Abstract
Ubiquitin-conjugating enzymes (UBC) catalyze the covalent attachment of ubiquitin to target proteins and are distinguished by the presence of a UBC domain required for catalysis. Previously identified members of this enzyme family are small proteins and function primarily in selective proteolysis pathways. Here we describe BRUCE (BIR repeat containing ubiquitin-conjugating enzyme), a giant (528-kD) ubiquitin-conjugating enzyme from mice. BRUCE is membrane associated and localizes to the Golgi compartment and the vesicular system. Remarkably, in addition to being an active ubiquitin-conjugating enzyme, BRUCE bears a baculovirus inhibitor of apoptosis repeat (BIR) motif, which to this date has been exclusively found in apoptosis inhibitors of the IAP-related protein family. The BIR motifs of IAP proteins are indispensable for their anti–cell death activity and are thought to function through protein–protein interaction. This suggests that BRUCE may combine properties of IAP-like proteins and ubiquitin-conjugating enzymes and indicates that the family of IAP-like proteins is structurally and functionally more diverse than previously expected.
The central importance of selective proteolytic systems in regulating cellular key events has been recognized recently. Progression through the eukaryotic cell cycle, for instance, is substantially regulated through a timed and coordinated degradation of cyclins and inhibitors of cyclin-dependent protein kinases (for reviews see Hochstrasser, 1996; King et al., 1996). Similarly, the shift from one transcriptional or developmental program to another is often achieved through regulated destruction of regulatory proteins. Unlike several other posttranslational events (e.g., phosphorylation), proteolysis is irreversible, and therefore proteolytic enzymes are usually used for controlling unidirectional cellular pathways.
Selective degradation in eukaryotes primarily requires the ubiquitin system that functions to mark proteins for degradation by the multicatalytic protease, the proteasome (for reviews see Ciechanover, 1994; Jentsch and Schlenker, 1995; Hochstrasser, 1996; Varshavsky, 1997). Proteins degraded by this pathway must first be recognized as substrates by components of the ubiquitin system. A cascade of reactions catalyzed by several classes of enzymes is required to form an isopeptide bond between the COOH terminus of ubiquitin and the ε-amino group of a lysine residue of an acceptor protein (for reviews see Jentsch, 1992; Ciechanover, 1994; Hochstrasser, 1996; Varshavsky, 1997). Ubiquitin-activating (E1)1 enzyme hydrolyses ATP and forms a high-energy thioester between a cysteine of its active site and the COOH terminus of ubiquitin. Activated ubiquitin is then passed on to ubiquitin-conjugating (E2) enzymes, which form thioester-linked complexes with ubiquitin in a similar fashion. Finally, ubiquitin is covalently attached to the substrate protein by the E2 enzymes or, alternatively, by ubiquitin-protein ligases (E3), which may possess substrate-binding properties (Scheffner et al., 1995). Successive rounds of ubiquitination result in the formation of multiubiquitin chains attached to proteolytic substrate proteins. Multiubiquitinated proteins are then recognized and degraded by the proteasome.
E2 enzymes are thought to provide substrate specificity to the proteolytic system and are encoded by large gene families by apparently all eukaryotes. In the yeast Saccharomyces cerevisiae, this family comprises 11 members (ubiquitin-conjugating enzymes [UBC]), which can be distinguished by their intracellular localization and cellular functions (Jentsch, 1992; Varshavsky, 1997). Known functions of the yeast E2s include DNA repair, cell cycle progression, sporulation, peroxisome biogenesis, and heat and heavy metal tolerance (Jentsch, 1992). Previously identified E2 enzymes are small proteins (16–35 kD) that bear a conserved ∼16-kD so-called UBC domain (Jentsch et al., 1990). The active-site cysteine residue required for the formation of a thioester-linked E2–ubiquitin complex is located within this domain. Some E2s consist solely of the UBC domain, whereas others possess short COOH-terminal extensions. Among the known substrates are cyclins, CDK inhibitors, transcription factors, subunits of trimeric G proteins, and aberrant proteins (Hochstrasser, 1996; Varshavsky, 1997). The majority of E2 enzymes are soluble proteins of the cytosol and the nucleus, but in yeast, three E2s are known to localize to intracellular membranes. UBC10 is a peripheral membrane protein of peroxisomes and is required for the biogenesis of this organelle (Wiebel and Kunau, 1992). UBC6 and UBC7 localize to the ER and are integral and peripheral membrane proteins, respectively. Both enzymes collaborate in ER-associated degradation of short-lived and aberrant proteins (Sommer and Jentsch, 1993; Biederer et al., 1997). From higher eukaryotes, however, no membrane-associated E2s have been reported previously.
Here we describe BRUCE (BIR repeat containing ubiquitin-conjugating enzyme), a strikingly novel ubiquitin-conjugating enzyme from mouse. BRUCE is a giant 528-kD protein that, in contrast to previously identified ubiquitin-conjugating enzymes, is associated with the Golgi compartment and the vesicular system. We show that the UBC domain of BRUCE can be charged with ubiquitin in vitro, indicating that the protein possesses ubiquitin-conjugating activity. Remarkably, BRUCE also possesses a baculovirus inhibitor of apoptosis repeat (BIR) motif, a hallmark of apoptosis inhibitors of the IAP (inhibitor of apoptosis protein) class. This emphasizes the structural diversity of IAP-like proteins and suggests that its new member, BRUCE, may function through its ubiquitin-conjugating activity.
