and the antibody used for immunoblotting is shown beneath the figures. Anti-Nup153 (380) immunoprecipitates a single major band of the expected size. (B) Soluble Xenopus egg extract (lane 1; 0.2 μl), and the membrane fraction of a Xenopus egg extract (lane 2; 0.2 μl of 10× membranes) were immunoblotted with anti-Tpr antibody. Anti-Tpr antibody recognizes a single band of the expected size in the soluble egg extract, which is not present in the membrane fraction.
Figure 3.
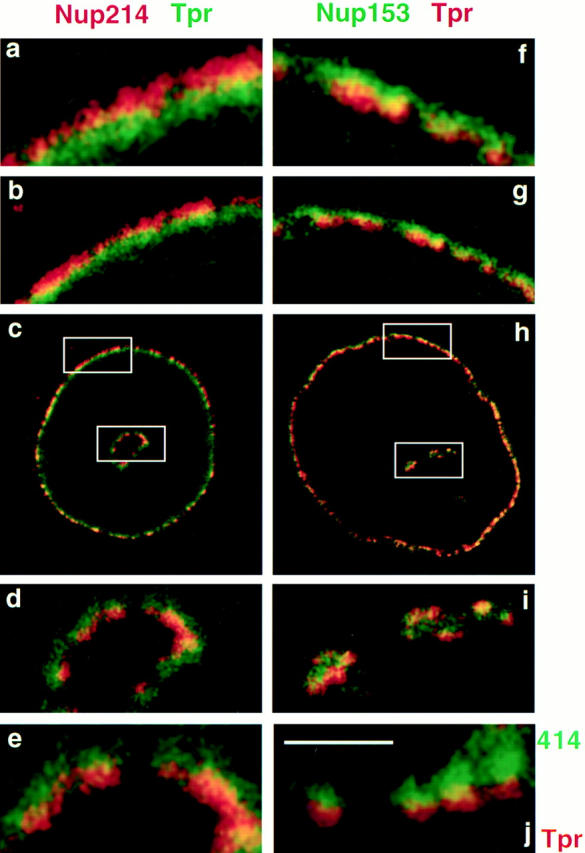
Distinct immunofluorescent localization of Nup153, Nup214, and Tpr. Anti-Nup153 (380) antibody was covalently linked to fluorescein, whereas anti-Tpr antibody was directly linked to rhodamine or fluorescein independently, using isothiocyanate derivatives (RITC and FITC, respectively) as described in Materials and Methods. For immunostaining and colocalization, anti-Nup214 was first used to immunostain formaldehyde fixed XL177 cells. After incubation of the cells with goat anti–rabbit-RITC antibody and subsequent blocking of this antibody with excess IgG, cells were then stained with FITC–anti-Tpr antibody, as described in Materials and Methods (a–e). The anti-Tpr stain is punctate and lies more to the nuclear interior than the anti–Nup214 stain. To compare localization of Nup153 and Tpr (f–i), XL177 cells were costained with FITC–anti-Nup153 (380) antibody and RITC–anti-Tpr antibody. Nup153 and Tpr show a more overlapping localization, although a large amount of Tpr staining still lies more intranuclearly than that of Nup153. A low magnification of the colocalization is shown in c and h, a 4× higher magnification view in panels immediately above and below (b, d, g, i), and an 8× higher magnified view lies above and below these (a, e, f). The nuclei shown contain internal tunnels of nuclear envelope, with magnified views of the tunnel shown in d, e, and i. Magnified views of the circumferential nuclear envelope are shown in a, b, f, and g. Note that the Nup214 staining (a–e) is always closer to the cytoplasm, whereas the Tpr staining (a–i) is always more proximal to the nuclear interior. j shows mAb414 (Oregon Green) and anti-Tpr (RITC) staining of a XL177 tissue culture cell, showing a segment of the nuclear envelope. Bar, ∼1 μm.
Next, we isolated a partial cDNA clone of Xenopus TPR and expressed a subfragment corresponding to aa 1668– 2203 in human TPR in E. coli for immunization of rabbits. The anti-Tpr antibody obtained specifically recognized a single prominent protein band in Xenopus egg extracts of ∼265–270 kD in size (Fig. 2 B, lane 1). This band was absent from the membrane fraction of a Xenopus egg extract (Fig. 2, lane 2). Immunofluorescence conducted on Xenopus XL177 cells with the anti-Tpr antibody yielded a punctate nuclear rim stain (Fig. 3, FITC in a–e, RITC in f–j).
Unexpectedly, during the course of immunofluorescence we were able to obtain a resolution of the nuclear pore at a level not previously predicted. Using anti-Tpr and anti-Nup214 antibodies, together with confocal immunofluorescence microscopy, we were able to observe a clear separation between the localization of different nuclear pore proteins. Tpr staining localized toward the nuclear side of the pores, whereas Nup214 staining localized more toward the cytoplasmic side (Fig. 3, a–c). Coimmunofluorescence of Xenopus cells with anti-Nup153 and anti-Tpr antibodies showed more overlap, although Tpr was still localized more proximally to the nuclear side of the pore than was Nup153 (Fig. 3, f–h). To ascertain that the distinction between FITC and RITC signals represented true differential localization of distinct nucleoporins, we searched for nuclei in the sample that contained nuclear “tunnels”, structures observable by immunofluorescence microscopy where the nuclear envelope contains a portion that has been pushed into the nucleus (Fricker et al., 1997). On these nuclear tunnels, an inversion of the antibody stains could be observed: Tpr stain was always toward the nuclear interior (Fig. 3, FITC in c–e), whereas the Nup214 stain lay closer to the cytoplasm in the center of the nuclear tunnel (Fig. 3, RITC in c–e). These immunofluorescence studies not only confirmed that the anti-Nup153 and anti-Tpr antibodies are suitable for characterization of Xenopus Nup153 and Tpr, but demonstrated that it is possible to obtain immunofluorescence localization to subregions on and around the nuclear pore. A striking example of the relative localization of mAb414 (Oregon Green), an mAb that reacts with FXFG nucleoporins, and anti-Tpr (RITC) staining on what we believe are likely four individual nuclear pores is shown in Fig. 3 j. It should be stressed that from this technique we can tell only relative localization of one nucleoporin to another, not absolute localization of a nucleoporin to the cytoplasmic fibrils, for example.
A Partner for Nup153: the Import Factor, Importin β
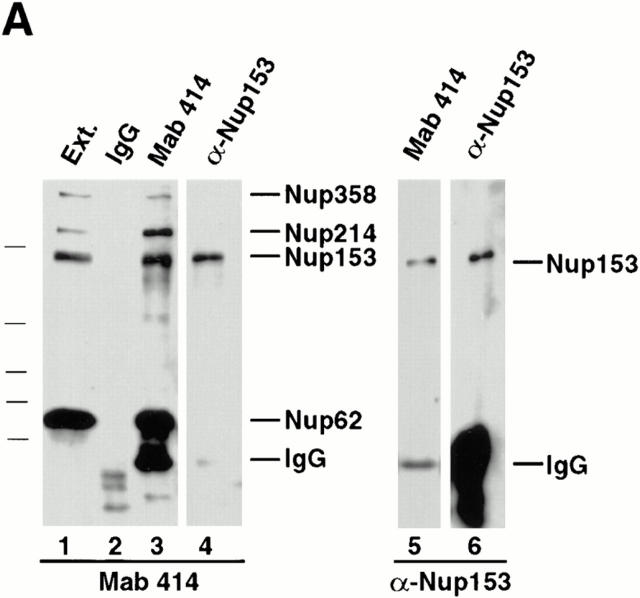
Importin β is known to arrest on or near the nuclear basket after a round of import before recycling to the cytoplasm (Gorlich et al., 1995b ). We set out to ask which, if any, proteins on the basket side of the nuclear pore interact with importin β in in vivo–like situations. We first addressed this question in Xenopus egg extracts that contain the components of ⩾30 million nuclear pores disassembled into subcomplexes (Finlay et al., 1991; Grandi et al., 1997). Immunoprecipitations from Xenopus egg extracts were performed using anti-nucleoporin antisera. The extracts were individually and quantitatively depleted of Nup93, Nup98, or Nup153. It was desirable in this experiment to immunoprecipitate all of these nucleoporins so that the entire population of each nucleoporin could be assayed for the presence of importin β. The immunodepleted extracts are shown in Fig. 4 A (lanes 1–3). For each, the extract can be seen to be quantitatively depleted of the appropriate nucleoporin. Upon analysis of the pellets, importin β was not present in the immunoprecipitate of Nup93 (Fig. 4, lane 5) or that of Nup98 (lane 6). However, importin β did coimmunoprecipitate with Nup153 (Fig. 4 A, lane 7; antiserum 361). This indicated that importin β is in a complex with Nup153 and that this complex is specific. It also indicated that two other vertebrate nucleoporins known to be present in the nuclear basket of the pore, Nup93 and Nup98, do not bind importin β in the Xenopus egg extract.
Figure 4.
