Abstract
A highly enriched spindle pole preparation was prepared from budding yeast and fractionated by SDS gel electrophoresis. Forty-five of the gel bands that appeared enriched in this fraction were analyzed by high-mass accuracy matrix-assisted laser desorption/ ionization (MALDI) peptide mass mapping combined with sequence database searching. This identified twelve of the known spindle pole components and an additional eleven gene products that had not previously been localized to the spindle pole. Immunoelectron microscopy localized eight of these components to different parts of the spindle. One of the gene products, Ndc80p, shows homology to human HEC protein (Chen, Y., D.J. Riley, P-L. Chen, and W-H. Lee. 1997. Mol. Cell Biol. 17:6049–6056) and temperature-sensitive mutants show defects in chromosome segregation. This is the first report of the identification of the components of a large cellular organelle by MALDI peptide mapping alone.
The mitotic spindle is a complex dynamic organelle that uses mechanochemical forces to separate chromosomes during cell division. Many proteins must be involved and a substantial proportion, in particular structural components of the centrosome and the kinetochore, remain to be identified. One of the most intensively studied spindles is that of Saccharomyces cerevisiae, where a combination of genetic and biochemical studies has identified a number of spindle components. Kinetochore components have been identified by purification of a centromere DNA-binding complex (Lechner and Carbon, 1991), and also from genetic screens principally based on chromosome loss (Doheny et al., 1993; Goh and Kilmartin, 1993; Strunnikov et al., 1995). Similar types of genetic screens have also identified mitotic kinesins (Hoyt et al., 1992; Roof et al., 1992), and a combination of genetic and biochemical screens (for reviews see Rose et al. [1993]; Winey and Byers [1993]; Pereira and Schiebel [1997]) have identified components of the spindle pole body (SPB),1 the functional equivalent of the centrosome in S. cerevisiae. A more complete description of the yeast mitotic spindle will probably require a combination of further genetic screens and more extensive biochemical analysis.
Major problems associated with biochemical analysis are the very low abundance of spindle components and the preparation of parts of the mitotic spindle pure enough to allow identification of individual components. We have previously described a preparation of yeast spindle poles (Rout and Kilmartin, 1990) that were ∼10% pure, sufficient to identify four SPB and spindle components from a monoclonal screen (Rout and Kilmartin, 1990, 1991). The three SPB components, Spc110p, Spc42p, and Spc98p (90-kD component) have essential roles in SPB function (Donaldson and Kilmartin, 1996; Geissler et al., 1996; Kilmartin and Goh, 1996; Stirling et al., 1996; Sundberg et al., 1996), and the fourth spindle component, Ndc80p, is described in this article. This spindle pole preparation was not pure enough to recognize gel bands that might correspond to less abundant spindle pole components, and in any case because of the limitations of gradient centrifugation, it was not possible to scale up sufficiently to give the quantities necessary for sequence identification by the Edman procedure.
Two recent developments have now allowed the identification of very small amounts of material in gel bands. First, with the completion of the S. cerevisiae genome sequencing project (Goffeau et al., 1996), the sequence of every yeast protein is now available in public databases. Second, peptide mass maps of very small amounts of enzymatically digested proteins obtained by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry are now sufficiently accurate to screen databases and identify proteins whose sequence is already known. With these developments in mind we have increased the purity of our previous spindle pole preparation so that we can now identify novel components of the yeast spindle pole by MALDI peptide mass mapping of SDS gel bands.
Materials and Methods
Preparation of Spindle Poles
Yeast spindle poles were prepared by a modification of the earlier method (Rout and Kilmartin, 1990). The isolation of nuclei was scaled up threefold and the spindle poles were enriched by sucrose velocity and equilibrium gradients followed by a modified Percoll gradient. Nuclei from 40 liters of Saccharomyces uvarum cells harvested at 2 × 107 cells/ml were pelleted (each Beckman Ty 70 tube [Becton Instruments, Inc., Palo Alto, CA] contained 250 OD260nm of nuclei which, for example, would correspond to 25 ml of nuclei with an OD of 10). Lysis buffer (Rout and Kilmartin, 1990) with 20 μg/ml RNase A was added (2.5 ml per tube) and vigorously whirlimixed to lyse the nuclei and release spindle poles. The pH was raised by addition of 0.25 ml of 0.1 M bis-tris (bt)-Cl, pH 6.5, buffer and then the tube was warmed to room temperature for 2–3 min to digest DNA and RNA. The tubes were spun at 2,000 rpm for 6 min in the Beckman Ty 70 rotor (Beckman Instruments, Inc.) and the supernatant was removed avoiding the floppy part of the pellet. A sucrose gradient was prepared from sucrose-bt solutions (Rout and Kilmartin, 1990) containing 0.01% Tween 20 (Sigma Chemical Co., St. Louis, MO) and DMSO supplements. The gradient was made by overlaying 10 ml of 0.8 M (20% DMSO), 1.2 M (15% DMSO), 1.6, and 2.0 M (10% DMSO) sucrose-bt in a SW28 tube (Beckman Instruments, Inc.), sealing the top with Parafilm (American National Can, Chicago, IL) and then placing it horizontally for 4 h at 4°C. Sufficient sucrose was removed from the top of this gradient to accommodate the supernatant from two Ty 70 tubes (Beckman Instruments, Inc.). The gradient was spun at 28,000 rpm for 50 min at 4°C. The spindle poles were visible as a broad band straddling the former 1.6/2.0 M sucrose interface. The sample was further purified and concentrated by a sucrose equilibrium gradient. The sample was incorporated into the solutions for the gradient, which, if there were no sample present, was prepared from overlaying 3.2 ml of 1.75, 2.0, 2.25, and 2.5 M sucrose-bt (with 0.01% Tween 20) in SW40 tubes (Beckman Instruments, Inc.), which were left horizontal for 4 h at 4°C to prepare the gradient. The velocity gradient sample was incorporated into this gradient by adjusting its sucrose concentration to replace one or more of the layers. The equilibrium gradient was spun at 38,900 rpm for 21 h at 4°C. The spindle poles banded at 2.22 M sucrose. They were further enriched on a modified Percoll gradient as previously described (Rout and Kilmartin, 1990), except that for each 2 ml of spindle poles, 9.7 ml 2.5 M sucrose, 2.4 ml Percoll, l and 1.5 ml DMSO were added. The concentrations of GTP, EGTA, solution P (90 mg phenylmethylsulfonylfluoride and 2 mg pepstatin in 5 mL absolute ethanol), and DTT were as before (Rout and Kilmartin, 1990). The gradient was spun in an angle head Ty 70 rotor (Beckman Instruments, Inc.) at 35,000 rpm for 3 h at 4°C.
MALDI Mass Spectrometry and Protein Identification
Enriched protein bands were excised and prepared for mass spectrometric analysis as described (Shevchenko et al., 1996; Wilm et al., 1996; Jensen et al., 1997b ) using a robotic workstation (Houthaeve et al., 1995). The data for the MALDI identifications for the 11 new spindle pole components are shown in Table I. The rest of the data are shown below. These are for the components already known to be at the spindle pole and for those gene products where epitope tagging did not show spindle pole staining under the particular fixation conditions used (Kilmartin et al., 1993). These data are shown as first the gene product name, then in parentheses are the number of tryptic peptides identified and then the percent of sequence covered by these peptides. These gene products in order of apparent size on the SDS gel were: Spc110p dimer (17 peptides, 23%), Stu1p (7, 6%), Pom152p (12, 12%), RNA pol I or Ypr010p (14, 17%), Kip1p (12, 17%), Spc110p (20, 27%), Stu2p (25, 30%), Spc97p (12, 20%), Ylr127p (12, 13%), Spc98p (15, 22%), Nic96p (15, 19%), Kar3p (22, 35%), Nup85p (12, 21%), acetyl CoA synthetase 2 or Ylr153p (14, 25%), Sgv1p (9, 24%), Ssa1p (19, 38%), Ssb1p (7, 18%), Tub1p (8, 30%), Tub2p (10, 32%), Nuf2p (9, 34%), EF1α or Ypr080p (9, 35%), Ynl011p (7, 22%), Spc42p (14, 38%), Ygl075p (6, 20%), Bim1p (11, 35%), L2A or Ybr031p (6, 25%), Yhr076p (6, 20%), Nip29p (5, 20%), ribosomal L7A-1 or Yll045p (10, 41%), ribosomal S4 or Yhr203p (9, 45%), phosphomannomutase or Yfl045p (7, 41%), Cdc31p (6, 31%), histone H4 or Ynl030p (7, 66%), and calmodulin or Cmd1p (4, 41%). References for these genes can be found by searching the Yeast Proteome home page (http://www.proteome.com).