Materials and Methods
DNA Techniques
Standard DNA techniques were used (Ausubel et al., 1994). Two degenerate primers (primers A and B from Matuschewski et al., 1996) corresponding to conserved sequences of ubiquitin-conjugating enzymes were used for amplification with genomic mouse DNA as a template. The obtained PCR fragment was used for screening a mouse brain cDNA library (CLONTECH Laboratories, Palo Alto, CA). One positive clone was isolated and sequenced, and 5′ sequences were used as a probe in a subsequent screen. After 11 similar rounds of screening, 15 overlapping cDNA clones encompassing a sequence of 15,475 bp with a single open reading frame (ORF) of 14,535 bp were each subcloned via the EcoRI site into pUC19 (Ausubel et al., 1994). All cDNA clones were sequenced at least twice in both orientations using a sequenase kit (Amersham Corp., Arlington Heights, IL). Northern blots of mouse multiple tissue and mouse embryo (CLONTECH Laboratories) were probed with gene-specific 5′ (bp 195–828) or 3′ sequences (bp 10865–15475), or with β-actin cDNA for loading control. Hybridization was carried out according to the manufacturer's protocol.
Antibodies and Western Analysis
Two polypeptides corresponding to the NH2-terminal (amino acids 356– 493) and to the COOH-terminal (amino acids 4439–4845) part of BRUCE fused to the His6-tag sequence were expressed and purified by the QIAexpress System (Qiagen, Chatsworth, CA). After purification by SDS-PAGE, rabbits were immunized with the protein, and two polyclonal antibodies, “N” and “C” (corresponding to the NH2- and COOH-terminal fragments, respectively), were obtained. Antibodies against synaptophysin, MAP2 (both from Boehringer Mannheim Corp., Indianapolis, IN), TGN38, and TAU (kindly provided by G. Banting [University of Bristol, UK] and R. Brandt [University of Heidelberg], respectively) are mouse monoclonals, and anti-PDI antibodies are rabbit polyclonal (kindly provided by B. Dobberstein [University of Heidelberg]). Dichlorotriazinyl-fluoresceine (DTAF)- or lissamine rhodamine sulfonyl chloride (LRSC)-labeled and peroxidase-conjugated goat anti–mouse and anti–rabbit antibodies were purchased from Dianova (Hamburg, Germany). For Western blots, the BRUCE protein of membrane fractions (see below) was separated by 4–14% gradient SDS-PAGE; all other proteins were separated by 12% SDS-PAGE and transferred to polyvinyl difluoride membranes.
Cell Fractionations
For the experiment shown in Fig. 3 B, rat PC12 cells were metabolically labeled with [35S]methionine and cracked in homogenization buffer (10 mM Hepes, pH 7.2, 0.25 M sucrose, 1 mM magnesium acetate, 1 mM EDTA, 1 mM PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin), and nuclei, organelles, and unbroken cells were pelleted by centrifugation at 1,000 g for 10 min. The postnuclear supernatant was centrifuged at 100,000 g for 60 min to obtain cytosolic and membrane fractions, and both fractions were adjusted to the same volume (yielding fractions S and P, Fig. 3 B, lanes 1 and 2). Two similarly prepared membrane fractions were further processed by extensive washes with either 0.5 M NaCl or PBS by passing through a 26-gauge needle, and the material was fractionated as above into soluble (S) and pellet (P) fractions (Fig. 3 B, lanes 3 and 4, and 5 and 6, respectively). BRUCE was detected in aliquots of these fractions by immunoprecipitation with antibodies C and N, and PDI and synaptophysin control proteins were detected by Western blotting. Immunoprecipitation of BRUCE with affinity-purified BRUCE-specific antibodies was carried out in a buffer containing 0.3% NP-40, 0.5 mM EDTA, 0.5 mM PMSF, 5 μg/ml aprotinin, and 5 μg/ml leupeptin for 1 h at 4°C. For the proteinase K sensitivity experiment shown in Fig. 3 C, postnuclear supernatant without protease inhibitors was incubated with or without 0.04 μg/ml proteinase K for 15 min on ice. After addition of protease inhibitors (2 mM PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin), membranes were pelleted by centrifugation at 100,000 g for 60 min, and BRUCE and PDI were detected by Western blot analysis.
Figure 3.