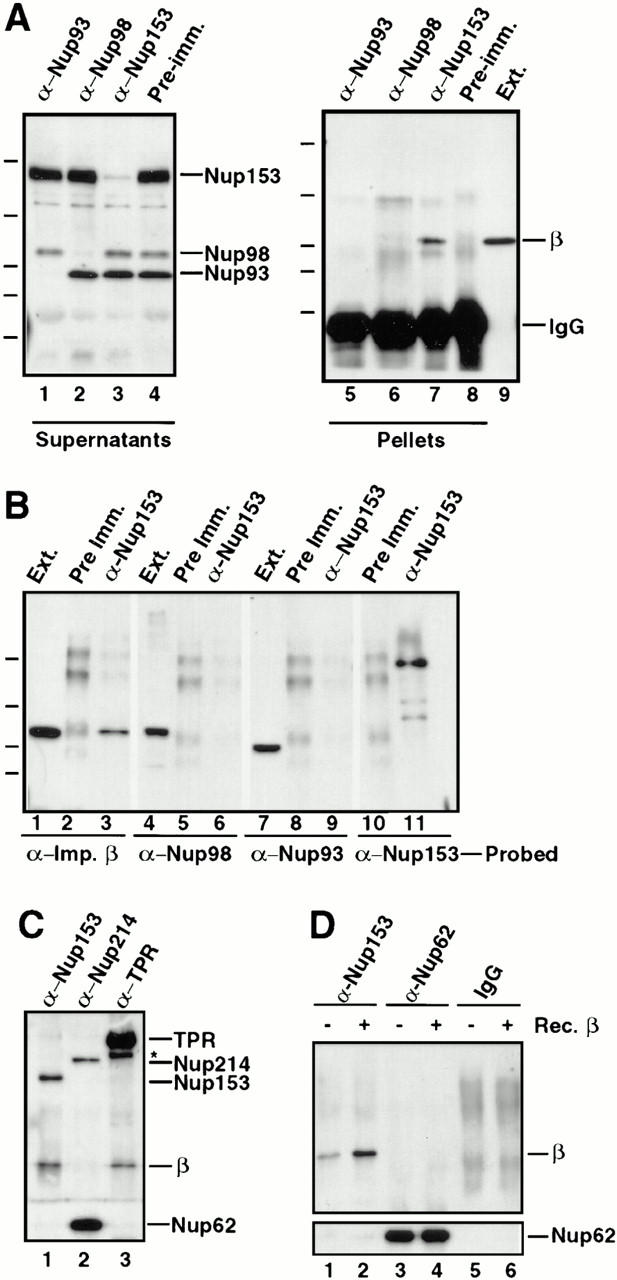
Importin β is in a complex with Nup153, but not Nup62, Nup93, Nup98, or Nup214. (A) Immunoprecipitations were conducted from Xenopus egg extract diluted in buffer RL using anti-Nup153 (361), protein A–purified 361 preimmune, anti-Nup93, and anti-Xenopus Nup98 antibodies. The antibody used for immunoprecipitation is shown at the top of each lane. The supernatants of the immunoprecipitates (lanes 1–4) were immunoblotted with a mixture of anti-Nup93, anti-Xenopus Nup98, and anti-rat Nup153 antibodies to demonstrate that each nucleoporin had been quantitatively immunoprecipitated. Lane 1 is depleted of Nup93, lane 2 is depleted of Nup98, and lane 3 is depleted of the great majority of Nup153. The proteins of the pellets immunoprecipitated from 2.5 μl Xenopus egg extract (lanes 5–8) were immunoblotted with anti–importin β antiserum. Soluble Xenopus egg extract (0.05 μl) is shown (lane 9) to indicate where importin β migrates. Importin β coimmunoprecipitates with Nup153 (lane 7), but not with Nup93 (lane 5) or Nup98 (lane 6). (B) Immunoprecipitation from soluble Xenopus egg extract diluted in buffer RL were performed using anti-Nup153 (380) and preimmune antisera bound to protein A–Sepharose. Immunoprecipitated protein was eluted with glycine and separated on an 8% SDS-PAGE gel along with aliquots of Xenopus egg extract (lanes 1, 4, and 7). Proteins were transferred to PVDF and analyzed by immunoblotting with the following antibodies: anti–importin β (lanes 1–3), anti-Xenopus Nup98 (lanes 4–6), anti-Nup93 (lanes 7–9), and anti-Nup153 (lanes 10 and 11). Note that importin β is detected in Nup153 immunoprecipitates and Xenopus egg extract (lane 1 and 3), but not in a preimmune immunoprecipitate (lane 2). Also note that Nup98 and Nup93 are detected in Xenopus egg extract (lanes 4 and 7), but not in preimmune or Nup153 immunoprecipitates (lanes 5, 6, 8, and 9). (C) Immunoprecipitates of Nup153, Nup214, and Tpr were separated, transferred, and probed with anti–importin β, anti-Tpr, and mAb414 to detect Nup214, Nup153, and Nup62. An asterisk marks a breakdown product of Tpr. Importin β immunoprecipitates with Nup153 (lane 1) and Tpr (lane 3), but not with Nup214 (lane 2). (D) Immunoprecipitates of Nup153, Nup62, and control IgG were separated, transferred, and probed with anti–importin β, and mAb414 to detect Nup62. Full-length recombinant importin β immunoprecipitates with Nup153 (lane 2), but not with Nup62 (lane 4) or nonspecific IgG (lane 6).
In a second experiment, a Nup153 immunoprecipitate performed using a different anti-Nup153 antiserum (380) was split into four aliquots, electrophoresed, blotted, and probed with either anti–importin β, anti-Nup98, anti-Nup93, or anti-Nup153. Once again, the Nup153 immunoprecipitate contained importin β (Fig. 4 B, lane 3). The immunoprecipitate did not contain Nup98 (lane 6) or Nup93 (lane 9), indicating that these nucleoporins are not present in the Nup153–importin β subcomplex. In other immunoprecipitates, we found that neither Nup214 (Fig. 4 C, lane 2) nor Nup62 (Fig. 4 D, lanes 3 and 4) immunoprecipitates contain importin β, although we did observe that Nup62 was present in an immunoprecipitated complex with Nup214 (Fig. 4 C, lane 2), as we had previously reported (Macaulay et al., 1995).
Immunoblotting of the Nup153-depleted supernatant with anti–importin β antibody revealed no detectable reduction in the amount of importin β present (data not shown), whereas Nup153 was almost quantitatively depleted (Fig. 4 A, lane 3). This indicates that, although Nup153 is in a complex with importin β, there is a large amount of importin β that is not in a complex with Nup153 in the egg extract.
Tpr Forms a Complex with Importin β
The pore-associated protein Tpr, which resides on intranuclear fibers extending from the basket of the pore in the nucleus, was next investigated for a possible interaction with importin β. Affinity-purified anti-Tpr antibodies coupled to protein A–Sepharose were used to immunoprecipitate Tpr from an egg extract. The immunoprecipitate was immunoblotted for Tpr and importin β. Tpr was efficiently immunoprecipitated from Xenopus egg extract with anti-Tpr antibody (Figs. 4 C, lane 3 and 5 B, lane 2), while no Tpr was detected in immunoprecipitates with nonspecific rabbit IgG (Fig. 5 B, lane 3). Interestingly, importin β was strongly detected in the anti-Tpr immunoprecipitation (Figs. 4 C, lane 3 and 5 B, lane 2), and not in the rabbit IgG immunoprecipitation (Fig. 5 B, lane 3). Approximately equivalent amounts of importin β were coimmunoprecipitated with anti-Tpr or anti-Nup153 (380) antibodies from an equal amount of Xenopus egg extract (Fig. 5 B, lanes 1 and 2). Thus, importin β is also present in a complex with Tpr in Xenopus egg extracts.
Figure 5.
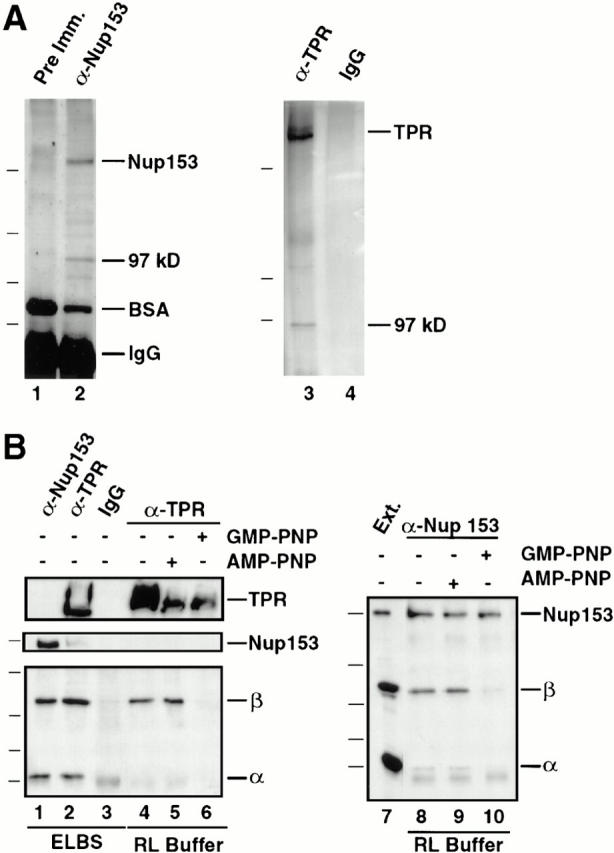
Stable independent complexes of Nup153 and Tpr with importin β exist. (A) Coimmunoprecipitation of a ∼97-kD band with Nup153 and with Tpr. Anti-Nup153 (380) and preimmune antibodies were bound to protein A–Sepharose (lane 1 and 2, respectively) and used for immunoprecipitation from 20 μl soluble Xenopus egg extract, as described in Materials and Methods. The immunoprecipitates were boiled with sample buffer, separated using SDS-PAGE (8%), and silver stained. In addition to Nup153, a prominent protein band of ∼97 kD immunoprecipitated with anti-Nup153 (lane 2). The markers for this gel are 193, 112, 86, and 70 kD. A separate experiment was performed with anti-Tpr antiserum; a band of ∼97 kD was observed to precipitate with Tpr (lane 3). The markers for this gel are 205, 116, and 97.4 kD. (B) Immunoprecipitations from soluble Xenopus egg extract were performed using anti-153 (380), anti-Tpr, or rabbit IgG antibodies coupled to protein A–Sepharose. Before antibody addition, the extract was diluted (1:100) in either ELBS buffer (lanes 1–3), or RL buffer (lanes 4 and 8). In other incubations Xenopus egg extract was preincubated with GMP-PNP (lanes 6 and 10) or AMP-PNP (lanes 5 and 9) before dilution with buffer RL and immunoprecipitation, as above. The immunoprecipitated proteins and an aliquot of soluble Xenopus egg extract (lane 7) were analyzed by gel electrophoresis and transfer to PVDF. The blot on the left was cut and probed by immunoblotting with anti-Tpr (top), anti-Nup153 (381; middle) or a mix of anti–importin α and β antibodies (bottom). The blot on the right was probed with a mix of anti-Nup153, importin β, and importin α antibodies. Importin α and β immunoprecipitate with anti-Nup153 (lane 1) and anti-Tpr (lane 2) antisera. The binding of importin β to Nup153 and Tpr is stable in both ELBS (lanes 1 and 2) and RL (lanes 4, 5, 8, and 9) buffers; however, the binding to importin α is disrupted in the more stringent RL buffer (lanes 4, 5, 8, and 9). GMP-PNP disrupts the complex of importin β with Tpr (lane 6) and Nup153 (lane 10). The differences in the amounts of TPR in Fig. 5, lanes 4–6 are due to inefficient transfer of the very large Tpr protein (∼270 kD) from the 8% gel used to simultaneously view importin α; reelectrophoresis of the samples revealed only small differences in the amounts of Tpr per lane (data not shown).