Table I.
Summary of Gene Products Found by MALDI Mass Spectrometry That Localize to the Yeast Spindle Pole
| Sequence name | Gene product | Other name | MW | MALDI | Coiled coil | Disruption | Location | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| peptides | seq | IF and/or GFP staining | ImmunoEM | |||||||||||||||
| (%) | ||||||||||||||||||
| YGL093w | Spc105p | 104.8 | 7 | 9 | weak | slow growth* | pole | pole | ||||||||||
| YOR373w | Nud1p | 94.1 | 13 | 23 | none | lethal | pole | SPB OP | ||||||||||
| YIL144w | Ndc80p | Tid3p | 80.5 | 18 | 38 | strong | lethal | pole + spindle | pole + spindle | |||||||||
| YAL047c | Spc72p | Spi6p | 72.1 | 16 | 27 | weak | slow growth | pole | SPB OP | |||||||||
| YNL225c | Cnm67p | 67.4 | 11 | 19 | strong | slow growth | pole | SPB OP | ||||||||||
| YPL255w | Bbp1p | 45.4 | 8 | 23 | weak | lethal | pole | — | ||||||||||
| YKR037c | Spc34p | 34.1 | 6 | 27 | weak | lethal | pole + spindle | pole + spindle | ||||||||||
| YPL124w | Spc29p | Nip29p | 29.3 | 5 | 20 | none | lethal | pole | — | |||||||||
| YER018c | Spc25p | 25.2 | 6 | 41 | weak | lethal | pole | pole | ||||||||||
| YMR117c | Spc24p | 24.6 | 5 | 28 | strong | lethal | pole | pole | ||||||||||
| YDR201w | Spc19p | 18.9 | 5 | 29 | strong | lethal | pole + spindle | pole + spindle | ||||||||||
The MALDI data is shown as number of peptides identified (peptides) and percentage of sequence covered (% seq). The coiled-coil data is classified using the Paircoil score (Berger et al., 1995) with strong defined as above a probability of 0.95 and weak as above 0.4. The disruptions of Spc72p, Cnm67p, and Bbp1p were done by S. Soues, Brachat et al. (1998), and Xue et al. (1996). Cnm67p was localized by immunofluorescence by Brachat et al. (1998). ImmunoEM for Ndc80p was published previously (Rout and Kilmartin, 1990). For the immunoEM results, pole staining was always on the nuclear side of the SPB. IF, immunofluorescence; MT, microtubule; OP, outer plaque.
Refer to Materials and Methods.
Epitope Tagging and Disruptions
Genes identified by MALDI mass spectrometry were tagged in the genome directly by PCR (Baudin et al., 1993). The cassette used for the PCR contained three tandem copies of the hemagglutinin (HA) epitope as a NotI fragment in pBluescript (Tyers et al., 1993). A stop codon was inserted by blunting the polylinker XbaI site, which should insert the sequence YPYDVPDYAGYPYDVPDYAGSYPYDVPDYAAQCGRSS* at the COOH terminus. Alternatively, a NotI fragment encoding GFP5 (Siemering et al., 1996) replaced the HA tag, which was designed to insert a short polypeptide linker AGAGA between the protein and the green fluorescent protein (GFP). Transformants were selected with an expression module containing the Saccharomyces pombe HIS5 gene (Wach et al., 1997) inserted as a EcoRI/HindIII fragment in the pBluescript polylinker. Transformants were checked with appropriate primers by colony PCR to show the presence of the tag and later the absence of the wild-type gene. A diploid strain K842 (Nasmyth et al., 1990) was transformed and sporulated to compare growth rates of tagged and untagged spores, and in all cases the histidine (His)+ marker segregated 2:2 and growth rates of the four spores were indistinguishable. In the case of GFP-tagged Spc72p and Cnm67p, the isogenic haploid strain K699 was transformed and both these tagged strains grew at normal rates. Strains that showed positive spindle pole staining by immunofluorescence (Kilmartin et al., 1993) were checked by immunoblotting to determine that the correctly sized HA-tagged protein was present. In all cases after subtraction of the 4.2 kD contributed by the tag, a band was detected within 3–15% of the size measured from the gel used to prepare the MALDI samples (see Fig. 3). Three of the proteins have discrepancies of greater than 15% between the measured and calculated molecular weights: Spc105p runs at an apparent size of 147 kD, Spc72p at 85 kD, and Spc19p at 23 kD.
Figure 3.
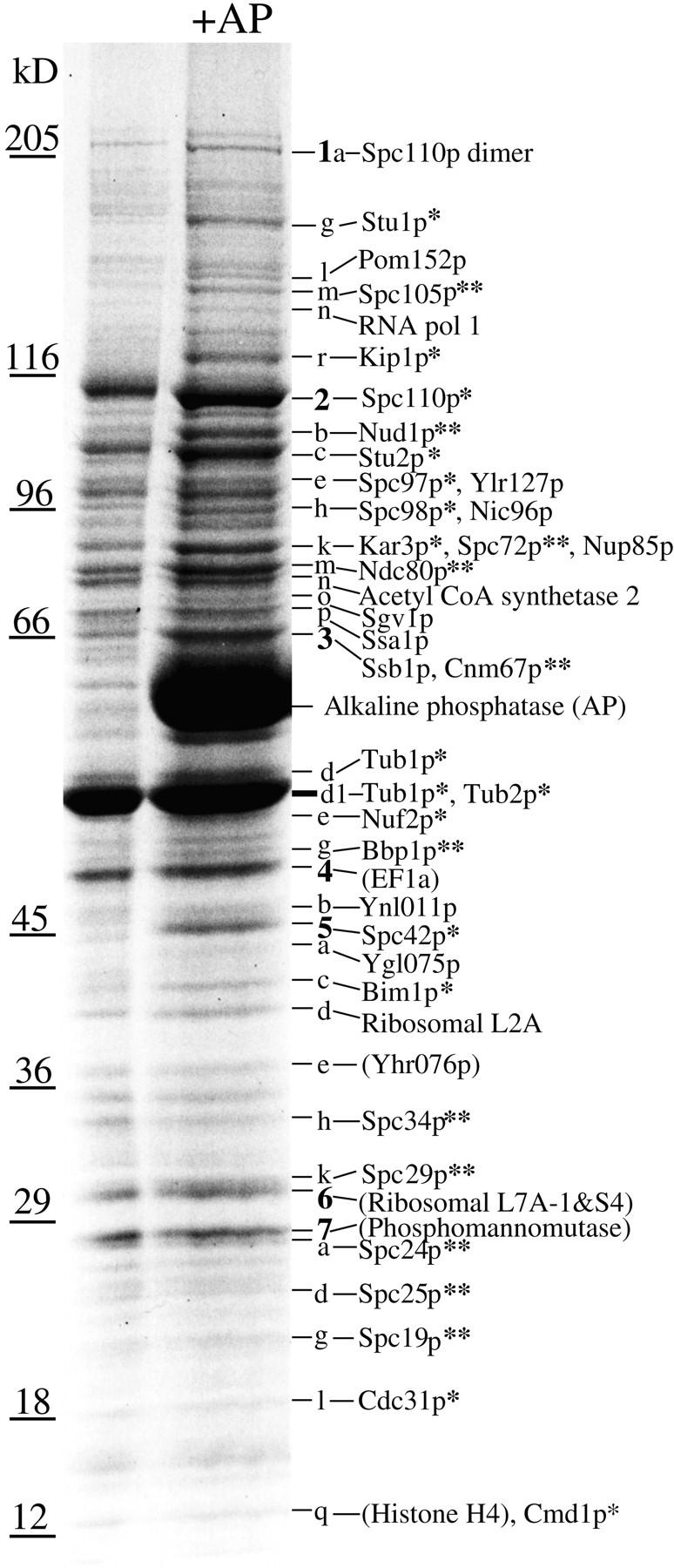
Identification of proteins in Coomassie-stained SDS gel bands from AP-treated yeast spindle poles by MALDI mass mapping. Single asterisk, a spindle pole component already identified; double asterisk, a new spindle pole component. Bands were labeled by giving all the prominent bands numbers (boldface) then bands underneath were given that number and a letter in order of decreasing size until the next prominent numbered band. Identifications in parentheses are from prominent bands that are not enriched in the spindle pole preparation. The faint Coomassie-stained bands between 205 and 116 kD contain ∼10–20 ng of protein by comparison with BSA standards.