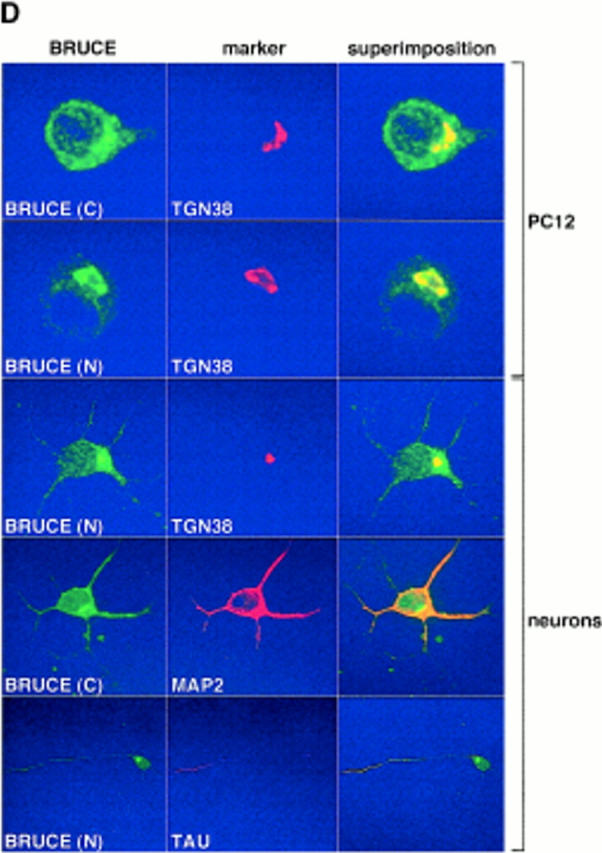
The BRUCE protein is associated with endomembranes. (A) Western blot of membrane fractions from mouse brain, mouse AtT20 (neuroendocrine), mouse N2a (neuroblastoma), and rat PC12 (neuroendocrine) cell lines probed with BRUCE-specific antibodies (antibody C). (B) Immunoprecipitation of in vivo–radiolabeled proteins with BRUCE-specific antibodies (antibodies C and N) from cytosolic (S, supernatant) and membrane (P, pellet) fractions (see Materials and Methods). BRUCE is enriched in membrane fractions (see below) and can be extracted by 0.5 M NaCl (compare lanes S and P) or buffer (PBS; compare lanes S and P). Antibodies specific for the ER-lumenal protein disulfide isomerase (PDI) and synaptophysin, an integral membrane protein of synaptic vesicles, were used for detection by Western blotting from the identical protein fractions. These proteins used as controls were found in the pellet fraction. A portion of lumenal PDI was also detectable in the cytosolic fraction (S, first lane), presumably because some microsomal vesicles were “inside-out” or disintegrated. (C) In contrast to lumenal PDI, BRUCE is sensitive to proteinase K treatment at membranes in vitro, indicating that BRUCE faces to the cytosol. (D) Confocal micrographs of rat PC12 cells and primary rat neurons stained with antibodies (indirect immunofluorescence) specific for BRUCE (green, antibodies C and N), and the marker proteins (red) TGN38 (marker for TGN; Humphrey et al., 1993), MAP2 (marker for dendrites), and TAU (marker for axons). Note that BRUCE appears in a punctated cytosolic distribution and is present in the TGN, axons, and dendrites. Colocalization of BRUCE with marker proteins is shown by superimposition (yellow). The BRUCE-specific antibodies used in these studies (A–D) are affinity-purified, and the signals are specific since they could be completely eliminated by competition with epitope-containing protein fragments (not shown).
Confocal Microscopy
Rat neuroendocrine PC12 cells and primary rat hippocampal neurons (kindly provided by A. Clement [University of Heidelberg]) were grown on coverslips, washed once with PBS, fixed, and permeabilized with precooled methanol for 5 min at −20°C. After drying, cells were washed with PBS and incubated with 10% goat serum for 20 min. Coverslips were incubated with the respective antibodies, washed, and stained with secondary antibodies (LRSC-labeled goat anti–mouse or DTAF-labeled goat anti– rabbit antibodies [1:200]) as described (Bos et al., 1993). Confocal immunofluorescence microscopy was performed using a Leica TCS4D microscope (Deerfield, IL) with a 63× 1.4 numerical aperture.
Thioester Assays
For thioester assays, ubiquitin was obtained by purifying a glutathione- S-transferase–ubiquitin fusion (containing a thrombin-cleavage and a protein kinase A phosphorylation site; kindly provided by M. Scheffner [German Cancer Center, Heidelberg, Germany]) from Escherichia coli cells expressing pGEXUBI using a glutathione-Sepharose column. The fusion protein was radiolabeled using protein kinase A and [γ32P]ATP and cleaved with thrombin, and thrombin was heat-inactivated. Radiolabeled SUMO-1 and NEDD8 were generated by a similar procedure (Schwarz et al., 1998). E. coli BL21(DE3) cells transformed with plasmid pET3a (Novagen, Madison, WI) expressing the COOH-terminal 406 amino acids from BRUCE (FUBC) or cells transformed with plasmid pQE9 (Qiagen) expressing an identical version NH2-terminally extended by additional six histidine residues (HFUBC) were generated. Extracts from these cells were incubated for 10 min at 25°C in the presence of a reticulocyte lysate (as a source for ubiquitin-activating activity; Stratagene, La Jolla, CA) and radiolabeled ubiquitin (or ubiquitin-like proteins) in a buffer containing 25 mM Tris, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 10 mM ATP, and 0.1 mM DTT. Extracts from cells expressing UbcH7 ubiquitin-conjugating enzyme were used as a positive control. The reaction products were incubated with sample buffer under nonreducing or reducing conditions (10 min boiling in the presence of 100 mM DTT) followed by SDS-PAGE and autoradiography.