To ask whether importin β is a prominent member of the Nup153 and Tpr subcomplexes, immunoprecipitates performed in different experiments using anti-Nup153 or anti-Tpr antiserum were electrophoresed and examined by silver staining. A distinct band of ∼97 kD was indeed seen in both the Nup153 (Fig. 5 A, lane 2) and Tpr (Fig. 5 A, lane 3) immunoprecipitates. For each, this band ran on gels at a size less than Nup98 and greater than Nup93. Thus, the silver stain results are consistent with the immunoblot results indicating that importin β is a major molecular partner of each of these nucleoporins.
The NLS Receptor Heterodimer, Importin α/β, Is Complexed to Nup153 and Tpr
In cytosol, it is known that importin β is often in a heterodimer with the NLS receptor subunit, importin α. This raised the question as to whether importin α was also present in the Nup153– and the Tpr–importin β complexes observed above. The IgG band would have obscured the presence of importin α in the silver-stained gels of Fig. 5 A. To test for the presence of importin α, affinity-purified anti-Nup153, anti-Tpr, and preimmune IgG were each covalently coupled to protein A–Sepharose. These were used for immunoprecipitations from soluble egg extract diluted either with RL buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin, 0.1% NP-40) or with a lower ionic strength buffer, ELBS (50 mM KCl, 2.5 mM MgCl2, 10 mM Hepes, pH 7.5, 1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin). The immunoprecipitates were electrophoresed, blotted, and then probed with a mixture of anti–importin α and β antisera to determine the presence of these import factors. Importin β was once again observed to specifically coimmunoprecipitate with anti-Nup153 (Fig. 5 B, lanes 1, 8, 9) and anti-Tpr antisera (Fig. 5 B, lanes 2, 4, 5). Importin α also coimmunoprecipitated with Nup153 and with Tpr (Fig. 5, lanes 1 and 2). However, importin α did so only when the Xenopus egg extract was diluted in the lower ionic strength buffer ELBS (Fig. 5 B, lanes 1 and 2), rather than in the higher ionic strength buffer RL (Fig. 5 B, lanes 4, 5, 8, 9). Because ELBS is a milder buffer, these results indicate that Nup153 and Tpr are in complexes with importin α and importin β in the extract, but that importin α is more weakly attached. More importantly, the results indicate that importin α is not required for the observed interaction between Nup153 and importin β and between Tpr and importin β.
Nup153, but Not Tpr, Can Bind to Importin α/β Carrying an NLS Cargo
A complex between proteins of the NLS receptor and a nucleoporin theoretically could form in the intact cell either (a) in the course of importing an NLS-bearing protein, (b) after release of the NLS-bearing protein, but before dissociation of importin β from the pore, or (c) after import, but in the course of recycling importin β to the cytoplasm. To distinguish between these possibilities, we analyzed the conditions required for importin α/β binding to a given nucleoporin. We began by asking whether Nup153 binds importin α/β that is bound to an NLS, as would be expected in (a) above. NLS peptide was covalently linked to HSA and added to Xenopus egg extract containing anti-HSA antibody–protein A–Sepharose beads. After incubation for 120 min, the NLS-HSA/antibody beads were pelleted and washed. NLS-HSA–bound proteins were analyzed by immunoblotting using antisera to importin α, importin β, and Nup153. Importin α and β, as expected, bound to the NLS-HSA beads (Fig. 6 A, lane 3), and did not bind to control HSA beads containing no NLS (lane 2). Importantly, Nup153 also bound to importin heterodimer complexed to the NLS-HSA beads (Fig. 6 A, lane 3), confirming the interaction of Nup153 with importin α and β, and consistent with the interaction occurring while the α/β heterodimer is recognizing an NLS substrate.
Figure 6.
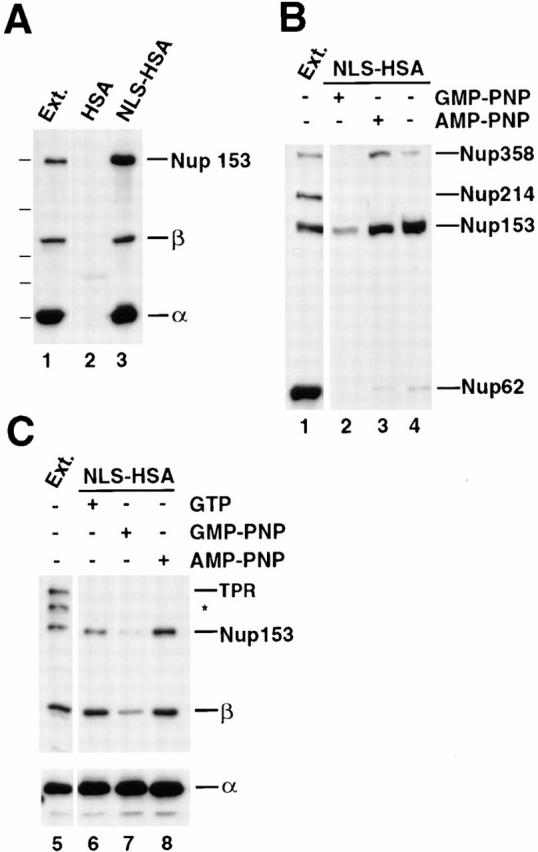
Nup153, but not Tpr, binds to importin α/β heterodimer in the NLS-bound form. (A) Anti-HSA antibody coupled to protein A–Sepharose was added to Xenopus egg extract (diluted 1:100 in buffer ELBS) containing 2.4 μg HSA (lane 2) or 1.9 μg NLS-HSA (lane 3). The proteins that bound to the NLS-HSA/anti-HSA/ Protein A beads were eluted and immunoblotted with a mix of anti–importin α and β and anti-Nup153 (381) antibodies. The proteins present in 0.2 μl of soluble Xenopus egg extract are shown in lane 1 for comparison. Importin α, importin β, and Nup153 all bind to an NLS-HSA affinity column (lane 3), but not to an HSA column (lane 2). (B) GMP-PNP disrupts Nup153 bound to importin α/β– NLS complexes. NLS-HSA/anti-HSA Sepharose beads were added to Xenopus egg extract pretreated with GMP-PNP, AMP-PNP, or no addition (lanes 2–4, respectively). Before bead addition, extracts were diluted with buffer ELBS and supplemented with 1.9 μg of NLS-HSA per aliquot. The NLS-HSA bead mixtures were incubated in extract for 2 h. Then bound proteins were eluted, electrophoresed, and immunoblotted with mAb414 (lanes 1–4). Soluble Xenopus egg extract (0.2 μl) was immunoblotted in parallel (lane 1) to indicate the nucleoporins recognized in the total extract by mAb 414 (Nup358, Nup214, Nup153, and Nup62). Much of the Nup358 and Nup153, but only a trace of Nup62, bind to the NLS-HSA affinity column (lanes 3 and 4). GMP-PNP disrupts the majority of this binding (lane 2), whereas AMP-PNP does not (lane 3). (C) Tpr does not bind to importin α/β–NLS complexes. The experiment in (A) was repeated, and the bound proteins were immunoblotted with a mix of anti-Tpr, anti-Nup153 (381), anti–importin α and β (lanes 5–8). Nup153 and importin α and β bind to the NLS-HSA beads (lane 6), but Tpr does not (lane 6). GMP-PNP releases the importin β and Nup153 from the beads (lane 7), whereas AMP-PNP does not (lane 8). (An asterisk denotes a breakdown product of Tpr.)