Disruptions were prepared by PCR by using appropriate primers to amplify the S. pombe HIS5 module (see above) and transforming K842 to disrupt one copy of the open reading frame. sporulation gave two viable (always His−) and two inviable spores. The inviable spores germinated to give between two and six cells or buds except for SPC105 which gave 20–30 cells or buds. With the exception of SPC105, the lethality was due to the presence of the disrupted gene since transformation of the disrupted diploid with the wild-type gene (isolated by gap repair or PCR) in the CEN URA3 plasmid pRS316, and sporulation now often gave four viable spores. Two of these were His+ and Ura+, showing that spores containing the disrupted gene were rescued by the plasmid (this experiment was not done for NUD1). These His+Ura+ progeny were dependent on the plasmid for growth since selection against the URA3 plasmid on 5-fluoroorotic medium gave no colonies. In the case of SPC105 some extremely slow growing colonies appeared. Colony PCR showed that the wild-type open reading frame was absent. When these cells were backcrossed to wild type and sporulated, it was clear that additional mutations were present. Because of these additional mutations and the extremely slow growth rate, this strain was not studied further.
Microscopy
Cells containing HA tags were processed for immunofluorescence using a rapid fixation method (Kilmartin et al., 1993). ndc80 cells were fixed in 3.7% formaldehyde for 1 h and then after processing for immunofluorescence (Kilmartin and Adams, 1984), the cells were stained with rabbit anti-Tub4p and rat anti-tubulin. Cells containing GFP tags were examined without fixation. All images were acquired with a cooled charge-coupled device camera (model RTEA/CCD-1800-Y, Princeton Instruments, Trenton, NJ).
For immunoEM the cells were fixed in formaldehyde for 20 min, the cell wall removed, and then the cells were incubated with either rabbit anti-GFP (a generous gift of K. Sawin, Imperial Cancer Research Foundation, London, UK) or mouse anti-HA mAb 12CA5 and then Nanogold Fab anti-rabbit IgG or anti-mouse IgG (all from Nanoprobes, Stony Brook, NY). Cells were glutaraldehyde-fixed, silver enhanced, treated with osmium and uranyl acetate, and then embedded.
Cloning and Mutagenesis of NDC80
Cell culture supernatant from the anti–80-kD spindle component mAb 34E12 (Rout and Kilmartin, 1990) was used to screen a λgt11 genomic DNA expression bank (this experiment was performed by A. Donaldson [Donaldson and Kilmartin, 1996]). Two sets of unrelated phage were isolated. One set was sequenced and found to encode an open reading frame of 80.5-kD, identical to Yil144p which we called Ndc80p. A deletion was made by inserting the LEU2 gene between bases 33 (using an ExoIII fragment) and an NsiI site inserted at the end of the open reading frame at 2076 (bases are numbered from the A of the putative initiator ATG). A linear fragment from −1002 to 4214 was transformed into K842 to disrupt one copy of NDC80 (Rothstein, 1983) and the correct integration was confirmed by Southern blotting. Sporulation of this strain gave two viable (all Leu−) and two inviable spores that germinated but arrested with large buds. We showed that this inviability was due to the disruption of NDC80 in the same way as the other disruptions (see above).
A triple HA epitope tag was inserted into the open reading frame between residues 116 and 117. A blunted SacI-ClaI fragment of the pBluescript polylinker containing a NotI fragment encoding a triple HA epitope was inserted in frame into the blunted StyI site at base 346. This tagged version of Ndc80p was able to rescue a deletion and maintain normal growth rates.
Temperature-sensitive (ts) mutations in NDC80 were prepared by mutagenic PCR (Foreman and Davis, 1993; Donaldson and Kilmartin, 1996) using suitable primers to amplify between an NdeI site inserted at −1 and the NsiI site at 2076. Two alleles were obtained, ndc80-1 and ndc80-2. The alleles were sequenced and ndc80-1 was found to have the following changes: Q2L, S73N, K122R, H313R, N321S, E389V, E429D, T473S, I490V, D556G, K613I, H648R, I658T; the A at 2023 was missing, causing a frameshift that together with an A2067T change and the base changes due to the NsiI site at 2076, would change the last 18 residues from IEELRNLEFETEHNVTNA* to LKSYEIWSLKLNITLQMHKINDI*. This frameshift was removed by PCR but this plasmid now no longer conferred the ts phenotype except for one transformant which, due to a PCR error, contained the additional change F598V (this allele was called ndc80-3). The allele ndc80-2 had the changes S56C, K305M, F308L, L406I, T473P, I596T, and E665D. Both the ndc80-1 and ndc80-2 mutations were transferred to the endogenous NDC80 locus, but this was not possible for the ndc10-3 allele. Here the mutagenized plasmid was integrated at the Trp1 locus in a strain containing a deletion of the NDC80 gene. Both ndc80-1 and ndc80-2 alleles were recessive, grew normally at 23°C and were fully complemented at 36°C by the NDC80 gene on a CEN plasmid. Synthetic lethal interactions were tested by crossing ndc80-1 and ndc80-2 with ndc10-1 (Goh and Kilmartin, 1993), ndc10-2 (Kopski and Huffaker, 1997), mps3-1 (a mutation in NDC10 from E. Siewert and M. Winey, University of Colorado, Boulder, CO), ctf14-42 and ctf13-30 (Doheny et al., 1993), and cep3-1 and cep3-2 (Strunnikov et al., 1995). In all cases spore viability was between 90 and 100% and the double ts mutants were recovered with the expected frequency.
Results
A Highly Enriched Yeast Spindle Pole Preparation
We described earlier a yeast spindle pole preparation that contained SPBs with nuclear or spindle microtubules attached (Rout and Kilmartin, 1990). This preparation was not pure enough to recognize gel bands that might correspond to individual spindle pole components even after further purification by extraction with DEAE-dextran or heparin (Rout and Kilmartin, 1990). Moreover, although these extraction procedures retain SPB components, they remove nuclear microtubules and thereby any microtubule-associated spindle pole components. Because we wished to identify both of these sets of components, it was important in any improved procedure to keep the nuclear microtubules attached but the dynamic nature of microtubules restricts the number of options available for further purification. We modified the previous spindle pole preparation by adding an extra velocity gradient step and by scaling up threefold to produce the quantities necessary to identify low abundance components (refer to Materials and Methods). Since many SPB components are phosphorylated (Donaldson and Kilmartin, 1996; Friedman et al., 1996; Knop et al., 1997) and thus would run heterogeneously on SDS gels, we treated the preparation with alkaline phosphatase (AP) to sharpen the bands on the 8–13% gradient SDS gels (Donaldson and Kilmartin, 1996). We then compared SDS gel tracks (Fig. 1) of two adjacent gradient fractions, one of which contained most of the spindle poles, to identify gel bands that might correspond to spindle pole components. A total of ∼80 gel bands were identified of which ∼45 seemed to be enriched in the spindle pole fraction. Two of the most abundant bands were identified by immunoblotting and contained Spc110p and Spc42p, two previously characterized SPB components (Kilmartin et al., 1993; Donaldson and Kilmartin, 1996). The gel demonstrates that Spc42p is highly phosphorylated (Donaldson and Kilmartin, 1996) and is not detectable by Coomassie staining in the absence of phosphatase treatment. If the Spc110p and Spc42p gel bands are reasonably clean, then we can estimate from the number of spindle poles loaded and the intensity of Coomassie staining that there are ∼1,000 copies each of Spc110p and Spc42p per diploid SPB.
Figure 1.
