Abstract
The mdm17 mutation causes temperature-dependent defects in mitochondrial inheritance, mitochondrial morphology, and the maintenance of mitochondrial DNA in the yeast Saccharomyces cerevisiae. Defects in mitochondrial transmission to daughter buds and changes in mitochondrial morphology were apparent within 30 min after shifting cells to 37°C, while loss of the mitochondrial genome occurred after 4–24 h at the elevated temperature. The mdm17 lesion mapped to MGM1, a gene encoding a dynamin-like GTPase previously implicated in mitochondrial genome maintenance, and the cloned MGM1 gene complements all of the mdm17 mutant phenotypes. Cells with an mgm1-null mutation displayed aberrant mitochondrial inheritance and morphology. A version of mgm1 mutated in a conserved residue in the putative GTP-binding site was unable to complement any of the mutant defects. It also caused aberrant mitochondrial distribution and morphology when expressed at high levels in cells that also contained a wild-type copy of the gene. Mgm1p was localized to the mitochondrial outer membrane and fractionated as a component of a high molecular weight complex. These results indicate that Mgm1p is a mitochondrial inheritance and morphology component that functions on the mitochondrial surface.
Keywords: mitochondrial inheritance, mitochondrial morphology, yeast, dynamin, outer membrane
M itochondria perform essential cellular functions yet cannot be synthesized de novo (Attardi and Schatz, 1988). Instead, these organelles are derived from preexisting mitochondria and specific cellular mechanisms act to ensure the faithful transmission of mitochondria to progeny. The molecular details of the inheritance process are largely unknown, but a growing list of key protein components is emerging from analysis of conditional mutants of the budding yeast Saccharomyces cerevisiae that are defective for mitochondrial distribution and morphology mutant (mdm)1 (Berger and Yaffe, 1996; Hermann and Shaw, 1998).
Thus far, most characterized MDM gene products have fallen into two basic categories. The first consists of cytoplasmic or cytoskeletal components such as: Mdm1p, an intermediate filament-like protein required for both mitochondrial and nuclear inheritance (McConnell and Yaffe, 1992, 1993); and Mdm20p, a protein of unknown function that is important for the structure of the actin cytoskeleton (Hermann et al., 1997). The second category consists of proteins of the mitochondrial outer membrane: Mdm10p, Mdm12p, and Mmm1p. These proteins play roles both in maintaining normal mitochondrial morphology and mediating mitochondrial inheritance (Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997). Genetic analysis suggests that a number of additional inheritance components remain to be identified. This report describes the isolation and characterization of a new mutant, mdm17, which displays conditional defects in mitochondrial inheritance, mitochondrial morphology, and maintenance of the mitochondrial genome.
Materials and Methods
Strains and Genetic Techniques
Yeast strains used in this study are listed in Table I. Culture and genetic analysis of yeast were performed by standard procedures (Rose et al., 1990). Semisynthetic lactate medium was prepared as described (Daum et al., 1982). Plasmid DNA was prepared from Escherichia coli strain DH5α.
Table I.
Yeast Strains
| Strain | Genotype | Source | ||
|---|---|---|---|---|
| MYY290 | MAT a, his3, leu2, ura3 | (Smith and Yaffe, 1991) | ||
| MYY291 | MATα, his3, leu2, ura3 | (Smith and Yaffe, 1991) | ||
| MYY298 | MAT a/α, his3/his3, leu2/leu2, ura3/ura3 | (Smith and Yaffe, 1991) | ||
| MYY900 | MATα, mdm13, his3, leu2, ura3 | This study | ||
| MYY971 | MATα, mdm17, his3, leu2, ura3 | This study | ||
| MYY972 | MAT a, mdm17, his3, leu2, ura3 | This study | ||
| MYY973 | MAT a/α, mgm1Δ::LEU2/MGM1, his3/his3, leu2/leu2, ura3/ura3 | This study | ||
| MYY974 | MAT a, mgm1Δ::LEU2, his3, leu2, ura3 | This study | ||
| MYY975 | MATα, mgm1Δ::LEU2, his3, leu2, ura3 | This study |
Isolation of mdm17
The mdm17 mutant was isolated in a screen for novel alleles of another mitochondrial inheritance gene, mdm13. Strain MYY900, containing the mdm13 lesion, was crossed to the SL collection of temperature-sensitive strains which were derived from strain MYY290, and the resulting diploids were tested for growth at 37°C on yeast extract/peptone/glucose (YPD) medium. Diploids that failed to grow were sporulated and the meiotic progeny were analyzed by backcrossing and allelism tests. The recovered mdm17 spores were backcrossed three times to the wild-type parental strain, and the temperature-sensitive growth and mitochondrial distribution defects were shown to cosegregate in a 2:2 pattern. The backcrossed strain no longer displayed nonallelic noncomplementation with mdm13 and that property apparently depended on the presence of a third, unlinked mutation in the original mdm17 mutant strain. The MDM13 gene has yet to be isolated.