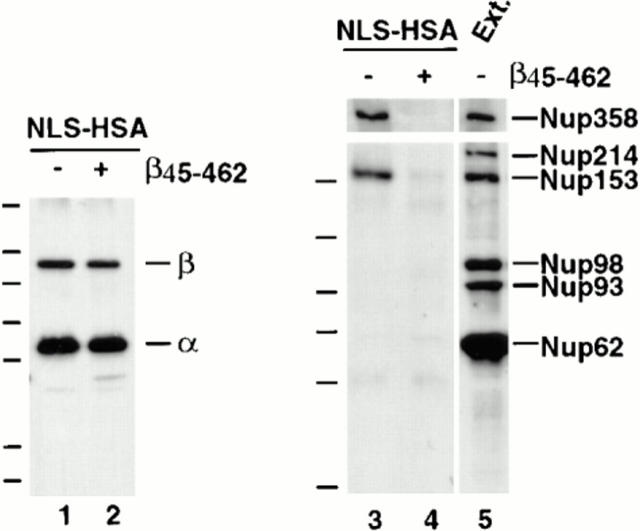
To test for the presence of other nucleoporins in importin α/β–NLS complexes, the experiment using NLS-HSA beads and egg extract incubation was repeated, but the proteins bound to the beads were probed this time with mAb 414 (an mAb that cross-reacts with Nup358, Nup214, Nup153, and Nup62 in Xenopus) and other nucleoporin antisera. We observed that not only Nup153, but also a nucleoporin derived from the cytoplasmic filaments of the pore, Nup358, bound to the NLS-HSA beads containing importin α and β (Fig. 6 B, lanes 3 and 4). Semiquantitative immunoblots indicated that ∼20–40% of the Nup153 and 10% of the Nup358 in the starting extract binds to the importin α/β–NLS-HSA beads (data not shown). In contrast, <1% of the Nup62 present in the extract and none of the nucleoporins Nup214, Nup98, or Nup93 were observed to bind to the NLS-HSA beads (Fig. 6 B, compare lane 4 to lane 1; Fig. 8, lane 3). Thus, only Nup153 and Nup358 of the above nucleoporins bind significantly to NLS-HSA beads in Xenopus egg extract.
Figure 8.
β45-462 removes nucleoporins Nup153 and Nup358 bound to an NLS-HSA column. Xenopus extract was diluted and supplemented with NLS-HSA (1.9 μg/immunoprecipitation). Importin β45–462 was added to half (lanes 2 and 4), while the equivalent amount of buffer was added to the other half (lanes 1 and 3). Anti-HSA antibody coupled to protein A–Sepharose was then added. Bound proteins were eluted from the NLS-HSA/anti-HSA beads with glycine. A portion of each was electrophoresed and immunoblotted with a mix of anti–importin α and β antisera (lanes 1 and 2), or a mix of mAb 414, anti-Nup214, anti-Xenopus Nup98, and anti-Nup93 (lanes 3–5). An aliquot of Xenopus egg cytosol (0.2 μl; lane 5) indicates that Nup358, Nup214, Nup153, Nup98, Nup93, and Nup62 would have been detected if bound to the NLS-HSA beads. Of these, only Nup358 and Nup153 bound (lane 3); both were displaced by added β45–462 (lane 4). This indicates that Nup153 and Nup358 bind to the NLS-HSA column through importin β.
Lastly, the presence of Tpr on the NLS-HSA beads after incubation with the extract was analyzed by immunoblotting with anti-Tpr antiserum. Surprisingly, no Tpr could be detected bound to the NLS-HSA beads (Fig. 6 C, lane 6), although Tpr was easily detectable in the extract (lane 5) and anti-Tpr antibodies immunoprecipitate importin α/β from the extract both under “+GTP” conditions (data not shown) and under the conditions shown where no GTP was added (Fig. 5 B, lane 2). These data demonstrate that the interaction between Tpr and the importin α/β complex is intact, but that this trimeric complex is unable to bind to an NLS column, strongly suggesting that the binding of Tpr to importin α/β is intrinsically different from the observed interaction of Nup153 with importin α/β. It would appear that Tpr cannot bind to an importin α/β heterodimer that is simultaneously occupied with an NLS.
Ran-GMP-PNP Disrupts the Interaction of Importin β with Both Nup153 and Tpr
RanGTP disrupts the binding of importin β to importin α, either by changing the conformation of the importin α binding site (Kutay et al., 1997) or by binding to a site on importin β that overlaps with the importin α binding site (Moroianu et al., 1996). If the Nup153/α/β complexes or the Tpr/α/β complexes observed in Xenopus egg extracts are mimicking steps in protein import, one might predict that RanGTP should disassociate these complexes. To test this, the nonhydrolyzable GTP analogue, GMP-PNP, was added to the type of assay performed above; GMP-PNP should lock Ran in a GTP-like form and cause it to bind to importin β. GMP-PNP was added to determine whether its presence caused the loss of nucleoporins Nup153 and Nup358 from the NLS-HSA beads. Addition of GMP-PNP (0.5 mM final) to the diluted Xenopus egg extract was found to greatly reduce the amount of importin β and Nup153 that bound to the NLS-HSA beads (Fig. 6 C, lane 7). The same reduction was also seen for Nup358 when GMP-PNP was added (Fig. 6 B, lane 2). The amount of importin α bound to the NLS-HSA/anti-HSA beads remained unchanged in the presence or absence of GMP-PNP (Fig. 6 C, lanes 6–8), indicating that GMP-PNP did not cause the dissociation of importin α from the NLS-HSA beads, only of importin β and its associated nucleoporins. The control analogue AMP-PNP caused no loss of importin β from the NLS-HSA beads (Fig. 6 B, lane 3 and C, lane 8). Importantly, if GMP-PNP was added to the pelleted NLS-HSA/anti-HSA beads and associated bound proteins after the unbound Xenopus cytosol was washed away, the GMP-PNP caused no loss of Nup153 and importin β from the NLS-HSA beads. This indicates that the presence of a soluble factor in the cytosol, presumably Ran, is required for the dissociation of Nup153 and importin β from the NLS beads (data not shown).
An equally important question is whether GMP-PNP promotes not only the dissociation of importin β from importin α, but also the disruption of Nup153 from importin β. Indeed, in the presence of GMP-PNP, but not AMP-PNP, importin β did not coimmunoprecipitate with Nup153 from soluble egg extract (Fig. 5 B, lanes 9 and 10). We found that GMP-PNP, but not AMP-PNP, also disrupted the complex of importin β with Tpr (Fig. 5 B, lanes 5 and 6). Thus, despite the fact that the binding of Tpr to importin β has distinct properties from the Nup153-importin β interaction, GMP-PNP disrupts both of these complexes.
Mapping of the Region of Importin β That Binds to Nup153
Importin β contains several domains that are involved in the binding of other proteins: an NH2-terminal domain that binds to the GTPase Ran, a partially overlapping domain that binds importin α, and a region that binds to the nuclear pore (Chi and Adam, 1997; Kutay et al., 1997). The pore-binding domain has been defined by adding fluorescent fragments of importin β to permeabilized cells and determining the minimal region of importin β that could bind to the pore (Kutay et al., 1997). aa 45–462 were shown to be necessary and sufficient for binding to the nuclear pore (Kutay et al., 1997). Moreover, such binding to the pore was irreversible, presumably because the importin β fragment was unable to bind RanGTP, which has been shown to disrupt the pore binding of full-length importin β. The presence of the Ran-binding region of importin β is necessary for efficient release from the nuclear pore and the return of importin β to the cytoplasm in the presence of GTP (Kutay et al., 1997; see Goldfarb, 1997, for review see Chi and Adam, 1997).
To determine whether the pore-binding region of importin β, β45–462, would bind to the nucleoporins Nup153 and Tpr, importin β45–462 (5 ng/μl final) was added to a Xenopus egg extract diluted 100-fold in RL buffer. Immunoprecipitation was performed using antibodies to Nup153, Tpr, or control rabbit IgG to look for β45–462 binding. The immunoprecipitated pellets were electrophoresed, blotted, and probed with anti–importin β antibody that reacts both with full-length importin β and with the β45–462 fragment. Importin β45–462 was found to be clearly present in a Nup153 immunoprecipitate (Fig. 7 A, lane 4), but absent in a control rabbit IgG immunoprecipitate (lane 6). Importin β45–462 did not coimmunoprecipitate with Nup62, Nup93, Nup98, or Nup214 in a Xenopus egg extract (data not shown). These results indicate that importin β45–462 specifically binds to Nup153. Moreover, addition of the β45–462 fragment caused loss of the endogenous full-length importin β from the anti-Nup153 immunoprecipitate (Fig. 7 A, compare lanes 3 and 4). It appears that the β45–462 fragment exchanges onto Nup153, replacing the endogenous full-length importin β.
Figure 7.