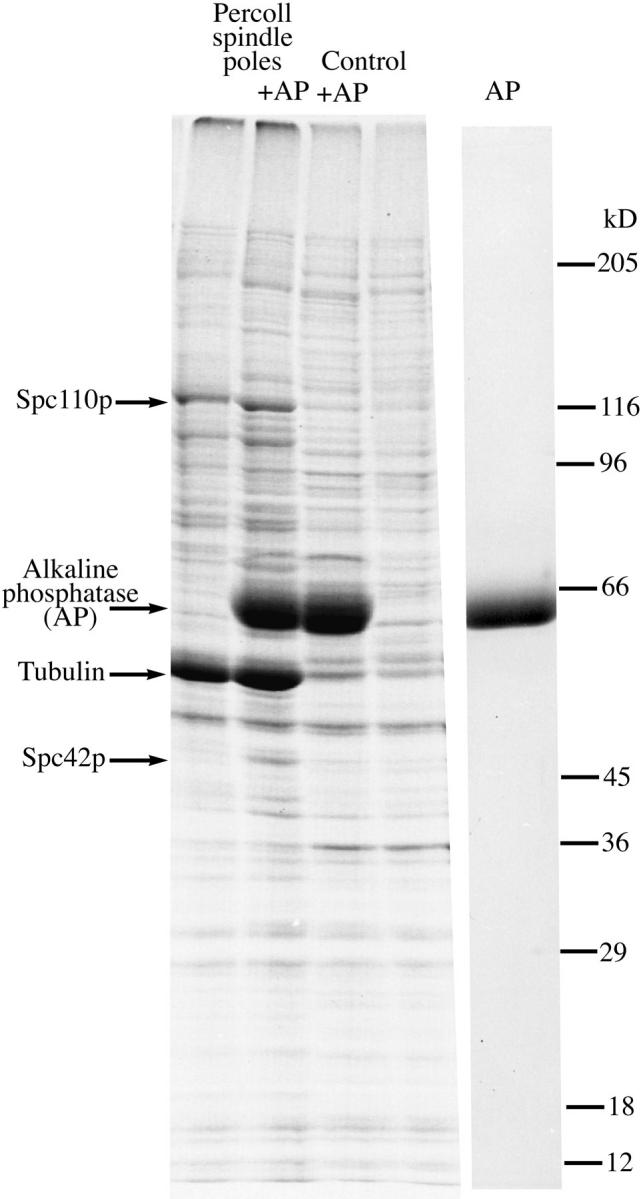
SDS gel of two adjacent fractions from the final Percoll gradient. The first two lanes contain most of the spindle poles treated with and without alkaline phosphatase (AP), the next two lanes labeled control contain the next gradient fraction which has less than 15% of the poles present in the first fraction. A large number of gel bands are specifically enriched in the spindle pole fraction. The last lane is AP alone.
MALDI Mass Spectrometric Analysis of the Yeast Spindle Poles
We applied high-mass accuracy MALDI peptide mass mapping to the analysis of the Coomassie-stained gel bands from the yeast spindle pole preparation. In this method no sequence information is obtained, instead the masses of a set of tryptic peptides derived from the proteins in the gel band are measured by MALDI and then compared with the calculated tryptic peptide masses for each entry in the database. The specificity of this approach depends mainly on the number of measured peptide masses and the peptide mass accuracy. We have previously shown that high-mass accuracy MALDI (10–25 parts per million [ppm] on average) results in the unambiguous identification of the vast majority of yeast proteins by their peptide mass maps alone (Shevchenko et al., 1996). Even simple protein mixtures present in a single gel band are resolved using an iterative sequence database search method (Jensen et al., 1997a ).
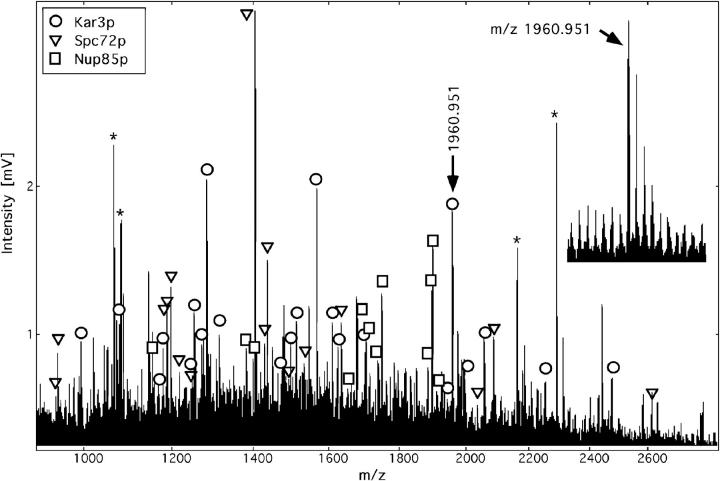
Band 2k from the gel in Fig. 3 will be used to demonstrate the approach for identification of a protein mixture. This 85-kD protein gel band generated after digestion a total of 87 peptide masses in a single analysis (Fig. 2). Searching the complete set of tryptic peptide masses in a comprehensive sequence database containing more than 250,000 entries from all species retrieved yeast Kar3p (84 kD) as the top candidate with 15 tryptic peptide matches within a mass error of 30 ppm. Detailed evaluation (second pass search) assigned a total of 22 peptides to the Kar3p sequence for an amino acid sequence coverage of 35%. This information and the fact that the protein was from yeast and in the right size range unambiguously identified it. The remaining masses were used to query the database again. This search now retrieved two further matches that were analyzed as above. These were Spc72p (Yal047p) with a total of 16 peptides matched and a sequence coverage of 27% and Nup85p with 12 peptides matched and a coverage of 21%. Although Spc72p has a calculated size of 72 kD, both it and the HA-tagged version have an apparent size close to 85 kD on SDS gels (refer to Materials and Methods). Further database searches did not retrieve any significant matches and we therefore concluded that band 2k contained three protein components, namely Kar3p, Spc72p, and Nup85p. All the identifications of bands marked in Fig. 2 were unambiguous since the other retrieved proteins identified had significantly fewer matching tryptic peptides, a different intact molecular weight, and were from a different species. Most of the database searches were performed with a mass accuracy of 30 ppm (refer to Table I and Materials and Methods) including in particular the protein mixtures, a few single bands were searched with a 50 ppm error. Using the iterative database search method (Jensen et al., 1997a ) as illustrated above, we identified two or three component protein mixtures in bands 2e, 2h, 2k, 3, 3d1, 6, and 7q. A total of 70 MALDI experiments were carried out on 49 gel bands. These included the 45 enriched bands described above and four of the more intensely stained bands which were probably contaminants. A total of 43 different proteins were identified (Fig. 3).
Figure 2.
Identification of the proteins Kar3p, Spc72p, and Nup85p in band 2k by MALDI peptide mass mapping and iterative database searching using a peptide mass error of less than 30 ppm. Asterisk, matrix or trypsin autolysis peaks used as internal calibration peaks; inset, data quality obtained for the ion signal at m/z 1960, mass resolution of 10,000 (full width at half maximum) allowed monoisotopic mass determination with a mass error of only 11 ppm. The measured masses (monoisotopic protonated peptide molecular weight) of the three proteins identified together with the deviations from the matched mass and any amino acid modifications in brackets were as follows: Kar3p: 992.627 (0.027), 1077.628 (−0.002), 1170.580 (0.034), 1178.627 (0.010), 1248.654 (0.011), 1252.675 (0.011), 1272.706 (−0.010), 1284.643 (−0.005, methionine sulphoxide), 1314.669 (−0.011), 1475.793 (−0.013), 1498.724 (0.014, methionine sulphoxide), 1513.832 (0.021), 1568.788 (0.029), 1610.862 (−0.003), 1626.839 (0.033, methionine sulphoxide), 1704.891 (0.021), 1945.031 (−0.004), 1960.951 (−0.021), 1997.982 (−0.014), 2057.999 (0.011), 2253.089 (−0.033), 2476.233 (−0.017, methionine sulphoxide); Yal047p/ Spc72p: 939.571 (0.009), 943.592 (−0.001), 1187.635 (−0.003), 1188.617 (0.032), 1196.685 (0.018), 1218.662 (−0.025), 1246.659 (−0.020), 1406.755 (0.028), 1429.754 (−0.010), 1437.828 (−0.018), 1486.812 (0.001), 1534.814 (−0.008), 1635.867 (−0.022), 2036.030 (0.037), 2087.030 (0.004), 2613.342 (0.053); Nup85p: 1147.631 (−0.008), 1383.750 (−0.006), 1399.740 (0.018), 1653.830 (−0.037), 1680.841 (0.014), 1705.901 (−0.026), 1738.873 (0.012), 1751.876 (−0.020), 1885.920 (−0.016), 1898.983 (−0.018), 1899.954 (0.002, S-acrylamidocysteine), 1934.916 (−0.049). The particular peptides identified in each protein can be obtained by entering the measured masses into the PeptideSearch software available at http://www.mann.embl-heidelberg.de
The MALDI analysis clearly shows that this spindle pole preparation is highly enriched since of the 33 enriched gel bands successfully analyzed, 25 contained one or more spindle pole components. Thus, of eight spindle components already shown to localize close to the pole: Stu1p (Pasqualone and Huffaker, 1994), Kip1p (Hoyt et al., 1992; Roof et al., 1992), Kar3p (Meluh and Rose, 1990; Saunders et al., 1997), Nuf2p (Osborne et al., 1994), Bim1p (Schwartz et al., 1997), Cin8p (Hoyt et al., 1992; Roof et al., 1992), Kip3p (Cottingham and Hoyt, 1997; DeZwaan et al., 1997), and Mhp1p (Irminger-Finger et al., 1996), we identified the first five. We have excluded the CBF3 or kinetochore components (Lechner and Carbon, 1991) even though one of these, Ndc10p/Cbf2p (Goh and Kilmartin, 1993; Jiang et al., 1993), localizes close to the pole because reactivity with anti-Ndc10p shows that it is absent in this preparation. There are nine SPB components currently identified: Spc110p (Kilmartin et al., 1993), Stu2p (Wang and Huffaker, 1997), Spc97p (Knop et al., 1997), Spc98p (Geissler et al., 1996), Spc42p (Donaldson and Kilmartin, 1996), Cdc31p (Spang et al., 1993), Cmd1p (Geiser et al., 1993; Stirling et al., 1994), Kar1p (Vallen et al., 1992; Spang et al., 1995), and Tub4p (Sobel and Snyder, 1995; Spang et al., 1996; Marschall et al., 1996), of which we identified the first seven. Some nuclear pore complex components were also identified: Pom152p (Wozniak et al., 1994), Nic96p (Grandi et al., 1993), and Nup85p (Goldstein et al., 1996; Siniossoglou et al., 1996), which probably reflects a slight contamination with pore complexes since both Pom152p and Nic96p are abundant components of the pore complex (Aitchison et al., 1995).