Gene Cloning and Mapping
mdm17 cells were transformed with a yeast genomic DNA library in the centromere-based LEU2 vector p366 (M. Hoekstra, ICOS Inc.). 10,000 Leu+ transformants were screened for complementation of the temperature-sensitive growth defect by replica plating to YPD medium and to yeast extract/peptone/glycerol (YPG) medium at 37°C. Four different complementing clones were isolated, and restriction analysis demonstrated that they contained overlapping yeast DNA inserts. The identity of the inserts was determined by nucleotide sequence analysis of the ends of the inserts and comparison to sequences in the Saccharomyces Genome Database.
A full-length version of the MGM1 gene was synthesized using PCR. The high-fidelity polymerase, Pfu (Stratagene), was used with primers 5′-CTCTCTAGAGTTCTTCTGCTCGCTAATGGTAAATG-3′ and 5′-CTCCTCGAGGCAAGAAGATGAGTTGGATGAAGG-3′ to amplify the MGM1 open reading frame together with ∼500 bp of flanking DNA sequences. The PCR product was phosphorylated with T4 DNA kinase and ligated into vector pRS313 (Sikorski and Hieter, 1989) that had been digested with SmaI and EcoRV and treated with calf intestinal phosphatase. The resulting plasmid was designated pRS313-MGM1.
For integrative mapping, a 2.6-kb HindIII fragment adjacent to the MGM1 locus was subcloned into the YIp5 vector (Struhl et al., 1979). The plasmid was linearized by digestion with HpaI and the DNA was transformed into strain MYY290. Ura+ transformants were crossed to strain MYY971 containing the mdm17 mutation. The meiotic progeny of the resulting diploid consisted of 19 parental ditype and 1 tetratype, mapping the mdm17 lesion to within 2.5 cM of the MGM1 locus.
Gene Replacement
A gene replacement cassette was created by PCR as described by Baudin et al. (1993). The oligonucleotides 5′-ATGAGTAATTCTACTTCATTAAGGGCCATCCCAAGAGTGGATTGTACTGAGAGTGCACC-3′ and 5′-TCATAAATTTTTGGAGACGCCCTTGTAGCTTTTCTTGAAAGTGCGGTATTTCACACCGC-3′ were used as PCR primers, and the LEU2 gene in plasmid pRS305 (Sikorski and Hieter, 1989) served as the template. The resulting PCR product was used to transform the diploid strain MYY298. Leu + transformants were screened by PCR to identify strains containing a replacement of one of two copies of MGM1 with LEU2. The heterozygous diploid strain was sporulated to obtain haploid segregants with the mgm1::LEU2 (mgm1-null) mutation.
Phenotypic Analysis
For phenotypic analysis, cultures were first grown overnight in YPD-liquid medium at 23°C and diluted to 0.5 A600/ml before incubation at 37°C for varying times. To evaluate respiration competence, cells were plated on YPD-agar medium, cultured at 23°C, and replica-plated to YPG medium. Colonies that failed to grow on YPG at 23°C were scored as having lost respiratory function. At least 700 colonies were scored for each time point.