Results
Identification of BRUCE and Expression Pattern
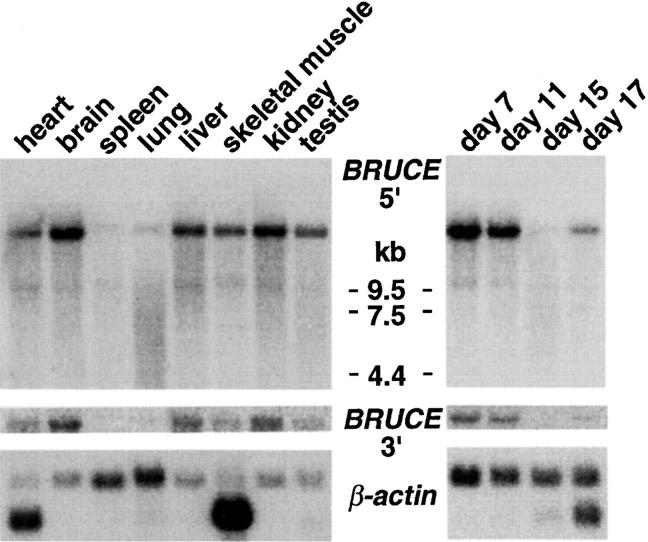
In the course of a homology-based screen for murine genes encoding ubiquitin-conjugating enzymes (Matuschewski et al., 1996), we identified a DNA fragment that hybridized to an exceptionally long mRNA. We obtained 15 overlapping complementary DNA (cDNA) clones after 12 rounds of screening, which identified a single large ORF. The DNA sequence of this ORF (14,535 bp) predicts a giant protein with 4,845 amino acid residues (528 kD; Fig. 1), which we termed BRUCE. Northern (RNA) analysis with probes corresponding to either NH2- or COOH-terminal sequences of BRUCE detected an identical ∼15-kb mRNA species, confirming that the isolated cDNAs corresponded to the same gene (Fig. 2). BRUCE is expressed in most tissues of adult mice, yet most prominently in the brain and the kidney. However, total mRNA levels vary strongly during mouse embryogenesis, possibly indicating that controlled levels of BRUCE are important during development (Fig. 2). Similar mRNAs could be detected in rat and man (not shown), indicating that BRUCE is ubiquitously expressed in mammals.
Figure 1.
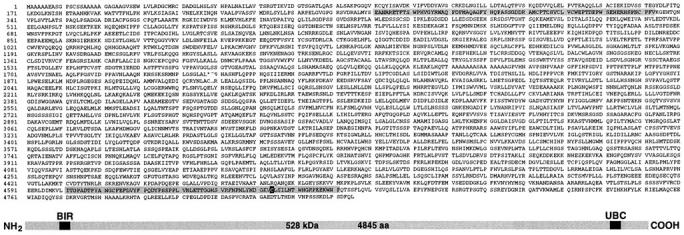
Primary structure of BRUCE predicted from the cDNA sequence. (Sequence data are available from GenBank/EMBL/ DDBJ under accession number Y17267.) The NH2-terminally positioned single BIR repeat and the UBC domain close to the COOH terminus are highlighted. The position of the presumed active-site cysteine residue required for thioester formation with ubiquitin is marked within the UBC domain. The presumed initiator methionine residue was deducted from the position of the first ATG codon found in the cDNA sequence. A schematic representation of the domain structure of BRUCE is shown at the bottom.
Figure 2.
Levels of BRUCE transcripts. Northern blots of oligo(dT)-purified mRNA from multiple tissues (left) and from total mouse embryos (right; days 7–17 postconception; both blots from CLONTECH Laboratories). Note that 3′- (bp 10865– 15475), as well as 5′-probes (bp 195–828) recognize the ∼15-kb transcript of BRUCE. The transcript is particularly abundant in brain and kidney, and levels vary during development. Actin mRNA was used for loading control. (The lower bands represent muscle-specific isoforms.)
Localization at Endomembranes
BRUCE is one of the largest proteins known to date, but unlike most other giant proteins, it is nonrepetitive in structure. The bulk of the protein exhibits no significant sequence similarity to other known proteins and displays a high percentage (∼35%) of hydrophobic amino acid residues. We therefore speculated that the protein might be membrane bound. Antibodies raised against NH2-terminal and COOH-terminal domains of BRUCE, respectively, identified an ∼500-kD protein in cell extracts, and fractionation studies indeed established a localization of the protein chiefly in membrane fractions (Fig. 3, A and B). The protein was extractable by salt, indicating that BRUCE is membrane associated but not embedded into the lipid bilayer (Fig. 3 B). As indicated by proteinase K sensitivity assays, the protein appears to localize to the cytosolic face of the membrane (Fig. 3 C). Interestingly, confocal microscopy identified BRUCE in a punctated cytosolic distribution and in a Golgi-related compartment, specifically the TGN, where the protein colocalized with TGN38 (Bos et al., 1993; Humphrey et al., 1993), a TGN-specific marker protein (Fig. 3 D). To refine the characterization of the intracellular distribution of BRUCE, we took advantage of the highly polarized organization of neuronal cells (Parton and Dotti, 1993). To this end, primary neurons were prepared and stained for BRUCE with affinity-purified BRUCE-specific antibodies. In these cells, besides its localization in the TGN of the cell body, BRUCE was also detectable in a dotted distribution in axons and dendrites, indicating that the protein is additionally associated with more distal vesicular structures (Fig. 3 D).
BRUCE Is Related to Ubiquitin-conjugating Enzymes
The predicted primary sequence indicates that BRUCE harbors a typical UBC domain close to the protein's COOH terminus (Fig. 1). UBC domains are hallmarks of E2 enzymes of the ubiquitin-dependent proteolytic system and related pathways (Jentsch et al., 1990). UBC domains are specific for E2 enzymes and carry the active-site cysteine residue required for thioester formation (Jentsch et al., 1990; and see introduction). It has been shown recently, however, that UBC domains are also found in E2 enzymes, which mediate the conjugation of ubiquitin-like proteins (Johnson and Blobel, 1997; Liakopoulos et al., 1998; Schwarz et al., 1998). The UBC domain of BRUCE is ∼37% identical to those of other ubiquitin-conjugating enzymes and harbors a putative active-site cysteine residue at a conserved position (Fig. 4 A).