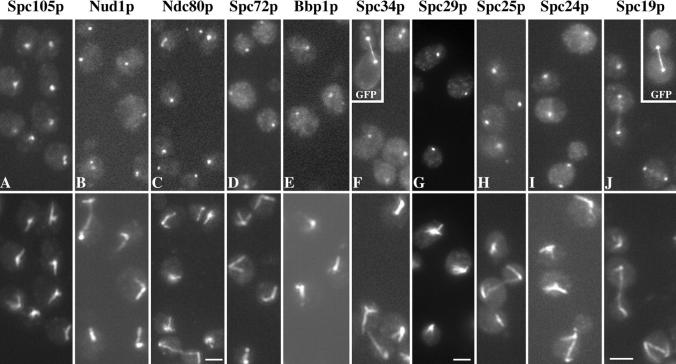
We also identified a number of proteins of unknown function and others not previously localized to the spindle pole. All of these with the exception of the Hsp70 proteins Ssa1p and Ssb1p were tagged with three copies of the HA epitope to determine whether they localized to the spindle pole by immunofluorescence. We used this epitope because the tagging has been very reliable and the sequence is short and thus less likely to affect the function of the protein. In all cases where the wild-type protein was replaced by the HA-tagged version, spores grew normally and immunoblots showed bands close to the size found in Fig. 3 (after subtraction of the contribution due to the tag, refer to Materials and Methods). Those that gave positive pole staining are listed in Table I and the results are shown in Fig. 4, with the exception of Cnm67p which was first localized to the pole by Brachat et al. (1998). Seven of the proteins appear to localize exclusively to the pole region: Spc105p, Nud1p, Spc72p, Bbp1p, Spc29p, Spc25p, and Spc24p. These staining patterns were confirmed by GFP labeling in unfixed cells for Nud1p, Spc72p, Bbp1p, Spc29p, Spc25p, and Spc24p (all GFP-labeled strains grew at normal rates). Three localize to both the pole and the spindle: GFP-labeled Spc34p gives faint spindle staining (Fig. 4 F, inset) that is not visible with HA-staining, Spc19p gives faint spindle staining with both GFP and HA (Fig. 4, J and inset), whereas Ndc80p gives more intense staining of some short spindles (Fig. 4 C, top left cell). All the unknown gene products with the exception of Ndc80p were given Spc names because of their predominant localization to the spindle pole. Ndc80p is identical to the 80-kD spindle component (Rout and Kilmartin, 1990) (refer to Materials and Methods) and as expected the staining pattern given by the HA-tagged gene product (Fig. 4 C) has the same features, particularly the staining of some short spindles, as found with the mAb (Rout and Kilmartin, 1990). NDC80 was also isolated as TID3 in a two-hybrid screen with Dmc1p as bait (Dresser et al., 1997), but the significance of this interaction is not yet clear. Spc72p was also isolated in a previous monoclonal screen as an uncharacterized 85-kD SPB component (Rout and Kilmartin, 1991).
Figure 4.
Immunofluorescent staining of HA-tagged spindle pole components with mouse anti-HA mAb 12CA5 (top) and rabbit anti-tubulin (bottom). Insets, F and I, unfixed diploid GFP-labeled cells at half magnification. Bars, 2.5 μm, except C and G where bar is 2 μm.
Immunoelectron Microscopy
Immunoelectron microscopy (immunoEM) was carried out using a preembedding method on the tagged components that still retained antigenicity after the 20 min of formaldehyde fixation. This included all of the components except Bbp1p and Spc29p (the immunoEM of Ndc80p has already been published [Rout and Kilmartin, 1990]). This period of fixation was necessary to preserve reasonable morphology of the spindle pole, but the cells did tend to break up during the procedure so long spindles were not preserved. We detected the antigen on serial sections using the Nanogold-silver intensification method (Vandre and Burry, 1992) with modifications which will be described later (Adams and Kilmartin, manuscript in preparation). GFP was used as the tag for all of the components examined except Spc105p where HA was used. As expected, some were integral SPB components whereas others were apparently spindle-associated. Nud1p, Spc72p, and Cnm67p localized to the outer plaque region with Spc72p possibly in a more peripheral position (Fig. 5, C–H). The other five components examined, Spc's 105p, 34p, 25p, 24p, and 19p, were all in the vicinity of nuclear microtubules or the spindle (Fig. 5, A, B, and I–O). Spc105p has a potential transmembrane domain (residues 851–868) and although some silver particles were close to the nuclear membrane, most were some distance from it as in Fig. 5, A and B. If Spc105p is a transmembrane protein it may have been displaced from the membrane by the mild fixation conditions necessary for immunoEM. There did appear to be differences between the individual staining patterns for these potentially spindle-associated components but it is not clear what the significance of this is, since these may reflect differences in the sensitivity of detection. Both Spc34p and Spc19p were associated with microtubules all along the spindle (Fig. 5, I, N, and O) in agreement with the GFP staining pattern (refer to Fig. 4, F and J). Spc's 105p, 25p, and 24p were mainly associated with the nuclear side of the pole (Fig. 5, A, B and J–M), and may be associated with some of the microtubules there; however, Spc105p did appear to have a more peripheral association. These immunoEM results for Spc25p and Spc24p are also in agreement with the GFP staining by light microscopy in that no connection was detected between the two GFP dots (data not shown).
Figure 5.
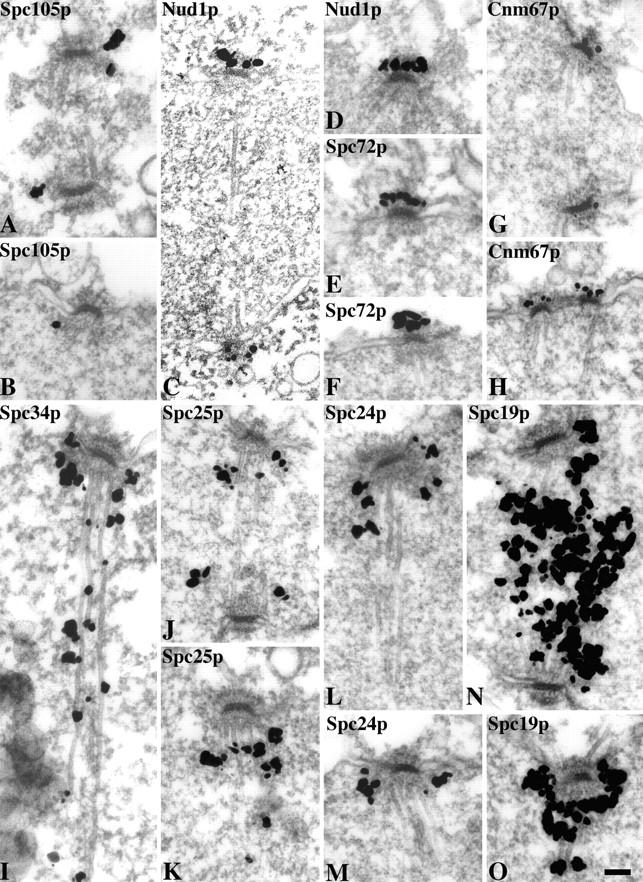
ImmunoEM using preembedding labeling and Nanogold-silver intensification of eight of the spindle pole components. Spc105p was tagged with HA and all the other components with GFP. Bar, 0.1 μm.