To characterize mitochondrial distribution and morphology in living cells, mitochondria were stained with the membrane potential–sensitive dye 2-(4-dimethylaminostryl)-1-methylpyridium iodide (DASPMI), and cells were examined by fluorescence microscopy (Yaffe, 1995). Indirect immunofluorescence microscopy was performed on fixed yeast cells as described (McConnell et al., 1990).
Antibody Preparation
Antibodies were raised against a peptide, CGGYKGVSKNL, of which the last eight residues correspond to the extreme COOH terminus of Mgm1p. Peptide synthesis, coupling of peptide to keyhole-limpet hemocyanin, and immunization of rabbits was carried out by Research Genetics, Inc. The antibodies were purified on an affinity column prepared by coupling the peptide antigen to Affigel 10 (Bio-Rad Laboratories).
Subcellular Fractionation and Analysis
Cells were grown in semisynthetic lactate medium (Daum et al., 1982), converted to spheroplasts, homogenized, and fractionated by differential centrifugation as previously described (Schauer et al., 1985; Yaffe, 1991). Mitochondrial subfractions were isolated as described (Daum et al., 1982). Mitochondria were subjected to trypsin treatment or extraction with sodium carbonate or sodium chloride as previously described (Sogo and Yaffe, 1994).
Pulse–Chase Analysis
Wild-type cells were grown in minimal medium lacking methionine and cysteine to an A600 = 0.5–0.8. Cells were harvested, resuspended in a solution of 40 mM KPi, pH 6.0, 1% glucose, and labeled for 5 min at 30°C by addition of 0.1 mCi [35S]-Translabel (ICN) per ml. The chase was initiated by adding unlabeled methionine and cysteine to 20 μM and, in some cases, cycloheximide to 1 μg/ml. Cells were collected and processed for immunoprecipitation as previously described (Yaffe, 1991).
Gel Filtration Analysis
Gel filtration analysis of Mgm1p was similar to that described by Rapaport et al. (1998). Briefly, 5 mg of mitochondria was pelleted and resuspended in 1 ml buffer A (150 mM potassium acetate, 1% Triton X-100, 4 mM magnesium acetate, 0.5 mM EDTA, 0.5 mM PMSF, and 30 mM Tris-HCl, pH 7.4). The resuspended mitochondria were sonicated for 15 min in an ice-water bath, and the mixture was centrifuged for 15 min at 90,000 g. The supernatant was loaded onto a 100-ml Sephacryl-300 column (Pharmacia Biotech Inc.) that had been equilibrated with buffer A and fractionated with an FPLC system (Pharmacia Biotech Inc.) at a flow rate of 0.5 ml/min. 1-ml fractions were collected, proteins were precipitated with TCA, and analyzed by SDS-PAGE and immunoblotting. Protein bands on the immunoblots were scanned and quantified using NIH Image software.
Characterization of the mdm17 Mutation
The mdm17 gene was amplified by PCR from genomic DNA isolated from mutant cells using Taq polymerase. DNA products were cloned into the TA vector (Invitrogen Corp.) and sequenced by the dideoxy method (Sanger et al., 1977). Additionally, portions of mdm17 were amplified from genomic DNA using Pfu polymerase (Stratagene) and sequenced directly using the CircumVent Kit (New England Biolabs, Inc.).
In Vitro Mutagenesis of MGM1
Site-directed mutagenesis to create point mutations in MGM1 was carried out using the “PCR SOEing” technique (Horton, 1995). The PCR products containing the mutations were used to replace corresponding wild-type sequences in pRS313-MGM1 via standard cloning techniques. DNA sequence analysis confirmed the presence of the novel mutations and the absence of any PCR-generated errors. Plasmids containing the mutant genes were transformed into the heterozygous (MGM1/mgm1::LEU2) diploid strain MYY973. Transformants were sporulated, tetrads dissected, and mgm1-null spores harboring each of the mutant constructs were identified and analyzed.