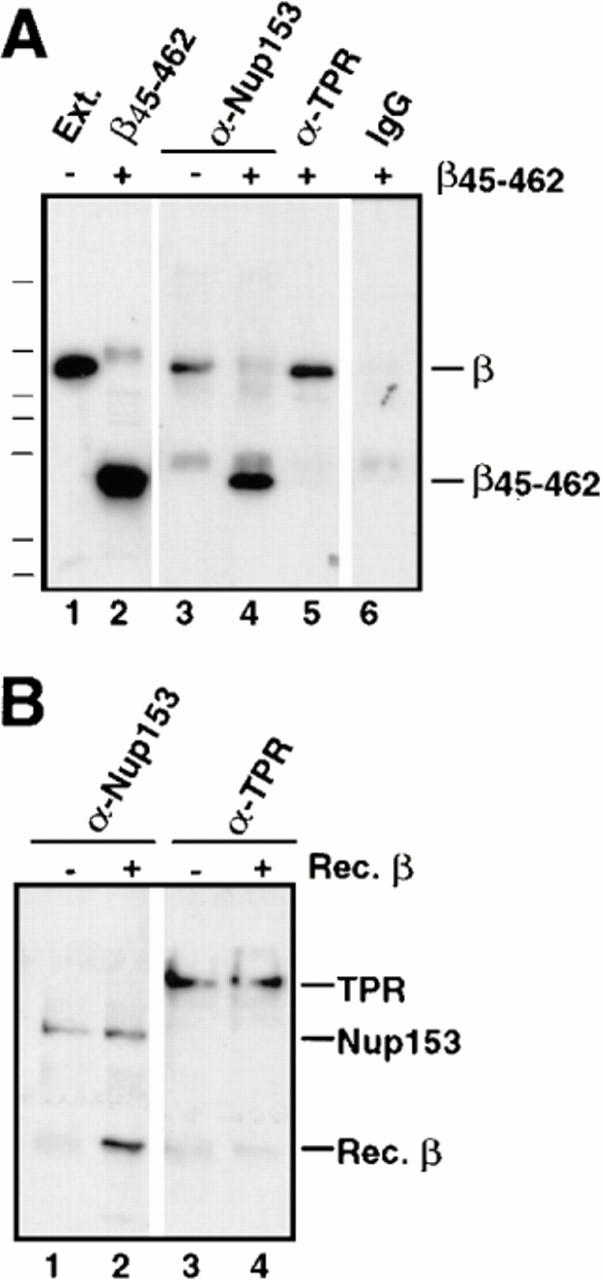
The effects of β45–462 on Nup153– and Tpr–importin β complexes. (A) Immunoprecipitations using anti-Nup153 (380), anti-Tpr, or rabbit IgG antibodies coupled to protein A–Sepharose were performed on soluble Xenopus egg extract diluted in buffer RL to which had been added the importin β fragment β45–462 buffer RL. The immunoprecipitates were eluted with glycine and immunoblotted with anti–importin β antiserum. Lane 1 shows an aliquot of egg extract and lane 2, an aliquot of recombinant importin fragment, β45–462; Importin β immunoprecipitates with Nup153 (lane 3) and Tpr (lane 5). Recombinant importin fragment β45–462 displaces full-length importin β on Nup153 (lane 4), but is unable to displace importin β on Tpr (lane 5). No binding of either form of importin β is observed on control rabbit IgG (lane 6). (B) Recombinant importin β is able to bind directly to Nup153, but not to Tpr. Anti-Nup153 (380) and anti-Tpr immunoprecipitates lacking full-length importin β were isolated from Xenopus egg extracts by pretreating the extract with GMP-PNP before immunoprecipitation and washing the beads with high salt. The washed immunoprecipitate beads from above were split into 2 tubes and either recombinant importin β in buffer RL (lanes 2 and 4), or buffer RL alone (lanes 1 and 3) was added. After incubation for 90 min, the beads were washed and bound proteins were electrophoresed, and immunoblotted with a mix of anti-Nup153 (381) and anti–importin β antibodies (lanes 1 and 2), or a mix of anti-Tpr and anti–importin β antibodies (lanes 3 and 4). Recombinant importin β was able to bind directly to Nup153 (lane 2), but not to Tpr (lane 4).
Interestingly, the importin β45–462 fragment failed to coimmunoprecipitate with Tpr, and did not displace endogenous full-length importin β from Tpr (Fig. 7 A, lane 5). These results again indicate that the Tpr–importin β complex that we observe must be intrinsically different from the Nup153/importin β complex and, once formed, is impervious to addition of recombinant β45–462 fragment.
To confirm that aa 45–462 of importin β are required for interaction with Nup153, NLS-HSA/anti-HSA beads were added to Xenopus egg extract in the presence or absence of the β45–462 mutant. Previously the nucleoporins Nup153, Nup358, and a trace of Nup62 bound to the NLS-HSA beads (Fig. 6 B, lanes 3 and 4; Fig 8, lanes 3 and 5), while Tpr, Nup214, Nup98, and Nup93 failed to bind. When the β45–462 fragment was added to an extract and the NLS-HSA beads were incubated, pelleted, and immunoblotted, Nup153 and Nup358 no longer bound to the NLS-HSA beads (Fig. 8, lane 4). This was true despite the fact that the β45–462 fragment caused no change in the total amount of endogenous importin α or β that bound to the HSA-NLS substrate (Fig. 8, lanes 1 and 2). Taken together, our results indicate that normally Nup153 and Nup358 can bind to the β subunit of an importin α/β heterodimer while it is in the process of recognizing an NLS moiety, such as the NLS-HSA beads. The β45–462 fragment can strip these nucleoporins off the column by binding to them and replacing the endogenous full-length importin β to which they are normally bound.
Do Nup153 and Tpr Bind Directly to Importin β?
The previous experiments clearly indicate that Nup153 and Tpr exist in endogenous complexes with importin β. However, it was not clear whether Nup153 and Tpr interact directly with importin β in these complexes or indirectly through intermediaries. To address this question, Nup153 and Tpr were immunoprecipitated in isolation from egg extracts supplemented with GMP-PNP (0.5 mM final), which dissociates β from these nucleoporins (as shown above). The GMP-PNP pretreated immunoprecipitates were washed with 0.5 M NaCl to remove any remaining traces of endogenous importin β. Nup153 and Tpr bound to the two sets of antibody beads were then mixed with recombinant full-length importin β in RL buffer and incubated for 90 min. To analyze whether recombinant importin β would bind directly to Nup153 or Tpr, the immunoprecipitates were immunoblotted with anti–importin β antiserum. It was found that recombinant importin β did bind to Nup153 and that importin β and Nup153 can interact with one another directly (Fig. 7 B, lane 2). In contrast, no significant amount of recombinant importin β could be detected bound to Tpr (Fig. 7 B, lane 4). Thus, recombinant importin β appears unable to bind directly to the antibody-bound Tpr. The recombinant importin β used here was found to be functional for binding to recombinant importin α: when recombinant importin β was coupled to Sepharose, it was functional for binding recombinant importin α (data not shown). In addition, the recombinant importin β was capable of binding to the nuclear pore: FITC-labeled recombinant importin β bound to the nuclear rim of reconstituted Xenopus nuclei (data not shown). These results argue that the inability of importin β to bind to Tpr directly is not due to a general defect in the recombinant importin β, but rather to some other difference.
In a separate experiment, excess amounts of recombinant importin β were added to diluted Xenopus egg extract to determine whether recombinant β would integrate into the endogenous Nup153 and Tpr complexes. It was found that recombinant importin β did bind to Nup153 and displaced the endogenous importin β from Nup153 (data not shown). However, recombinant importin β could not displace endogenous importin β from Tpr (data not shown). This result indicates that lack of binding of Tpr to recombinant importin β in the direct experiment of Fig. 7 B may not be due simply to the absence of a bridging factor, since one would expect such a factor to be present in the extract. Rather, importin β binding to Tpr may be regulated at a different level: binding could, for example, require posttranslational modification of the recombinant importin β. The nature of this binding will be an area of future interest.
Mapping of the Importin β/Nup153 Interaction Domain
To characterize the interaction of Nup153 with importin β more precisely, experiments were performed to identify the region of Nup153 that interacts directly with importin β, at least in vitro. The strategy employed was to produce radiolabeled fragments of Xenopus Nup153 in a reticulocyte lysate and then to assay the binding of the individual Nup153 fragments to importin β beads. The fragments of Xenopus Nup153 cDNA are shown in Fig. 9 A. Recombinant importin β (or control HSA) was covalently coupled to CNBr Sepharose and incubated with the above translation mixes diluted in RL buffer. No binding of the Nup153 fragments or of luciferase was detected to the control HSA beads (Fig. 9 B, HSA beads, lanes 1–4). Similarly, no binding of luciferase to importin β beads was detected. However, the importin β beads showed efficient binding (80–90%) of the Nup153 constructs that contained more than two FXFG repeats (Fig. 9 B, importin β beads, Constructs 1, 4, 5, 6, 7, and 8 in lanes of the same number). Construct 2 with two FXFG repeats and the majority of the Zn finger bound weakly (Fig. 9, lane 2). The non-FXFG fragment of Nup153 containing more NH2-terminal aa and one Zn finger domain did not bind to importin β (Fig. 9, Construct 3; importin β beads, lane 3). These results indicate in vitro that the COOH-terminal portion of Nup153, which is the FXFG repeat containing domain, is a site of direct interaction of Nup153 with importin β. The binding of Constructs 1, 4, and 8, which lack ⩾110 aa at the COOH terminus of Nup153 (Fig. 9 B, importin β beads) further indicates that the entire COOH-terminal portion is not needed for the observed importin β binding. Indeed, the binding of Constructs 4–8 demonstrates that the COOH-terminal third of Nup 153 contains redundant or cooperative β−binding domains.
Because the above experiment contained reticulocyte cytosol, we wished to determine if the Nup153 constructs can interact with recombinant importin β alone in solution. For this, two fragments of recombinant Xenopus Nup153, corresponding to Constructs 1 and 3, were produced in E. coli, purified, and assayed for their ability to bind to recombinant importin β. The proteins, 900 ng of Nup153 Construct 1 or 3 fragment and 300 ng of importin β, were incubated in RL buffer for 2 h. Only Construct 1, which contains FXFG repeats, interacted with importin β (data not shown). This result confirms that Nup153 can directly interact with importin β without the requirement for any other cytosolic protein and, moreover, that this interaction is indeed mediated through the FXFG-repeat containing region of Nup153.