Homologous Proteins
Mitosis in S. cerevisiae has many of the features of mitosis in higher eukaryotes (Winey et al., 1995; Straight et al., 1997) and thus there should be homologues of some of these spindle pole proteins in other eukaryotes. So far the proportion of yeast spindle pole components with published homologues has not been high. Of the seventeen components already known (see above), and if kinesins, calmodulin, and Tub4 are excluded, then of the remaining 11 only three have potential homologues: Cdc31p (Lee and Huang, 1993; Errabolu et al., 1994), Bim1p (Beinhauer et al., 1997; Schwartz et al., 1997), and Stu2p (Wang and Huffaker, 1997). The proportion of homologues identified among the eleven new components described here is even lower. After removal of the potential coiled-coil regions and application of the BLAST search program (Altschul et al., 1990), clear homologues were found for only Ndc80p. Over the first 235 amino acids, Ndc80p shares 30% identity and 55% similarity (Fig. 6 A) with an unknown S. pombe 72-kD protein (Q10198) and 26% identity and 45% similarity with HEC, a human protein of 72-kD identified from a two-hybrid screen with retinoblastoma protein as bait (Chen et al., 1997). Although these levels of identity and similarity are not particularly high, the fact that they are usually shared between the three proteins (Fig. 6 A) suggests that all three are related. In addition, all three proteins have a similar coiled-coil domain structure (Fig. 6 B).
Figure 6.
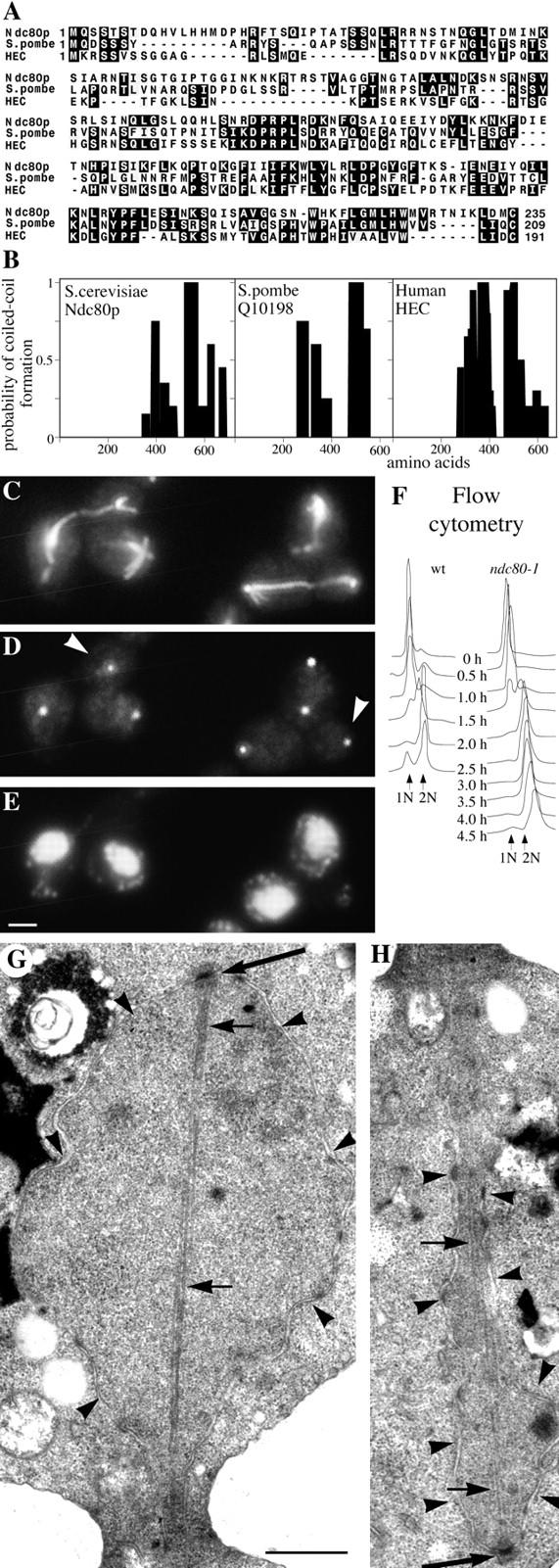
Characterization of Ndc80p. (A) Homology between the NH2-terminal domains of Ndc80p, S. pombe Q10198, and human HEC; (B), comparison of the coiled-coil domains (determined by Paircoil) of the same three proteins; (C–E), immunofluorescent staining of ndc80-1 after 2 h at 36°C with anti-tubulin (C); anti-Tub4p to stain the SPBs (D); and DAPI to stain DNA (E). (D) White arrowheads, SPBs not associated with chromosomes; (F) flow cytometry to measure DNA replication in wild-type and ndc10-1 cells synchronized by elutriation and released at 36°C; (G and H), electron micrographs of serial thin sections from the same large budded ndc80-1 cell synchronized by α-factor and released at 36°C for 2.5 h. G shows one end of a postanaphase spindle and H the other end of the same spindle in the adjacent section. Large arrows, SPBs; small arrows, microtubules; arrowheads, nuclear pores and nuclear membrane. Bars: (C–E) 2 μm; (G and H) 0.5 μm.
Functional Studies of Some of the Spindle Pole and Spindle Proteins
Perhaps the most interesting of the proteins for functional studies is Ndc80p because of its homology with human HEC protein. Ts mutants were made and the phenotype of ndc80-1 is shown in Fig. 6, C–H. The two other alleles showed similar but less severe phenotypes. ndc80-1 cells pass through S phase normally (Fig. 6 F) but anaphase is defective; poles are segregated but most of the DNA, which appears somewhat disperse, remains at one pole (Fig. 6, C–E). Consequently there is an accumulation of aploid large buds containing SPBs but little DNA. The phenotype was confirmed by EM of cells synchronized to enrich for post-anaphase spindles (Fig. 6, G and H). Here the spindle pole regions of the same cell are shown in two panels: Fig. 6 G shows one pole of the spindle associated with a large nuclear mass containing most of the DNA whereas the other pole (Fig. 6 H) lies in an isthmus of nuclear envelope, presumably containing very little DNA. Nuclear microtubules appear to connect the two poles, although at later stages of the block microtubules did tend to dissociate from the SPB with few chromosomes. We are calling this gene product Ndc80p because the phenotype is very similar to a ts mutant in Ndc10p (Goh and Kilmartin, 1993) that encodes a component of the yeast kinetochore (Doheny et al., 1993; Jiang et al., 1993). Because of this similarity in phenotype and because Ndc80p is a potential homologue of human HEC protein which localizes to the centromere (Chen et al., 1997), we examined whether Ndc80p might be a component of the yeast kinetochore. We have not yet been able to reach a firm conclusion on this. We detected no genetic interactions with genes encoding three of the CBF3 components, NDC10/CBF2/ CTF14, CBF3/CEP3, and CTF13 (refer to Materials and Methods). Ndc80p is not detected in the centromere DNA-binding complex CBF3 (Lechner and Carbon, 1991), and although it partly copurifies with the factors which bind kinetochores to microtubules (Sorger et al., 1994), it is absent from the final fraction (Severin and Hyman, unpublished results). In fixed cells there was no consistent colocalization of anti-Ndc80p staining and GFP- labeled centromeres on metaphase spindles (Straight et al., 1997). Thus, although both the phenotype and the immunofluorescence staining pattern of Ndc80p are consistent with localization to the kinetochore (Rout and Kilmartin, 1990), it seems likely that any association if present is indirect. However, it is clear that the phenotype observed is consistent with localization of Ndc80p to the spindle pole.