Results
mdm17 Cells Display Conditional Defects in Mitochondrial Inheritance, Mitochondrial Morphology, and Maintenance of the Mitochondrial Genome
The mdm17 mutant was isolated in a screen for novel alleles of mdm13, an uncharacterized gene that is required for normal mitochondrial inheritance and morphology. This screen involved crossing a haploid strain harboring the temperature-sensitive mdm13 lesion to a collection of temperature-sensitive strains and analyzing growth of the resulting diploids at the nonpermissive temperature (37°C). One diploid strain displayed temperature-sensitive growth, and analysis of its meiotic progeny indicated that one of two mutations conferring temperature-sensitive growth mapped to a genetic locus unlinked to mdm13. Therefore, rather than being an allele of mdm13, the new mutation defined a distinct gene. Analysis of cells harboring only the new mutation (after its genetic isolation from mdm13) by staining with the mitochondria-specific, vital dye DASPMI and fluorescence microscopy revealed defects in mitochondrial distribution and morphology after incubation at the nonpermissive temperature (Fig. 1). Complementation and allelism tests indicated that the new mutation was unlinked to previously characterized mdm mutations. Genetic analysis further demonstrated that the defects in growth at 37°C, mitochondrial distribution, and mitochondrial morphology were caused by a single nuclear mutation. This novel mutation was designated mdm17.
Figure 1.

mdm17 cells are conditionally defective for mitochondrial inheritance and mitochondrial morphology. Wild-type (MYY291) and mdm17 (MYY972) cells were grown at 23°C in YPD-liquid medium and incubated at (A) 23°C or (B) 37°C for 4 h. Mitochondria were stained with DASPMI and visualized by fluorescence microscopy. Bar, 2 μm.
To quantify the effect of mdm17 on mitochondrial distribution and morphology, cells were stained with DASPMI and examined by fluorescence microscopy. In wild-type cells incubated at either 23°C or 37°C, mitochondria appeared as extended, tubular structures, distributed throughout the cytoplasm of both mother and bud portions of the cell (Fig. 1, A and B). Mitochondria in cells harboring the mdm17 mutation appeared very similar to wild-type at 23°C (Fig. 1 A) but displayed dramatic changes in mitochondrial distribution and morphology after incubation at 37°C (Fig. 1 B). In particular, mitochondrial aggregation and empty daughter buds were apparent in many cells. These alterations in mitochondrial distribution and morphology were confirmed by indirect immunofluorescence microscopy (data not shown).
To examine the development of mutant phenotypes at 37°C, populations of mutant cells were analyzed by indirect immunofluorescence microscopy at various times after shifting to the nonpermissive temperature (Fig. 2, A and B). After 1 h at 37°C, most cells (87%) contained aggregated mitochondria, and a significant fraction of cells (18%) possessed buds devoid of mitochondria. Some of the cells with empty buds still possessed mitochondria with normal tubular morphology (data not shown). By 2 h, 90% of cells displayed aggregated mitochondria and a majority (56%) possessed empty buds. These results demonstrate that the mdm17 mutation rapidly causes defects in mitochondrial distribution and morphology after shifting cells to the nonpermissive temperature.
Figure 2.
Quantitation of mitochondrial inheritance, morphology, and respiration defects in mdm17 cells. Wild-type (MYY291) and mdm17 (MYY972) cells were grown at 23°C, incubated at 37°C for 0, 1, 2, or 4 h, fixed, and processed for indirect immunofluorescence microscopy. Mitochondria were visualized with antibodies against OM14, a mitochondrial outer membrane protein. To quantify the defects, at least 300 budded cells at each time were examined, and the percentages of cells displaying aggregated mitochondria (A) and cells with buds devoid of mitochondria (B) were determined. (C) Wild-type (MYY291) and mdm17 (MYY972) cells were grown in glucose medium (YPD) at 37°C for 0, 1, 2, 4, 16, or 24 h, and plated onto YPD-agar plates. Cells were cultured at 23°C, colonies were replica plated to glycerol medium (YPG), and incubated at 23°C. The percentage of colonies that failed to grow on YPG was scored as having lost respiration competence.