The Nup153– and the Tpr–Importin β Interactions Occur in Extracts of Fully Assembled Nuclear Pores
Because the Xenopus egg extract is a mixture of disassembled pore subcomplexes and cytosolic proteins, we wished to investigate whether the Nup153– and Tpr–importin β complexes observed in egg extracts also exist in fully assembled pores. Rat liver nuclei, which were the source of the nuclear proteins previously analyzed in blot overlay assays by others, were isolated and treated with PBS 2% Triton X-100 to extract subcomplexes of assembled pores, as was done originally for the Nup62 subcomplex (Finlay et al., 1991). Immunoprecipitation from this nuclear extract was then performed using anti-Nup153, anti-Tpr, anti-Nup62, anti-Nup93, anti-rat Nup98 (Vaso, S., and D. Forbes, unpublished observation), and control rabbit IgG antibodies (Finlay et al., 1991; Powers et al., 1995; Grandi et al., 1997). (Nup214 could not be assessed in assembled pores due to lack of an appropriate antibody.) The individual immunoprecipitated pellets were then electrophoresed, transferred to membrane, and immunoblotted for the presence of importin β. No importin β was detected in Nup62, Nup93, Nup98, or rabbit IgG immunoprecipitates (Fig. 10 A, lanes 2, 5, and 6 and C, lane 3), even though all the nucleoporins were immunoprecipitated by their respective antisera (Fig. 10 B, lanes 5 and 6 and C, lane 3). However, importin β was detected in immunoprecipitates of both Nup153 and Tpr from rat liver nuclei (Fig. 10 A, lanes 3 and 4 and C, lanes 1 and 2). These results indicate that complexes of Nup153/importin β and of Tpr/importin β do indeed exist in extracts of the assembled nuclear pores of rat liver nuclei, mirroring the results found with Xenopus egg extract.
Discussion
A major question in the field of nuclear import is the molecular mechanism by which import takes place. Inherent to this question is the need to determine the in vivo docking sites for the import complex on the pore as that complex is actively transported through the pore. In vitro overlay assays have shown most of the known FXFG repeat-containing proteins to be capable of binding importin β on blots (Radu et al., 1995a ; Moroianu et al., 1995). In vitro solution binding assays have similarly observed interactions between a subset of these proteins and importin β (Radu et al., 1995b ; Rexach and Blobel, 1995; Percipalle et al., 1997). Importantly, there has been no way to illuminate what the actual situation is in vivo, specifically to probe which critical proteins of the assembled pore interact in vivo with the NLS-substrate–importin α/β complex. To address these questions, we analyzed the interactions that occur between nucleoporins and the NLS receptor within the context of a Xenopus egg extract, and then assessed the conclusions derived within the context of assembled nuclear pores themselves.
The Xenopus egg extract contains nuclear pore subcomplexes maintained in an assembly competent state, and thus contains complexes very close to an in vivo–like state. Using this extract, we set out to search for the partners of importin β by analyzing the majority of the known proteins of the vertebrate pore, Nup62, Nup93, Nup98, Nup153, Nup214, Nup358, and Tpr. Surprisingly, immunoprecipitation revealed that the nucleoporin Nup153 and the pore-associated protein Tpr each exist tightly complexed within the extract to importin β. Little to no binding of importin β to the FXFG-containing proteins Nup62 and Nup214/CAN, or to the GLFG-containing protein Nup98, either in the extract or in assembled nuclear pores, was observed. (We were unable to assess the binding of Nup214 in extracts of assembled pores due to lack of an appropriate antibody.) Similarly, no interaction of importin β with Nup93 was observed. Importin β did associate with Nup358, an interaction previously noted in Xenopus egg extracts (Saitoh et al., 1996) and HeLa cell extracts (Chi et al., 1996). In the latter study, importin α was not present in that complex, the authors speculate because extraction conditions were relatively harsh. However, we found that Nup358 binds to importin α/β and does so when the receptor is bound to NLS beads. A model of the interactions of importin β observed here is presented in Fig. 11, with β indicating interaction with importin β in the extract and in assembled pores, and the lack of β indicating no interaction in these situations. As can be seen, importin β interacts with a protein of the cytoplasmic filaments of the pore, Nup358, a protein of the basket, Nup153, and a protein of the (adjacent) pore-associated nuclear filaments, Tpr. We also saw a very small amount (⩽1%) of Nup62, a protein of the central transporter region, interacting with importin β, which leads to the speculation that this reflects an authentic interaction, but a more transient one than that which occurs with Nup153, Nup358, and Tpr.
The Nup153– and Tpr–importin β complexes were found here to be fundamentally different in their ability to exchange endogenous importin β for added recombinant β or β45–462 fragment. Another striking difference between these two importin β–binding nucleoporins lies in the ability of the α/β heterodimer to bind to the nucleoporins when simultaneously carrying its NLS-cargo. While an α/β–NLS-substrate complex can clearly interact with Nup153, importin β binding to Tpr is mutually exclusive with its participation in an α/β–NLS–protein complex. Despite these distinct characteristics, importin β binding to both Tpr and Nup153 can be disrupted by GMP-PNP.
The observed differences in the abilities of nucleoporins in the Xenopus egg extract to bind to importin β, or to importin α/β/NLS-HSA, allows one to begin to speculate upon a hierarchy of affinity of nucleoporins for importin β, with the internal basket protein Nup153 having the highest affinity. The location of Nup153 on the basket and the fact that two other basket nucleoporins, Nup98 and Nup93, as they exist in Xenopus egg extract, have no detectable affinity for importin β or for importin α/β/NLS complex, makes Nup153 a strong candidate for the termination site of protein import. The sensitivity of the interaction between Nup153 and importin β to the nonhydrolyzable GTP analogue, GMP-PNP, is consistent with a role for Ran-GTP dissociation of importin–NLS complexes from the nuclear pore after import (Rexach and Blobel, 1995; Gorlich et al., 1996; Chi and Adam, 1997; Goldfarb, 1997; Kutay et al., 1997).
In our assay, Nup358 also showed significant binding to importin–NLS complexes through importin β (Fig. 6 B). That Nup358 interacts specifically through importin β is suggested by its binding to the NLS-HSA column and its subsequent removal from this column by the β45–462 fragment (Fig. 8). This behavior and its localization in the pore are consistent with Nup358 being the initial binding site of the importin–NLS complex on the cytoplasmic fibrils extending from the pore, as suggested previously for Nup358 (Melchior et al., 1995; Chi et al., 1996), and confirmed by a recent study of Nup358 that was published after the submission of this manuscript (Delphin et al., 1997). Although Nup358 shows significant binding to importin–NLS complexes in our studies, a greater percent of Nup153 is associated with these complexes in Xenopus egg extract. This suggests a tighter binding constant for Nup153 to importin β, implying that there could indeed exist an importin β binding gradient on the pore with the strongest interaction occurring on the basket of the nuclear pore complex at Nup153. This would make sense in that the final destination of importin β, while it is complexed with an NLS protein and importin α, is the inside of the nucleus (Gorlich et al., 1995b ). Once in the nucleus, import is believed to be terminated by the binding of Ran-GTP to importin β, disrupting the importin–NLS complex. We provide direct evidence in this report that the GTP analogue GMP-PNP lowers the affinity of importin β for Nup153.
Interestingly, the pore proteins Nup214, Nup98, and Nup62 showed little affinity for the importin α/β–NLS complex or for importin β in Xenopus egg extract when compared with Nup153, Nup358, and Tpr, despite the fact that they have been shown to bind importin β in ligand blot and solution binding systems (Moroianu et al., 1995; Radu et al., 1995a,b; Rexach and Blobel, 1995; Percipalle et al., 1997). This might indicate that these proteins are unable to compete with Nup153 and Tpr for importin β in egg extract. However, the complete depletion of Nup153 from egg extract does not cause a detectable loss in the amount of importin β remaining in the extract, indicating there is a large excess of importin β relative to Nup153. Alternative explanations might suggest either that these nucleoporins, unlike Nup153 and Tpr, participate in interactions with importin β that are very transient, or that they are prevented from interacting with importin β in the extract and in the pore by their normal in vivo molecular partners (see below).
If there is an importin β binding gradient that exists on the pore with Nup153 having the highest affinity, there must be a distinction in the importin β–binding region of Nup153 compared with that of the other FXFG proteins. Nup153 interacts with importin β through its FXFG domain, but importin β shows no affinity in vitro for the FXFG region of Nup62 (Percipalle et al., 1997). Instead, importin β interacts in vitro with the coiled-coil region of Nup62 (Percipalle et al., 1997). In vitro Nup358, Nup214, and Nup98, all of which contain FG repeats of some type, interact with importin β on ligand blots, but Nup214, Nup98, and Nup62 do not do so here. The tacit assumption has been that the interaction seen on ligand blots is through their FG regions, since that is the primary common feature of these proteins; the GLFG region of Nup98 also can act as an affinity resin for importin β in vitro (Radu et al., 1995b ). The explanation for these discrepancies is not obvious, but a unique binding domain may explain the ability of Nup153 to bind importin β in the Xenopus egg extract.