Figure 4.
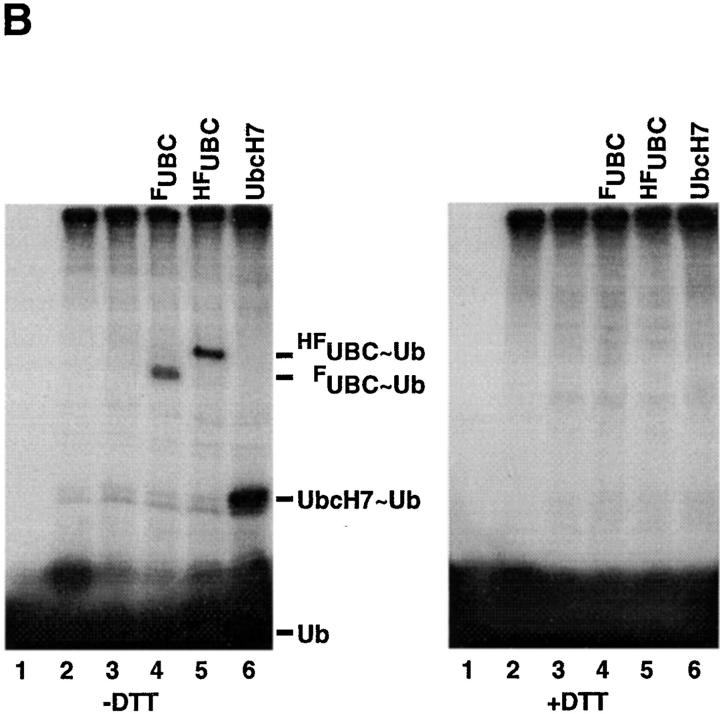
BRUCE is a ubiquitin-conjugating enzyme. (A) Sequence comparison of the UBC domain of BRUCE (amino acids 4601–4671) with those of other ubiquitin-conjugating enzymes (S. cerevisiae UBC1, -2, -3, -5, -6, and -7; mouse UbcM2; human UbcH7 and -8; and Drosophila UbcD1). Residues identical in at least four domains (majority) are shaded. The position of the presumed active-site cysteine residue required for thioester formation with ubiquitin is marked by an asterisk. (B) Thioester complex formation between ubiquitin and fragments of BRUCE (FUBC, HFUBC) consisting of the COOH-terminal 406 amino acid residues of BRUCE, which include the complete UBC domain. Assay reactions contained ATP, 32P-labeled ubiquitin, reticulocyte lysate as a source for E1 activity, and crude extracts from bacteria expressing FUBC, HFUBC, or UbcH7 (Nuber et al., 1996; human ubiquitin-conjugating enzyme as positive control). Reactions were analyzed by SDS-PAGE under nonreducing (−DTT) or reducing (+DTT) conditions (Nuber et al., 1996). Note that only in the presence of FUBC or HFUBC were additional radiolabeled complexes with sizes of ∼55 and ∼60 kD formed (−DTT, lanes 4 and 5), consistent with the sizes of adducts between ubiquitin and FUBC or HFUBC, respectively. These complexes are thioester-linked since they disappear after boiling under reducing conditions (+DTT, lanes 4 and 5). Control reactions containing labeled ubiquitin incubated with either bacterial extract (lane 1), reticulocyte lysate (lane 2), or reticulocyte lysate plus extracts of bacteria harboring the vector plasmid (lane 3) are shown. Complex formation of ubiquitin with added UbcH7 (lane 6) or UbcH7 present in the reticulocyte lysate (lanes 2–5) is used as positive control. Bands corresponding to free 32P-labeled ubiquitin and the respective thioester adducts are indicated.
Ubiquitin-conjugating Activity of BRUCE
Because of the presence of a UBC domain, we speculated that BRUCE possesses ubiquitin-conjugating activity. To test this hypothesis, we asked whether the protein can be charged in vitro with ubiquitin in a reaction depending on ubiquitin-activating enzyme, E1. Because of the size of the protein and the absence of a cDNA clone encompassing the complete 14,535 bp of the BRUCE open reading frame, we expressed COOH-terminal fragments of BRUCE in E. coli for enzyme assays. Two variants were constructed, one corresponding to the COOH-terminal 406 amino acid residues of BRUCE containing its complete UBC domain, and a similar version, extended by an additional poly-histidine tag. Bacterial protein extracts expressing either of the two fragments were assayed in vitro for thioester formation of the recombinant polypeptides with radiolabeled ubiquitin in the presence of E1 activity and ATP. Only in the presence of these bacterial extracts (Fig. 4 B) were additional radiolabeled protein complexes with sizes of ∼55 and ∼60 kD formed, consistent with the sizes of adducts between ubiquitin and the shorter or longer fragments of BRUCE's UBC domain, respectively (Fig. 4 B, −DTT, lanes 4 and 5). Complex formation depended on ATP and the presence of E1 activity. The complexes were sensitive to boiling under reducing conditions, indicating that the protein fragments are indeed linked via a thioester bond (Fig. 4 B, +DTT, lanes 4 and 5). This reaction was specific for ubiquitin, as neither SUMO-1 nor NEDD8, two mammalian ubiquitin-like proteins (Kumar et al., 1992; Johnson and Hochstrasser, 1997), were able to form adducts with the UBC domain of BRUCE under similar conditions (data not shown).