The fraction of cells in the mutant population that showed mitochondrial staining with the potential-dependent dye DASPMI decreased during incubation at the nonpermissive temperature, suggesting that the cells were becoming respiration deficient. To analyze this phenotype, mdm17 cells were incubated at 37°C for various times, plated on glucose (YPD) medium, and cultured at 23°C. Colonies were tested for respiratory competence by replica plating onto glycerol (YPG) medium. At early times, almost all cells were respiration competent (Fig. 2 C). Significant numbers of respiration-deficient cells began appearing in the population only after 4 h at 37°C. After 24 h, >95% of the cells were respiration deficient (Fig. 2 C). When cells from the 16-h time point were stained with the DNA-binding dye 4,6-diamino-2-phenylindole and examined by fluorescence microscopy, mitochondrial DNA staining was no longer evident in a majority of cells (data not shown), indicating that the respiration deficiency reflected a loss of mitochondrial DNA.
mdm17 Is Allelic to MGM1, a Gene Implicated in Maintenance of the Mitochondrial Genome
To identify the molecular basis for the mdm17 phenotypes, the wild-type MDM17 gene was cloned by complementation of the temperature-sensitive growth defect of mdm17 mutant cells. From 10,000 transformants, four strains harboring complementing plasmids were isolated. Restriction mapping of the yeast genomic DNA inserts in these plasmids indicated that all contained overlapping clones. DNA sequence analysis and comparison with sequences in the Saccharomyces Genome Database indicated that the genomic inserts in the plasmids corresponded to a region of chromosome XV. Deletion analysis of the plasmids localized complementing activity to a single open reading frame corresponding to the previously identified MGM1 gene (Jones and Fangman, 1992). Transformation of mdm17 cells with a centromere-based plasmid containing only a single copy of MGM1 complemented all of the mutant phenotypes. Finally, the mdm17 mutation was localized to the MGM1 locus by integrative transformation and mapping (described in Materials and Methods). These results demonstrate that mdm17 is a mutant allele of MGM1.
Previous studies reported that mgm1 mutant cells readily lost mitochondrial DNA and contained aggregated mitochondria but showed normal mitochondrial distribution to buds (Guan et al., 1993). This latter property differs from the phenotype of the mdm17 mutant, where a large fraction of cells exhibited buds devoid of mitochondria. Accordingly, the phenotype of a null mutant in mgm1 generated in the same strain background as the mdm17 mutant was examined. Indirect immunofluorescence microscopy revealed many mgm1-null cells with buds devoid of mitochondria, even in cells cultured at 23°C (Fig. 3). As previously described (Guan et al., 1993), mitochondria in the mgm1-null strain were extensively aggregated.
Figure 3.
mgm1-null cells display defects in mitochondrial inheritance as well as aberrant mitochondrial morphology. mgm1-null cells (MYY974) were grown in YPD at 23°C, fixed, and processed for indirect immunofluorescence. Mitochondria were visualized as described for Fig. 2. Staining of microtubules using an antitubulin antibody demonstrates accessibility of buds to antibodies. Two representative cells are shown. Bar, 2 μm.
Mgm1p Is Localized to the Mitochondrial Outer Membrane
Previously reported phenotypes of the mgm1 mutant, together with the resemblance of the protein's predicted NH2 terminus to a mitochondrial targeting sequence, suggested that Mgm1p is a mitochondrial protein (Guan et al., 1993). However, direct evidence of the protein's subcellular location was lacking. To determine the functional location of Mgm1p, antibodies were generated that were specific for a peptide corresponding to the extreme COOH terminus. These antibodies were used to detect the protein in subcellular fractions. Immunoblotting of total cellular extracts indicated that the affinity-purified antibodies recognized two polypeptides of ∼100 and 90 kD (Fig. 4 A). Neither of these species was apparent in mgm1-null cells (Fig. 4 A), indicating that both polypeptides are products of the MGM1 gene. Furthermore, both species displayed substantially increased levels in cells harboring a multicopy (2 μ) plasmid encoding MGM1 (Fig. 4 A). These increased levels had no apparent effect on mitochondrial distribution and morphology (data not shown).
Figure 4.