What feature of the FG repeat region of Nup153 forms a binding site for importin β? Although it had been reported that yeast importin β binds to FXFG repeats in vitro, but not to yeast GLFG-containing nucleoporins (Rexach and Blobel, 1995), a larger sampling of yeast nucleoporins found that importin β does bind to certain yeast GLFG nucleoporins in vitro, the latter finding supported by genetic studies (Iovine et al., 1995; Iovine and Wente, 1997). We analyzed the FXFG region of Nup153 more closely in an attempt to establish rules for the interaction of this protein with importin β. The FG repeat domain on Xenopus Nup153 contains 38 FXFG-related repeats that are for the most part very regularly spaced. Interestingly, the repeats in the early part of the FG domain (aa 557–798 in Fig. 1) are separated by spacers that are highly charged, whereas the repeats in the latter part of the domain are separated by spacers that are largely uncharged with A, T, S, G, P, Q, and N residues. Human and rat Nup153 mirror this organization (Sukegawa et al., 1993; McMorrow et al., 1994). We find that either type of FXFG-spacer region can form an efficient binding site for importin β (Constructs 1 and 7, for example), suggesting that it is the short FXFG sequence itself, rather than the spacers, that mediates binding of importin β, at least in vitro. From our analysis, it is reasonable to hypothesize that importin β, specifically the aa 45–462 (Kutay et al., 1997), has affinity for the FXFG repeats of Nup153. Further, it is possible to propose that the affinity of importin β for the FXFG-related repeats of Nup153 is disrupted by the binding of Ran-GTP to importin β (Kutay et al., 1997).
Comparison of this region with the FXFG regions in human Nup62, Nup98, Nup214, and Nup358 reveals some differences, but no smoking gun. Nup62 with 13 FXFG-like repeats, rat Nup98 with 42 (GL)FG-related repeats, and human Nup214 with 38 repeats all contain spacers consisting almost entirely of noncharged residues. Nup358 contains FXFG repeats scattered widely throughout portions of its sequence, often separated by long stretches of unique sequence or short charged regions; there are almost no uncharged spacers. A simple analysis does not reveal major differences between the FXFG regions of Nup153 and the other nucleoporins.
Perhaps more relevant is the fact that the nucleoporins that fail to bind importin β are involved in interactions with other proteins in vivo and may be permanently or transiently unavailable to importin β. These interactions in one case involve, not importin β, but an importin β–related protein, exportin 1. Specifically, Nup214 is immunoprecipitated from an extract of human cells in complex with the newly discovered nucleoporin, Nup88, and the importin β–related export receptor, exportin 1 (Bastos et al., 1997; Fornerod et al., 1997a,b). Nup62 in rat is found in a tight complex with the nucleoporins, Nup58, Nup54, and Nup45 (Finlay et al., 1991; Kita et al., 1993; Guan et al., 1995; Buss and Stewart, 1995); a similar complex exists in the Xenopus egg extract. In the Nup62 complex, these other proteins complex with the non-FXFG region, and may cause the Nup62 complex to bind importin β much less strongly than is possible with Nup153 or Nup358, preventing it from forming a stable complex with importin β in the extract and in assembled pores. Nup98 is contained in a complex with an additional novel protein, although the binding site of this protein on Nup98 has not yet been established (Powers, M., S. Vasu, and D. Forbes, manuscript in preparation). A future understanding as to why an interaction between the nucleoporins Nup214, Nup98, Nup62 with importin β can be observed on ligand blots, but not on the pore or in Xenopus egg extracts, may further reveal molecular details of the mechanisms by which different nucleoporins contribute to nuclear import.
What is the previous evidence for Nup153 involvement in nuclear import or export? Bastos et al. (1996) found that the overexpression of full-length Nup153 in tissue culture cells resulted in the accumulation of poly(A)+ RNA. Expression of the FXFG region of Nup153 caused the same effect, even when the expressed protein was cytoplasmically localized. Minimally, these Nup153 results imply that a factor needed for export binds to Nup153 and is titrated by FXFG overexpression. No effect on the nuclear import of glucocorticoid receptor was observed in that study, thus one might conclude from these results that Nup153 is not involved in nuclear import. We find here that importin α/β binds to Nup153 and, from the NLS-HSA results, that this interaction is consistent with involvement in nuclear import. We find that importin β is in large excess over endogenous Nup153 in egg extracts. Thus, it is important to note that in the tissue culture studies of Bastos et al. (1996), unless the Nup153 fragments were present in excess over importin β, a fact that was not determined, one cannot conclude that Nup153 is uninvolved in nuclear import.
Very recently, a separate study done on recombinant fragments of Nup153 in solution found that the very COOH-terminal 17 aa of Nup153 can interact on a column with a fragment of human importin α2 in the absence of importin β (Moroianu et al. 1997). In contrast, we find that Nup153 interacts with an NLS–importin complex through importin β in a Xenopus egg extract. This was demonstrated by our removal of Nup153 from an NLS-HSA column by addition of β45–462, which cannot interact with either importin α or Ran (Fig. 8, lane 4). Moreover, addition of GMP-PNP to NLS beads removed both importin β and Nup153 from the beads, while leaving the amount of importin α bound to the NLS beads relatively unchanged (Fig. 6 C, lane 7). Direct interaction between Nup153 and importin β is also supported by the finding that importin α is removed from a Nup153 immunoprecipitate with the addition of a buffer containing a modest amount of salt and detergent, while importin β remains bound to Nup153 (Fig. 5 B). Lastly, Nup153 lacking up to 391 aa at the COOH terminus can interact in vitro with importin β on beads (Fig. 9). Thus, although it is possible that Nup153 can also interact with importin α weakly in a NLS–importin α/β complex, it appears that the burden of the interaction of Nup153 with the NLS receptor complex falls on importin β.
Tpr, the second major pore-associated protein we found to interact with importin β, is a 270-kD protein found on long filaments or hollow tubular cables extending from the basket of the pore into the nucleus (Byrd et al., 1994; Cordes et al., 1997; Ris, 1997; Zimowska et al., 1997). Unexpectedly, we found that confocal microscopy could clearly distinguish the localization of Tpr from that of nucleoporins such as Nup214 and even Nup153. We had assumed that the pore was too minute a structure in terms of light microscopy to distinguish individual regions, but clearly this is not the case. A more detailed resolution than the one achieved here may well be possible in the future. Tpr contains a predicted coiled-coil domain (aa 50–1630) in the NH2-terminal two-thirds of the protein, terminated by a highly acidic tail domain of ∼700 aa, and has no FXFG repeats (Byrd et al., 1994). The mechanistic role of Tpr and of the novel complex that we observe between Tpr and importin β is unknown. We speculate that Tpr could be a pore-binding site for importin β downstream from Nup153, a site that would have to be used only after importin α/β had dropped its NLS cargo. Alternatively, Tpr may bind importin β during its recycling in the initial steps of the export of importin β from the nucleus; however, if this were the case, one would not expect importin α to still be bound.
The inability of recombinant importin β, which we show is functional for binding to importin α, Nup153, and nuclear pores, to bind to Tpr suggests several possibilities. One is that the Tpr–importin β complex is extremely stable; however, if this is the explanation one would have to also explain why salt-stripped Tpr cannot rebind recombinant importin β. A second possibility is the potential existence of a population of modified endogenous importin β in the extract. This population could possess a modification required for Tpr binding that the recombinant version lacks. Future characterization of Tpr's interaction with importin β may reveal further interesting regulation of this transport factor in nuclear protein import.
In summary, we have identified components of the nuclear pore and its attached fibrils, Nup153 and Tpr, that form strong interactions with importin β under multiple physiological and in vivo–like conditions, including assembly competent Xenopus egg extracts and extracts of isolated rat liver nuclei. Nup153 and Tpr can complex to importin β alone, the importin α/β heterodimer and, in the case of Nup153, but not Tpr, the α/β heterodimer complexed to an NLS-substrate. This latter finding is consistent with a role for Nup153 in nuclear import, as is our finding that GMP-PNP disrupts the complex, as if it were a terminal complex in import, rather than an initiating complex in export. We think it highly likely that Nup153 and Tpr will also play roles in export, both from their localization and from the dominant negative effects of certain Nup153 fragments on export (Bastos et al., 1996). In the course of this study, we also found evidence for the interaction of Nup358 with importin β in Xenopus egg extract. We could not find evidence for interaction between Nup62, Nup93, Nup98, or Nup214 with importin β, we speculate either because of the potential transient nature of such complexes (in the case of Nup62) or because the nucleoporins play other roles in the nuclear pore, such as Nup98 in RNA export (Powers et al., 1997) or Nup93 in pore assembly (Grandi et al., 1997). The primary role of Nup214/CAN may be in export (or recycling) as this pore protein has been found in a complex with exportin 1, rather than with importin β, when assembled nuclear pores were analyzed (Fornerod et al., 1997a ). It will be interesting in the future to dissect the exact roles of Nup153 and Tpr in import and to determine what dual roles they may play in nuclear export. It will be equally interesting to determine whether the FG regions of other nucleoporins each serve as unique interaction sites, or landing pads, for individual members of the recently discovered extended family of importins and exportins.
Acknowledgments
We wish to thank Dr. Dirk Gorlich for the kind gift of importin α and β clones and antisera and Dr. Thomas Joos for the variant integrin cDNA clone, α5tr. We thank Drs. Brian Miller, Katherine Ullman, and Sanjay Vasu for helpful reading of the manuscript, Drs. Maureen Powers and Brian Miller for helpful discussions, and Bismarck Oh for technical help.
Abbreviations used in this paper
- aa
amino acid
- NLS
nuclear localization sequence
- PVDF
polyvinylidene difluoride
Footnotes
S. Tugendreich was a Damon Runyon-Walter Winchell Cancer Society Postdoctoral Fellow and S. Shah was a Gann Predoctoral Fellow. This work was supported by National Institutes of Health (GM33279) and American Cancer Society (CB no. 199) grants to D. Forbes.
Dr. Tugendreich's present address is Iconix Pharmaceuticals, Inc., Mountain View, CA.