The phenotypes of depletion or disruption of three of the other components are also consistent with localization to the pole or spindle. Depletion of Bbp1p causes a mitotic phenotype and may partly disconnect the SPB from chromosomes (Xue et al., 1996). Disruption of one of the nonessential genes, SPC72, causes nuclear migration defects probably due to the reduced cytoplasmic microtubules in this mutant (Souès, S., manuscript in preparation). This reduction in cytoplasmic microtubules is consistent with the immunoEM localization of Spc72p to the SPB outer plaque (refer to Fig. 5, E and F). Disruption of one of the other nonessential genes, CNM67, also causes nuclear migration defects and cells lacking Cnm67p have severely reduced SPB outer plaques (Brachat et al., 1998). This is also in agreement with the immunoEM localization of Cnm67p to the outer plaque region (refer to Fig. 5, G and H).
Discussion
We describe in this paper a preparation of spindle poles from S. cerevisiae, their analysis by MALDI mass spectrometry, and the identification of 10 novel gene products encoding components of the spindle pole, and one, Bbp1p, is not previously localized to the pole; eight of these have been localized to particular parts of the SPB and spindle by immunoEM (Figs. 5 and 7). In addition, five spindle components and seven SPB components that had been previously described were also identified. The success of this approach was dependent on several factors: first, the spindle poles, though not completely pure, were sufficiently enriched for successful MALDI analysis; second, phosphatase treatment focused the bands corresponding to the substantial proportion of phosphoproteins present in this preparation; and third, the sensitivity and mass accuracy of the MALDI method is now high enough to identify several proteins in one gel band. Two additional factors are the speed of the MALDI analysis, typically 20–30 min per band, and the speed and simplicity of the insertion of epitope tags into the yeast genome to check the cellular localization of proteins identified by the MALDI analysis. We should emphasize that on this occasion we did not attempt to be comprehensive in our identifications. We were unable to identify all of the enriched bands and because of shortage of material, we only used a one-dimensional gel for separation. Also due to problems with the dynamic range of MALDI, some of the less abundant components may not have been detected because of comigration with more abundant components, particularly if the abundance ratio is greater than 5 or 10:1. This might explain why the SPB components Kar1p and Tub4p were not detected since they may comigrate with the abundant Tub1p and Tub2p bands. Because 75% of the enriched bands from which proteins were successfully identified contained a pole component, we suspect that further components will be identified by a more comprehensive analysis involving further scaling up of the preparation and a separation step before the final SDS gel.
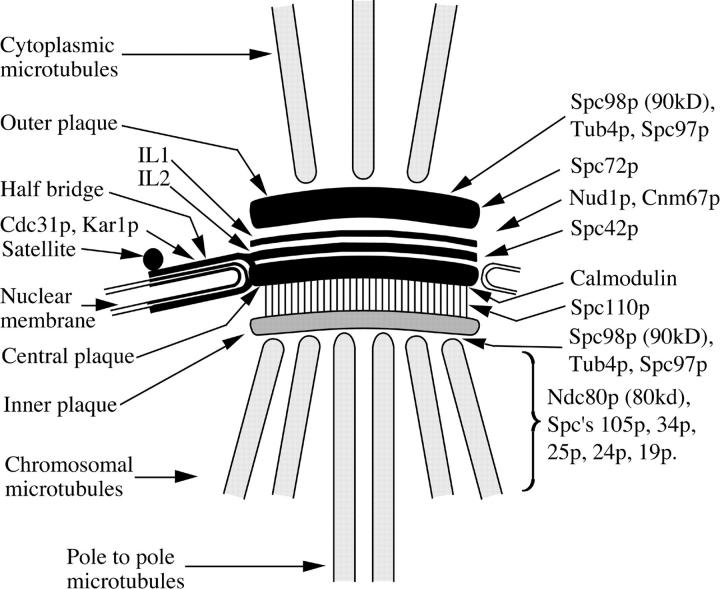
Figure 7.
Diagram of the localizations of spindle pole and SPB components described in this article. Other spindle pole components already localized by immunoEM are also shown. The localizations of the components Ndc80p and Spc's 105p, 34p, 25p, 24p, and 19p have not been established precisely relative to the nuclear side of the SPB. Ndc80p and Spc's 34p and 19p are probably spindle-associated since they can localize along the length of the spindle. IL1 and IL2, intermediate lines 1 and 2 (Bullitt et al., 1997). In thin sections IL2 is not visible since it appears fused to the central plaque.
Ndc80p and HEC
Ndc80p and HEC may be members of a new family of proteins involved in chromosome segregation. Both have a similar phenotype in that cells microinjected with a mAb against HEC failed to form normal mitotic spindles, the microtubules were disorganized in relation to the centromeres, and the chromosomes segregated randomly into the daughter cells (Chen et al., 1997). In ndc80-1 there is an almost complete failure of chromosome segregation into the daughter cell. There are differences in their behavior: HEC is only present in mitotic cells whereas Ndc80p localizes to the spindle pole throughout the cell cycle and along some short spindles (Rout and Kilmartin, 1990). HEC shows a complex pattern of staining in mitotic cells with predominant localization to the cytoplasm and a smaller fraction of HEC at the centromere (Chen et al., 1997). Presently the precise function of these two proteins is not clear, but it seems unlikely that they have a direct role at the kinetochore. They might have some indirect role in the organization of microtubules at the kinetochore.
Coiled-coil Proteins in Complex Organelles
A high proportion of the components localized to the yeast spindle pole region are potential coiled-coil proteins. Of the 27 identified so far (17 identified previously and listed in the Results section together with the 11 identified in Table I), 19 are potential coiled-coil proteins. Excluding the four kinesins that are a special class, this high proportion presumably reflects the structural role of these proteins. There may also be a similarly high proportion of potential coiled-coil proteins in the centrosome (Doxsey et al., 1994; Heuer et al., 1995; Bouckson-Castaing et al., 1996). Overexpression studies in yeast show that two coiled-coil SPB components, Spc42p and Spc110p, form unusual structures. Spc42p forms a large dome (Donaldson and Kilmartin, 1996) whereas Spc110p forms a polyhedron composed of balls and spokes (Kilmartin and Goh, 1996). Both these polymers reflect the structural arrangement of the two proteins in the SPB (Kilmartin and Goh, 1996; Bullitt et al., 1997). It seems probable that some of the other potential coiled-coil proteins may also form unusual polymeric structures probably of varying sizes, which may act as building blocks in the assembly of parts of the yeast spindle pole and the centrosome. Similar principles may also apply to the assembly of other complex eukaryote organelles.
Our identification of ten new gene products encoding spindle pole components and one, Bbp1p, not previously localized to the pole shows the potential of MALDI peptide mass mapping. Unambiguous identifications were obtained from MALDI mass mapping alone. This method is fast and can now identify mixtures of proteins in gel bands from organelle fractions that do not need to be completely pure. It seems likely that the application of MALDI methodology will greatly facilitate the identification of components of organelles in eukaryotes whose genome is sequenced.
Acknowledgments
We thank T. Hyman for bringing the two groups together and S. Dyos, D. Kershaw (both from MRC), A. Shevchenko (EMBL Protein and Peptide Group) and A. Kaja for technical assistance. M.S. Robinson and I. Adams (all three from MRC) are thanked for discussion. We are grateful to K. Sawin for anti-GFP antibodies and A. Straight (University of California at San Francisco, San Francisco, CA) for providing the GFP-CEN and GFP-tubulin yeast strain.
Abbreviations used in this paper
- SPB
spindle pole body
- AP
alkaline phosphatase
- MALDI
matrix-assisted laser desorption/ionization
- HA
hemagglutinin
- GFP
green fluorescent protein
- ppm
parts per million
- ts
temperature sensitive
Footnotes
O.N. Jensen was the recipient of a European Union Biotechnology Programme postdoctoral fellowship.
P.A. Wigge and O.N. Jensen contributed equally to this work.