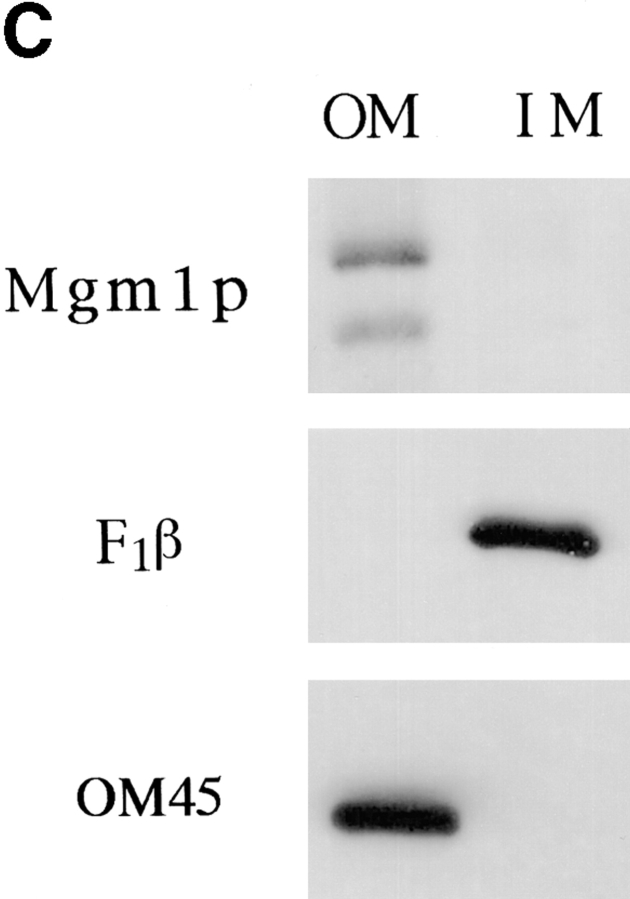
Mgm1p is localized to the mitochondrial outer membrane. (A) Wild-type cells (strain MYY290) (MGM1) and mgm1-null cells (strain MYY974) (mgm1Δ) were grown on YPD at 30°C. Wild-type cells (strain MYY290) harboring a multicopy plasmid encoding MGM1 (p423-MGM1) (2 μ, MGM1) were grown at 30°C on minimal glucose medium without histidine. Extracts of total cellular proteins were prepared by glass bead lysis and subjected to SDS-PAGE and immunoblot analysis using antibodies specific for the COOH terminus of Mgm1p. The mobilities of molecular mass markers are indicated in kilodaltons at the left. (B) Subcellular fractions were prepared by differential centrifugation of homogenate from wild-type cells (MYY291) grown on semisynthetic lactate medium at 30°C. Proteins were separated by SDS-PAGE and analyzed by immunoblotting. Fractions were probed with anti-Mgm1p antibodies (top) or with antibodies specific for the mitochondrial outer membrane protein, OM45 (bottom). Subcellular fractions are all of the following: T, total cell homogenate; L, low speed pellet; M, mitochondrial fraction; I, intermediate speed pellet; H, high speed pellet; and C, cytosolic fraction. (C) Mgm1p is enriched in the mitochondrial outer membrane. Mitochondria from wild-type cells (strain MYY290) were fractionated by osmotic shock followed by sucrose density gradient centrifugation to separate outer and inner membranes. Equivalent amounts (20 μg) of protein from outer (OM) and inner (IM) membrane fractions were analyzed by SDS-PAGE and immunoblot analysis. Fractions were analyzed with anti-Mgm1p antibodies (top), anti-F1β antiserum (middle), and anti-OM45 (bottom). (D) Mitochondria isolated from wild-type cells (MYY290) grown on semisynthetic lactate medium at 30°C were treated with varying concentrations of trypsin for 10 min at 4°C in the presence (+) or absence (−) of 1% Triton X-100. Samples were analyzed by SDS-PAGE and immunoblotting with antibodies against Mgm1p (top), Tom70p, a protein of the mitochondrial outer membrane (middle), and Mas2p, a matrix protein (bottom).