References
- Adam EJ, Adam SA. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J Cell Biol. 1994;125:547–55. doi: 10.1083/jcb.125.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW, Radermacher M. Architecture of the Xenopusnuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Ribas de Pouplana L, Enarson M, Bodoor K, Burke B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J Cell Biol. 1997;137:989–1000. doi: 10.1083/jcb.137.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Stewart M. Macromolecular interactions in the nucleoporin p62 complex of rat nuclear pores: binding of nucleoporin p54 to the rod domain of p62. J Cell Biol. 1995;128:251–261. doi: 10.1083/jcb.128.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd DA, Sweet DJ, Pante N, Konstantinov KN, Guan T, Saphire AC, Mitchell PJ, Cooper CS, Aebi U, Gerace L. Tpr, a large coiled coil protein whose amino terminus is involved in activation of oncogenic kinases, is localized to the cytoplasmic surface of the nuclear pore complex. J Cell Biol. 1994;127:1515–1526. doi: 10.1083/jcb.127.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam SA. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol Biol Cell. 1997;8:945–956. doi: 10.1091/mbc.8.6.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Visser GD, Adam SA. RanBP1 stabilizes the interaction of Ran with p97 nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson WD, Kent HM, Stewart M. Separate binding sites on nuclear transport factor 2 (NTF2) for GDP-Ran and the phenylalanine-rich repeat regions of nucleoporins p62 and Nsp1p. J Mol Biol. 1996;263:517–524. doi: 10.1006/jmbi.1996.0594. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. The NTF2 gene encodes an essential, highly conserved protein that functions in nuclear transport in vivo. J Biol Chem. 1996;271:18477–18484. doi: 10.1074/jbc.271.31.18477. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LC, Laskey RA. DNA replication occurs at discrete sites in pseudonuclei assembled from purified DNA in vitro. Cell. 1991;66:271–275. doi: 10.1016/0092-8674(91)90617-8. [DOI] [PubMed] [Google Scholar]
- Dabauvalle MC, Loos K, Scheer U. Identification of a soluble precursor complex essential for nuclear pore assembly in vitro. Chromosoma. 1990;100:56–66. doi: 10.1007/BF00337603. [DOI] [PubMed] [Google Scholar]
- Dasso M, Newport JW. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. . Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- Dasso M, Dimitrov S, Wolffe AP. Nuclear assembly is independent of linker histones. Proc Natl Acad Sci USA. 1994;91:12477–12481. doi: 10.1073/pnas.91.26.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI. The nuclear pore complex. Annual Review of Biochemistry. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- Delphin C, Guan T, Melchior F, Gerace L. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol Biol Cell. 1997;8:2379–2390. doi: 10.1091/mbc.8.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? . Trends Biochem Sci. 1991;16:478–81. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Dworetzky SI, Lanford RE, Feldherr CM. The effects of variations in the number and sequence of targeting signals on nuclear uptake. J Cell Biol. 1988;107:1279–1287. doi: 10.1083/jcb.107.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: Transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Boer J, van Baal S, Jaegle M, von Lindern M, Murti KG, Davis D, Bonten J, Buijs A, Grosveld G. Relocation of the carboxy-terminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene. 1995;10:1739–1748. [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj I. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997a;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. Eur Mol Biol Organ J. 1997b;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker M, Hollinshead M, White N, Vaux D. Interphase nuclei of many mammalian cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol. 1997;136:531–544. doi: 10.1083/jcb.136.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Allen TD. High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J Cell Biol. 1992;119:1429–1440. doi: 10.1083/jcb.119.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DS. Whose finger is on the switch? . Science. 1997;276:1814–1816. doi: 10.1126/science.276.5320.1814. [DOI] [PubMed] [Google Scholar]
- Gorlich D. Nuclear protein import. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995a;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995b;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Panté N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homolog of yeast Nic96, forms a complex with a novel 205 kD protein and is required for correct nuclear pore assembly. Mol Biol Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Muller S, Klier G, Pante N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol Biol Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 726 pp.
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kD component of nuclear pore-targeting complex in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Wente SR. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos TO, Whittaker CA, Meng F, DeSimone DW, Gnau V, Hausen P. Integrin alpha 5 during early development of Xenopus laevis. . Mech Dev. 1995;50:187–199. doi: 10.1016/0925-4773(94)00335-k. [DOI] [PubMed] [Google Scholar]
- Kita K, Omata S, Horigome T. Purification and characterization of a nuclear pore glycoprotein complex containing p62. J Biochem (Tokyo) 1993;113:377–382. doi: 10.1093/oxfordjournals.jbchem.a124054. [DOI] [PubMed] [Google Scholar]
- Kraemer D, Wozniak RW, Blobel G, Radu A. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci USA. 1994;91:1519–1523. doi: 10.1073/pnas.91.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO (Eur Mol Biol Organ) J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey RA, Leno GH. Assembly of the cell nucleus. Trends Genet. 1990;6:406–410. doi: 10.1016/0168-9525(90)90301-l. [DOI] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- McMorrow I, Bastos R, Horton H, Burke B. Sequence analysis of a cDNA encoding a human nuclear pore complex protein, hnup153. Biochim Biophys Acta. 1994;1217:219–223. doi: 10.1016/0167-4781(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. (Published erratum appears in J. Cell Biol. 1994. 124:217.) . J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Guan T, Yokoyama N, Nishimoto T, Gerace L. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J Cell Biol. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. Eur Mol Biol Organ. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moore MS, Blobel G. Purification of a Ran-interacting protein that is required for protein import into the nucleus. Proc Natl Acad Sci USA. 1994;91:10212–10216. doi: 10.1073/pnas.91.21.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1/β and α2/β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. Nuclear protein import: Ran-GTP dissociates the karyopherin alphabeta heterodimer by displacing alpha from an overlapping binding site on beta. Proc Natl Acad Sci USA. 1996;93:7059–7062. doi: 10.1073/pnas.93.14.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. RanGTP-mediated nuclear export of karyopherin alpha involves its interaction with the nucleoporin Nup153. Proc Natl Acad Sci USA. 1997;94:9699–9704. doi: 10.1073/pnas.94.18.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer DD, Forbes DJ. Nuclear import can be separated into distinct steps in vitro: nuclear pore binding and translocation. Cell. 1988;52:641–653. doi: 10.1016/0092-8674(88)90402-3. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Finlay DR, Forbes DJ. In vitro transport of a fluorescent nuclear protein and exclusion of non-nuclear proteins. J Cell Biol. 1986;103:2091–2102. doi: 10.1083/jcb.103.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, Spann T. Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987;48:219–230. doi: 10.1016/0092-8674(87)90425-9. [DOI] [PubMed] [Google Scholar]
- Pante N, Aebi U. The nuclear pore complex. J Cell Biol. 1993;122:977–984. doi: 10.1083/jcb.122.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P, Clarkson WD, Kent HM, Rhodes D, Stewart M. Molecular interactions between the importin α/β heterodimer and proteins involved in vertebrate nuclear protein import. J Mol Biol. 1997;266:722–732. doi: 10.1006/jmbi.1996.0801. [DOI] [PubMed] [Google Scholar]
- Pollard VM, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995a;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995b;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Mills AD, Dilworth SM, Laskey RA, Dingwall C. Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell. 1988;52:655–664. doi: 10.1016/0092-8674(88)90403-5. [DOI] [PubMed] [Google Scholar]
- Ris H. Three-dimensional imaging of cell ultrastructure with high resolution low voltage SEM. Inst Phys Conf Ser. 1989;98:657–662. [Google Scholar]
- Ris H. The three dimensional structure of the nuclear pore complex as seen by high voltage electron microscopy and high resolution low voltage scanning electron microscopy. Electron Microsc Soc Am. 1991;21:54–56. [Google Scholar]
- Ris H. High-resolution field-emission scanning electron microscopy of the nuclear pore complex. Scanning. 1997;19:368–375. doi: 10.1002/sca.4950190504. [DOI] [PubMed] [Google Scholar]
- Rush MG, Drivas G, D'Eustachio P. The small nuclear GTPase Ran: how much does it run? . Bioessays. 1996;18:103–112. doi: 10.1002/bies.950180206. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Cooke CA, Burgess WH, Earnshaw WC, Dasso M. Direct and indirect association of the small GTPase ran with nuclear pore proteins and soluble transport factors: studies in Xenopus laevisegg extracts. Mol Biol Cell. 1996;7:1319–1334. doi: 10.1091/mbc.7.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. C. Nolan, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Ullman KS, Forbes DJ. RNA polymerase III transcription in synthetic nuclei assembled in vitro from defined DNA templates. Mol Cell Biol. 1995;15:4873–4883. doi: 10.1128/mcb.15.9.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshawsky D, Miller L. Tissue-specific in vivo protein-DNA interactions at the promoter region of the Xenopus63 kD keratin gene during metamorphosis. Nucleic Acids Res. 1995;23:4502–4509. doi: 10.1093/nar/23.21.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP. Replication timing and Xenopus5S RNA gene transcription in vitro. Dev Biol. 1993;157:224–231. doi: 10.1006/dbio.1993.1126. [DOI] [PubMed] [Google Scholar]
- Wong DH, Corbett AH, Kent HM, Stewart M, Silver PA. Interaction between the small GTPase Ran/Gsp1p and Ntf2p is required for nuclear transport. Mol Cell Biol. 1997;17:3755–3767. doi: 10.1128/mcb.17.7.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- Zimowska G, Aris JP, Paddy MR. A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci. 1997;110:927–944. doi: 10.1242/jcs.110.8.927. [DOI] [PubMed] [Google Scholar]