BRUCE Is Related to IAP Apoptosis Inhibitors
Intriguingly, database searches identified in addition to the UBC domain a single BIR repeat close to the NH2 terminus of the BRUCE protein. BIR repeats are cysteine rich, putative zinc-binding domains of about 70 amino acid residues in length, which to this date have exclusively been found in IAP-related apoptosis inhibitors. Members of the family of IAP-related cell death inhibitors are characterized by the presence of up to three NH2-terminally positioned BIR repeats, followed by distinct COOH-terminal sequences (Birnbaum et al., 1994; Hay et al., 1995; Rothe et al., 1995; Roy et al., 1995; Ambrosini et al., 1997; Clem and Duckett, 1997). Previously known members fall into three classes, distinguished by their protein domain structure. The first class is represented by the anti-apoptotic IAP proteins of baculovirus, insects, and mammals, which are small proteins with up to three NH2-terminal BIR repeats, linked to a COOH-terminal RING finger (Birnbaum et al., 1994; Hay et al., 1995; Rothe et al., 1995; Clem and Duckett, 1997). The second class is defined by neuronal apoptosis inhibitor protein (NAIP), a human 140-kD protein with three NH2-terminal BIR repeats and a long COOH-terminal tail that lacks a RING finger motif (Roy et al., 1995; Liston et al., 1996). Survivin, a small human cell death inhibitor, which essentially consists of a single BIR motif, defines the third class (Ambrosini et al., 1997).
The BIR motif of BRUCE is ∼40% identical in sequence to those of other IAP relatives and bears all known consensus elements of this domain, including a characteristic cysteine/histidine motif, which is thought to function in zinc binding (Fig. 5). The structure of BRUCE is unprecedented, however, as the protein has a single BIR repeat (similar to survivin) plus an exceptionally long tail, which exhibits no homology to the extensions of previously known IAP relatives, i.e., the tail of NAIP and the RING fingers of the IAP proteins. BRUCE is the first example of an IAP-like protein with a known enzymatic activity. Interestingly, the BIR motifs of BRUCE and survivin are unique since they contain an insertion of three amino acid residues (one of which is a proline residue) in between the two conserved halves of the BIR motif, which we term the FY (rich in phenylalanine and tyrosine residues) and the CH (rich in cysteine and histidine residues) regions, respectively. In the mouse genome, the regions encoding the FY and CH domains of BRUCE are separated by an intron (Pyrowolakis, G., and S. Jentsch, unpublished results). This suggests that the complete BIR motif has probably evolved by exon shuffling and that the FY and CH may represent functionally distinct folding domains separated by a linker sequence.
Figure 5.
Sequence comparison of the BIR motif of BRUCE (amino acids 261–333) with those from other IAP relatives. Residues identical in at least four motifs (majority) are shaded. Individual BIR repeats found in a single protein are numbered (I–III). The BIR motif of BRUCE is ∼40% identical in sequence to those of other IAP relatives, with a conserved FY (left part) and CH region (right part; see text).
Discussion
Apoptotic cell death is a highly regulated process of multicellular organisms through which superfluous or harmful cells are eliminated (for review see White, 1996). During this process, apoptotic cells go through sequential steps of disintegration that include chromatin cleavage, organelle breakdown, and the fragmentation of cellular material into membrane-surrounded vesicles that are rapidly ingested by neighboring cells. Cell death can be triggered by a variety of stimuli, but once initiated, apoptosis appears to proceed through a common pathway. The basic cell death machine comprises a family of related cysteine proteases, termed caspases, which cleave proteins COOH-terminal of aspartate residues (for review see Salvesen and Dixit, 1997). Caspases are synthesized as inactive proenzymes that are activated by proteolytic cleavage. The processing sites themselves match the consensus cleavage specificity of caspases, and it is thus assumed that the apoptotic suicide program is orchestrated through a cascade of auto- and trans-cleavages of different caspases. Activated caspases can cleave further targets, including components of the cytoskeleton, nuclear lamins, poly(ADP-ribose) polymerase, DNA-dependent protein kinase, and inhibitor of caspase-activated DNase (Salvesen and Dixit, 1997; Enari et al., 1998; Sakahira et al., 1998). The cleavage of these downstream targets is thought to execute the final cell death program with the characteristic morphological changes.
The cellular suicide program can be blocked by a variety of proteins, and many of these factors are known to interfere with caspase function. Bcl-2, a membrane protein of primarily the outer membrane of mitochondria, appears to inhibit apoptosis by blocking the efflux of cytochrome c from mitochondria (White, 1996; Reed, 1997). This in turn is thought to impede caspase-9 activation because maturation from its inactive proenzyme form is stimulated by cytochrome c in conjunction with the Apaf-1 protein (Li et al., 1997). In contrast, direct inhibition of caspase function is achieved by the cell death inhibitor p35 from baculovirus, which seems to act through caspase binding (Clem et al., 1991; Bump et al., 1995; Xue and Horvitz, 1995).
Another group of anti-apoptotic proteins is the IAP family of apoptosis inhibitors (for review see Clem and Duckett, 1997). Members of this protein family are distinguished by the presence of up to three BIR motifs. BIR motifs are required, and often sufficient, for the anti-apoptotic function of IAP-related proteins and are known to act through protein–protein interactions (Clem and Duckett, 1997). Several binding proteins have been found previously, which include TRAF signaling molecules (Rothe et al., 1995) and the proteins Doom and Reaper from flies (Harvey et al., 1997; Vucic et al., 1997). Recent data indicated that a principal anti-apoptotic function of BIR-containing proteins may be their ability to bind and inhibit specific caspases (Devereaux et al., 1997; Roy et al., 1997; Seshagiri and Miller, 1997).