The exact relationship between the two Mgm1p species is unclear, but the 90-kD form appears to be derived from the 100-kD form by proteolysis. The relative amounts of the two species varied randomly in different experiments, and pulse–chase analysis in intact cells failed to reveal a biosynthetic precursor-product relationship (data not shown). However, prolonged incubation of subcellular fractions in vitro (even at 0°C in the presence of a cocktail of protease inhibitors) led to conversion of the larger species to the smaller form (data not shown). These results suggest that the 90-kD form is a product of the partial proteolytic degradation of the 100-kD species.
Earlier characterization of MGM1 failed to identify which of five NH2-terminal methionine residues in the predicted open reading frame actually constituted the translational start of the protein (Jones and Fangman, 1992; Guan et al., 1993). To resolve this uncertainty, in vitro mutagenesis was used to generate versions of mgm1 in which individual methionine codons were converted to alanine codons. These mutant versions were tested for their complementation of the phenotypes of mgm1-null cells. No complementation was observed (and the protein was not detected) when methionine-2 was changed to alanine (data not shown). Mutation of the remaining four methionines did not affect complementation of the mutant defects, and both 100- and 90-kD forms of Mgm1p were detected in cells harboring these genes. These results indicate that the authentic translational start residue is methionine-2 (referred to henceforth as residue 1 of the coding region).
The subcellular distribution of Mgm1p was characterized by immunoblotting of proteins extracted from subcellular fractions isolated from wild-type cells. Mgm1p was localized predominantly to a subcellular fraction enriched in mitochondrial proteins (Fig. 4 B). Analysis of purified submitochondrial fractions revealed that Mgm1p was associated with the mitochondrial outer membrane (Fig. 4 C). Furthermore, Mgm1p was susceptible to protease treatment of intact mitochondria under conditions where internal proteins, such as the matrix-localized Mas2p, were protected (Fig. 4 D).
To examine the nature of Mgm1p association with the outer membrane, isolated mitochondria were extracted with sodium chloride and with sodium carbonate. The latter treatment strips peripheral proteins but leaves integral proteins embedded in the membrane (Fujiki et al., 1982). Mgm1p was resistant to extraction with 1 M NaCl (Fig. 5), indicating that the protein was tightly associated with the membrane. After carbonate treatment, the 90-kD form of Mgm1p was recovered in the soluble fraction, while the 100-kD form remained associated with the membrane (Fig. 5). This result indicates that the full-length Mgm1p is an integral membrane protein. The behavior of the 90-kD fragment suggests that Mgm1p is embedded in the membrane via the putative membrane-spanning domain in the NH2 terminus, since both Mgm1p species are recognized by the antibody specific for the extreme COOH terminus.
Figure 5.
Mgm1p is an integral membrane protein. Mitochondria were isolated from wild-type (MYY290) cells and treated with 0.1 M sodium carbonate, 1 M NaCl, or buffer. Samples were separated into supernatant and pellet fractions by centrifugation at 150,000 g for 1 h, and equivalent amounts were subjected to SDS-PAGE and immunoblot analysis. Supernatant (S) and pellet (P) fractions were analyzed with antibodies against Mgm1p (top) and OM45, an integral protein of the mitochondrial outer membrane (bottom).
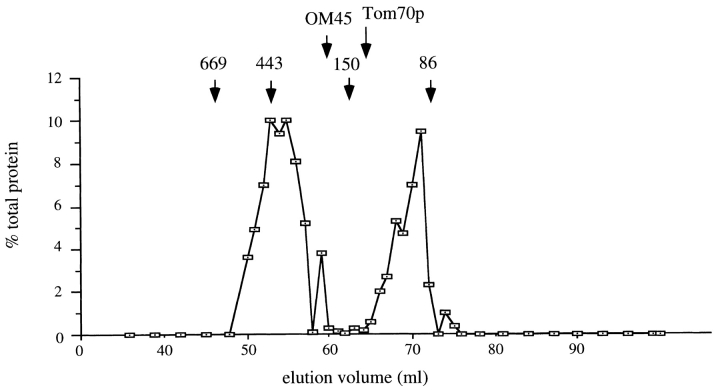
The state of Mgm1p in the mitochondrial outer membrane was further characterized by gel filtration analysis of mitochondrial proteins solubilized in 1% Triton X-100. About 60% of Mgm1p eluted in a peak corresponding to a molecular mass of ∼400 kD, while the remainder of the protein eluted in a peak appropriate to the monomeric size (90–100 kD) (Fig. 6). The 90-kD (fragment) form of the protein eluted in a distinct peak with an apparent molecular mass of ∼120 kD (data not shown). The elution behavior of Mgm1p was distinct from that of two other integral proteins of the mitochondrial outer membrane, Tom70p and OM45 (Fig. 6). These results suggest that Mgm1p is part of a high molecular mass complex either in a homo-oligomeric state or complexed with other proteins on the mitochondrial surface.