Address all correspondence to J.V. Kilmartin, MRC Laboratory of Molecular Biology, Hills Rd., Cambridge, CB2 2QH, UK. Tel.: (44) 1223 402242. Fax: (44) 1223 412142. E-mail: jvk@mrc-lmb.cam.ac.uk and O.N. Jensen, Department of Molecular Biology, Odense University, DK-5230 Odense, Denmark. Tel.: (45) 65 57 23 68. Fax: (45) 65 93 27 81. E-mail: jensen@pr-group.ou.dk
S. Holmes's present address is Institut Curie, 26 rue d'Ulm, 75248 Paris, France.
References
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup155p: Complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozier K, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. . Nucl Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckson-Castaing V, Moudjou M, Ferguson DJP, Mucklow S, Belkaid Y, Milon G, Crocker PR. Molecular characterization of ninein, a new coiled-coil protein of the centrosome. J Cell Sci. 1996;109:179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- Brachat, A., J.V. Kilmartin, A. Wach, and P. Philippsen. 1998. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half bridge-organized microtubules. Mol. Biol. Cell. In press. [DOI] [PMC free article] [PubMed]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Chen Y, Riley DJ, Chen P-L, Lee W-H. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiaeis accomplished by antagonistically acting microtubule motors. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin- related KIP3 of Saccharomyces cerevisiaeis required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiaekinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Kilmartin JV. Spc42p: a phosphorylated component of the S. cerevisiaespindle pole body (SPB) with an essential function during SPB duplication. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Dresser ME, Ewing DJ, Conrad MN, Dominguez AM, Barstead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errabolu R, Sanders MA, Salisbury JL. Cloning of a cDNA encoding human centrin, an EF-hand protein of centrosomes and mitotic spindle poles. J Cell Sci. 1994;107:9–16. doi: 10.1242/jcs.107.1.9. [DOI] [PubMed] [Google Scholar]
- Foreman PK, Davis RW. Point mutations that separate the role of Saccharomyces cerevisiaecentromere binding factor 1 in chromosome segregation from its role in transcriptional activation. Genetics. 1993;135:287–296. doi: 10.1093/genetics/135.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, Sundberg HA, Huang EY, Davis TN. The 110-kD spindle pole body component of Saccharomyces cerevisiae is a phosphoprotein that is modified in a cell cycle-dependent manner. J Cell Biol. 1996;132:903–914. doi: 10.1083/jcb.132.5.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Sundberg HA, Chang BH, Muller EGD, Davis TN. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. . Mol Cell Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soués S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiaeat the sites of microtubule attachment. EMBO (Eur Mol Biol Organ) J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SB. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Goh P-Y, Kilmartin JV. NDC10: A gene involved in chromosome segregation in Saccharomyces cerevisiae. . J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, Snay CA, Heath CV, Cole CN. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol Biol Cell. 1996;7:917–934. doi: 10.1091/mbc.7.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with ‘GLFG' nucleoporins and a novel nuclear pore protein NIC96. EMBO (Eur Mol Biol Organ) J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer, J.G., K. Li, and T.C. Kaufman. 1995. The Drosophila homeotic target gene centrosomin (cnn) encodes a novel centrosomal protein with leucine zippers and maps to a genomic region required for midgut morphogenesis. Development (Camb.). 121:3861–3876. [DOI] [PubMed]
- Houthaeve T, Gausepohl H, Mann M, Ashman K. Automation of micro-preparation and enzymatic cleavage of gel electrophoretically separated proteins. FEBS (Fed Eur Biochem Soc) Lett. 1995;376:91–94. doi: 10.1016/0014-5793(95)01242-7. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiaekinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irminger-Finger I, Hurt E, Roebuck A, Collart MA, Edelstein SJ. MHP1, an essential gene in Saccharomyces cerevisiaerequired for microtubule function. J Cell Biol. 1996;135:1323–1339. doi: 10.1083/jcb.135.5.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O, Podtelejnikov A, Mann M. Identification of the components of simple protein mixtures by high-accuracy peptide mass mapping and database searching. Anal Chem. 1997a;69:4741–4750. doi: 10.1021/ac970896z. [DOI] [PubMed] [Google Scholar]
- Jensen, O.N., A. Shevchenko, and M. Mann. 1997b Protein analysis by mass spectrometry. In Protein Structure: A Practical Approach. 2nd edition. T.E. Creighton, editor. IRL Press, Oxford University, Oxford, UK. 29–57.
- Jiang W, Lechner J, Carbon J. Isolation and characterization of a gene (CBF2)specifying a protein component of the budding yeast kinetochore. J Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. . J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Goh P-Y. Spc110p: Assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO (Eur Mol Biol Org) J. 1996;15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. A spacer protein in the Saccharomyces cerevisiaespindle pole body whose transcript is cell- cycle regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccahromyces cerevisiaeand functions in microtubule organization and spindle pole body duplication. EMBO (Eur Mol Biol Org) J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopski K, Huffaker TC. Suppressors of the ndc10-2 mutation: A role for the ubiquitin system in Saccharomyces cerevisiaekinetochore function. Genetics. 1997;147:409–420. doi: 10.1093/genetics/147.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Lee VD, Huang B. Molecular cloning and centrosomal localization of human caltractin. Proc Natl Acad Sci USA. 1993;90:11039–11043. doi: 10.1073/pnas.90.23.11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast γ-tubulin–like protein: Implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Adolf G, Lydall D, Sneddon A. The identification of a second cell cycle control on the HOpromoter in yeast: cell cycle regulation of SWI5 nuclear entry. Cell. 1990;62:631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- Osborne MA, Schlenstedt G, Jinks T, Silver PA. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J Cell Biol. 1994;125:853–866. doi: 10.1083/jcb.125.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualone D, Huffaker TC. STU1, a suppressor of a beta-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Schiebel E. Centrosome-microtubule nucleation. J Cell Sci. 1997;110:295–300. doi: 10.1242/jcs.110.3.295. [DOI] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Biggins S, Satterwhite LL. Unravelling the tangled web at the microtubule-organizing center. Curr Opin Cell Biol. 1993;5:105–115. doi: 10.1016/s0955-0674(05)80015-8. [DOI] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Yeast spindle pole body components. Cold Spring Harbor Symp Quant Biol. 1991;56:687–691. doi: 10.1101/sqb.1991.056.01.077. [DOI] [PubMed] [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiaekinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K, Richards K, Botstein D. BIM1encodes a microtubule-binding protein in yeast. Mol Biol Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, Mortensen P, Shevchenko A, Boucherie H, Mann M. Linking genome and proteome by mass spectrometry: Large scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemering KR, Golbik R, Haseloff J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biology. 1996;6:1653–1663. doi: 10.1016/s0960-9822(02)70789-6. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent γ-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. . J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Severin FF, Hyman AA. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiaeis a component of the half bridge of the spindle pole body. J Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Grein K, Matzner M, Schiebel E. The Cdc31p-binding protein Kar1p is a component of the half bridge of the yeast spindle pole body. J Cell Biol. 1995;128:863–877. doi: 10.1083/jcb.128.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-Tubulin-like Tub4p of Saccharomyces cerevisiaeis associated with spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling DA, Welch KA, Stark MJR. Interaction with calmodulin is required for the function of Spc110p, an essential component of the yeast spindle pole body. EMBO (Eur Mol Biol Org) J. 1994;13:4329–4342. doi: 10.1002/j.1460-2075.1994.tb06753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling DA, Raynor TF, Prescott AR, Stark MJR. Mutations which block the binding of calmodulin to Spc110p cause multiple mitotic defects. J Cell Sci. 1996;109:1297–1310. doi: 10.1242/jcs.109.6.1297. [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Kingsbury J, Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. . J Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg HA, Goetsch L, Byers B, Davis TN. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M, Tokiwa G, Futcher B. Comparison of the S. cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO (Eur Mol Biol Organ) J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Scherson TY, Roberts T, van Zee K, Rose MD. Asymmetric mitotic segregation of the yeast spindle pole body. Cell. 1992;69:505–515. doi: 10.1016/0092-8674(92)90451-h. [DOI] [PubMed] [Google Scholar]
- Vandre DD, Burry RW. Immunoelectron microscopic localization of phosphoproteins associated with the mitotic spindle. J Histochem Cytochem. 1992;40:1837–1847. doi: 10.1177/40.12.1453002. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. . Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wang PJ, Huffaker TC. Stu2p: A microtubule-binding protein that is an essential component of the yeast spindle pole body. J Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiaemitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Blobel G, Rout MP. POM152is an integral protein of the pore membrane domain of the yeast nuclear envelope. J Cell Biol. 1994;125:31–42. doi: 10.1083/jcb.125.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Shan X, Sinelnikov A, Melese T. Yeast mutants that produce a novel type of ascus containing asci instead of spores. Genetics. 1996;144:979–989. doi: 10.1093/genetics/144.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]