In this paper we identify BRUCE, an unusual membrane-associated protein from mouse that combines properties of IAP-like proteins with ubiquitin-conjugating enzymes. Although the cellular function of BRUCE is not known at present, the intriguing modular design of the protein with an NH2-terminal BIR repeat and an active ubiquitin-conjugating enzyme domain at the protein's COOH terminus suggests a role in coupling anti-apoptosis pathways to the ubiquitin/proteasome proteolytic machinery. It seems attractive to speculate that BRUCE may be regulated by specific BIR motif–binding proteins or, alternatively, may target these proteins (e.g., caspases) for degradation. In contrast to a possibly reversible inhibition of caspases by other IAP relatives, proteolytic inactivation of apoptosis regulators would allow an irreversible shut-down of specific cell death pathways.
BRUCE is structurally strikingly different from previously known E2 enzymes, which are small proteins (14–34 kD) that either consist of the UBC domain alone (class I) or possess additional short COOH- (class II) or NH2-terminal (class III) extensions (Jentsch et al., 1990; Matuschewski et al., 1996). BRUCE, which bears a long NH2-terminal and a short COOH-terminal extension, defines a new class of E2 enzymes, termed class IV. Interestingly, the overall domain structure of BRUCE resembles more a class of E3 enzymes (ubiquitin ligases) of the ubiquitin system that possess COOH-terminal, so-called HECT domains involved in thioester formation with ubiquitin (Huibregtse et al., 1995; Scheffner et al., 1995, Rosa et al., 1996). These E3 enzymes typically bear within their NH2-terminal domains binding sites for proteolytic substrates and for cofactors that alter the enzyme's substrate specificity (Huibregtse et al., 1995; Scheffner et al., 1995). This modular structure enables the enzyme to bind substrates directly and to catalyze their ubiquitination via its COOH-terminal catalytic domain. It is conceivable that BRUCE may function analogously and that the BIR motif of BRUCE constitutes either a binding site for a substrate or a regulator.
BRUCE also differs from previously known E2 enzymes with respect to its intracellular localization. BRUCE is the first example of a membrane-associated E2 enzyme from mammalian cells, and its localization at a Golgi-related compartment and the vesicular system is unprecedented for this enzyme class. Remarkably, only one other enzyme of the ubiquitin system has been detected at a Golgi-related compartment previously. This is p532, a very large putative E3 ubiquitin ligase with a COOH-terminal HECT domain and an expression pattern similar to BRUCE (Rosa et al., 1996). This enzyme bears several WD and RCC1 repeats, binds clathrin, and stimulates guanine nucleotide exchange on ARF1 and Rab proteins, which are important for membrane fusion and trafficking (Rosa et al., 1996; Rosa and Barbacid, 1997). How this function is mechanistically tied to the ubiquitin system is not known at present, but it will be important to test whether BRUCE as an E2 enzyme can charge this large putative E3 enzyme with ubiquitin. If so, it seems attractive to speculate that their cooperative function is linked to the membrane fusion events that occur during apoptosis. Further studies require the assembly of the complete ∼15-kb cDNA of BRUCE into a single clone for gene transfer studies, which are expected to answer if and how BRUCE is involved in preventing apoptosis.
Acknowledgments
This work was supported by grants of the Deutsche Forschungsgemeinschaft and Fonds der Chemischen Industrie to S. Jentsch.
Abbreviations used in this paper
- BIR
baculovirus inhibitor of apoptosis repeat
- BRUCE
BIR repeat containing ubiquitin-conjugating enzyme
- E1
ubiquitin-activating enzyme
- IAP
inhibitor of apoptosis protein
- NAIP
neuronal apoptosis inhibitor protein
- ORF
open reading frame
- UBC
ubiquitin-conjugating enzyme
Footnotes
The work by H.-P. Hauser was done in the former laboratory of S. Jentsch in Tübingen. We thank T. Hoppe for important contributions to this study; M. Scheffner, G. Banting, R. Brandt, B. Dobberstein, and A. Clement for providing materials; M. Scheffner for help and advice for thioester experiments; F. Weinreich for help with DNA sequencing; E. Löser and P. Hubbe for technical assistance; and H. Ulrich for comments on the manuscript.
H.-P. Hauser and M. Bardroff contributed equally to this work.
Address all correspondence to Dr. Stefan Jentsch, ZMBH, Zentrum für Molekulare Biologie, Universität Heidelberg, Im Neuenheimer Feld 282, D-69120 Heidelberg, Germany. Tel.: +49 6221 548716. Fax: 49 6221 545891. E-mail: jentsch@zmbh.uni-heidelberg.de
The present address of Hans-Peter Hauser is Dade Behring, Inc., M 106, P.O. Box 1149, 35001 Marburg, Germany.
The present address of Michael Bardroff is MorphoSys GmbH, Am Klopferspitz 19, 82152 Martinsried/Munich, Germany.
References
- Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidmna, J.A. Smith, and K. Struhl. 1994. Current Protocols in Molecular Biology. Green and Wiley, New York.
- Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1808. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- Birnbaum MJ, Clem RJ, Miller LK. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol. 1994;68:2521–2528. doi: 10.1128/jvi.68.4.2521-2528.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos K, Wraight C, Stanley KK. TGN38 is maintained in the trans-Golgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO (Eur Mol Biol Organ) J. 1993;12:2219–2228. doi: 10.1002/j.1460-2075.1993.tb05870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bump NJ, Hackett M, Hugunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, et al. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome system. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Clem RJ, Duckett CS. The iapgenes: unique arbitrators of cell death. Trends Cell Biol. 1997;7:337–339. doi: 10.1016/S0962-8924(97)01088-X. [DOI] [PubMed] [Google Scholar]
- Clem RJ, Fechheimer M, Miller LK. Prevention of apoptosis by the baculovirus gene during infection of insect cells. Science. 1991;254:1388–1390. doi: 10.1126/science.1962198. [DOI] [PubMed] [Google Scholar]
- Devereaux QL, Takahashi R, Salvesen GS, Reed JR. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyamo H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Harvey AJ, Bidwai AP, Miller LK. Doom, a product of the Drosophila mod(mdg4)gene, induces apoptosis and binds to baculovirus inhibitor-of-apoptosis proteins. Mol Cell Biol. 1997;17:2835–2843. doi: 10.1128/mcb.17.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophilahomologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley P. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey JS, Peters PJ, Yuan LC, Bonifacino JS. Localization of TGN38 to the trans-Golgi network: involvement of a cytoplasmic tyrosine-containing sequence. J Cell Biol. 1993;120:1123–1135. doi: 10.1083/jcb.120.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Schlenker S. Selective protein degradation: a journey's end within the proteasome. Cell. 1995;82:881–884. doi: 10.1016/0092-8674(95)90021-7. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Seufert W, Sommer T, Reins H-A. Ubiquitin-conjugating enzymes: novel regulators of eukaryotic cells. Trends Biol Sci. 1990;15:195–198. doi: 10.1016/0968-0004(90)90161-4. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Hochstrasser M. SUMO-1: ubiquitin gains weight. Trends Cell Biol. 1997;7:408–413. doi: 10.1016/S0962-8924(97)01132-X. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters LM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/ caspase-9 complex inhibits an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO (Eur Mol Biol Organ) J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton HG, Farahani R, McLean M, Ikeda JE, MacKenzie A, Korneluk RG. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAPgenes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- Matuschewski K, Hauser H-P, Treier M, Jentsch S. Identification of a novel family of ubiquitin-conjugating enzymes with distinct amino-terminal extensions. J Biol Chem. 1996;271:2789–2794. doi: 10.1074/jbc.271.5.2789. [DOI] [PubMed] [Google Scholar]
- Nuber U, Schwarz S, Kaiser P, Schneider R, Scheffner M. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and RSP5. J Biol Chem. 1996;271:2795–2800. doi: 10.1074/jbc.271.5.2795. [DOI] [PubMed] [Google Scholar]
- Parton RG, Dotti CG. Cell biology of neuronal endocytosis. J Neurosci Res. 1993;36:1–9. doi: 10.1002/jnr.490360102. [DOI] [PubMed] [Google Scholar]
- Reed JC. Cytochrome c: can't live with it—can't live without it. Cell. 1997;91:559–562. doi: 10.1016/s0092-8674(00)80442-0. [DOI] [PubMed] [Google Scholar]
- Rosa JL, Barbacid M. A giant protein that stimulates guanine nucleotide exchange on ARF1 and Rab proteins forms a cytosolic ternary complex with clathrin and Hsp70. Oncogene. 1997;15:1–6. doi: 10.1038/sj.onc.1201170. [DOI] [PubMed] [Google Scholar]
- Rosa JL, Casaroli-Marano RP, Buckler AJ, Vilaro S, Barbacid M. p619, a giant protein related to the chromosome condensation regulator RCC1, stimulates guanine nucleotide exchange on ARF1 and Rab proteins. EMBO (Eur Mol Biol Organ) J. 1996;15:4262–4273. [PMC free article] [PubMed] [Google Scholar]
- Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- Roy N, Mahadevan MS, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X, et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- Roy N, Devereaux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO (Eur Mol Biol Organ) J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakahira H, Enari M, Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 1998;391:98–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Dixit VM. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Schwarz SE, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzymes. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshagiri S, Miller LK. Baculovirus inhibitors of apoptosis (IAP) block activation of Sf-caspase-1. Proc Natl Acad Sci USA. 1997;94:13606–13611. doi: 10.1073/pnas.94.25.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin-conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system. Trends Biol Sci. 1997;22:383–387. doi: 10.1016/s0968-0004(97)01122-5. [DOI] [PubMed] [Google Scholar]
- Vucic D, Kaiser WJ, Harvey AJ, Miller LK. Inhibition of Reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs) Proc Natl Acd Sci USA. 1997;94:10183–10188. doi: 10.1073/pnas.94.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebel FF, Kunau WH. The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature. 1992;359:73–76. doi: 10.1038/359073a0. [DOI] [PubMed] [Google Scholar]
- White E. Life, death, and the pursuit of apoptosis. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- Xue D, Horvitz HR. Inhibition of Caenorhabditis eleganscell-death protease CED-3 by a CED-3 cleavage site in baculovirus p35 protein. Nature. 1995;377:248–251. doi: 10.1038/377248a0. [DOI] [PubMed] [Google Scholar]