Figure 6.
Mgm1p participates in a high molecular mass complex. Purified mitochondria were solubilized in 1% Triton X-100 and the mixture was centrifuged to remove insoluble debris. The supernatant was loaded onto a Sephacryl-300 column and subjected to gel filtration chromatography. Proteins in collected fractions were precipitated with TCA and analyzed by SDS-PAGE and immunoblotting. The percentage of total Mgm1p recovered was plotted for each fraction. Arrows indicate peak fractions for marker proteins: 669 kD, thyroglobulin; 443 kD, apoferritin; 150 kD, carbonic anhydrase; and 86 kD, bovine serum albumin. Also shown are elution peak fractions for two other outer membrane proteins, OM45 and Tom70p. The latter protein was reported to be in a complex with Tom37p (Mas37p) with an apparent molecular mass of ∼110 kD (Gratzer et al., 1995).
Mgm1p Function Depends on the Conserved GTP-binding Domain
To explore further the function of Mgm1p, the identity of the mdm17 mutation was determined. The mutant gene contained a single change: a transition of G to A at nucleotide 880, resulting in a change of Glu294 to Lys. This lesion mapped near the conserved, tripartite GTP-binding motif, suggesting a role for this site in Mgm1p function. To test the importance of the GTP-binding site, a mutant form of MGM1 incorporating a change in a conserved residue, S224N, was created (the numbering of this residue is based on the designation of the second methionine in MGM1 as the NH2-terminal residue as described above). The equivalent mutation in the H-ras-GTPase (S17N) eliminates the protein's binding of GTP (Feig and Cooper, 1988), and a corresponding mutation in rat dynamin (S45N) blocks the protein's function in endocytosis (Herskovits et al., 1993). Immunoblot analysis of cellular extracts detected Mgm1p in mgm1-null cells harboring the mutant gene (Fig. 7 A). However, the mgm1-S224N gene failed to complement any of the mutant phenotypes of a cell containing an mgm1-null lesion (Fig. 7 B and data not shown). In addition, the mgm1-S224N gene did not complement the growth or mitochondrial distribution and morphology defects of the mdm17 mutant cells (Fig. 7 C). These results indicate that an intact GTP-binding domain is essential for Mgm1p function.
Figure 7.
Mgm1p with a mutated GTP-binding site fails to complement growth or mitochondrial inheritance defects of mgm1-null cells. (A) Detection of Mgm1p expressed from the S224N mutant allele. Yeast cells with an mgm1-null mutation (derived from strain MYY973) harboring plasmids encoding either wild-type MGM1 (left) or mgm1-S224N (right) were grown in minimal medium without histidine. Cells were homogenized by glass-bead lysis, and proteins were analyzed by SDS-PAGE and immunoblotting with antibodies specific for Mgm1p. (B) Cells with an mgm1-null mutation (derived from strain MYY973) harboring plasmids encoding wild-type MGM1 (pRS313-MGM1), mgm1-S224N (p313-MGM1-S224N), or vector alone (pRS313) were cultured in different sectors of a YPG-agar plate at 30°C for 48 h. (C) mdm17 cells (strain MYY972) were transformed with plasmids encoding wild-type MGM1 (pRS313-MGM1) or the S224N mutant (pMGM1-S224N) and cultured in minimal medium without histidine at 23°C. Cells were incubated at 37°C for 1 h, stained with DASPMI, and examined by phase-contrast (left of each pair) and fluorescence microscopy (right of each pair). Two representative cells of each type are shown. Bar, 2 μm.
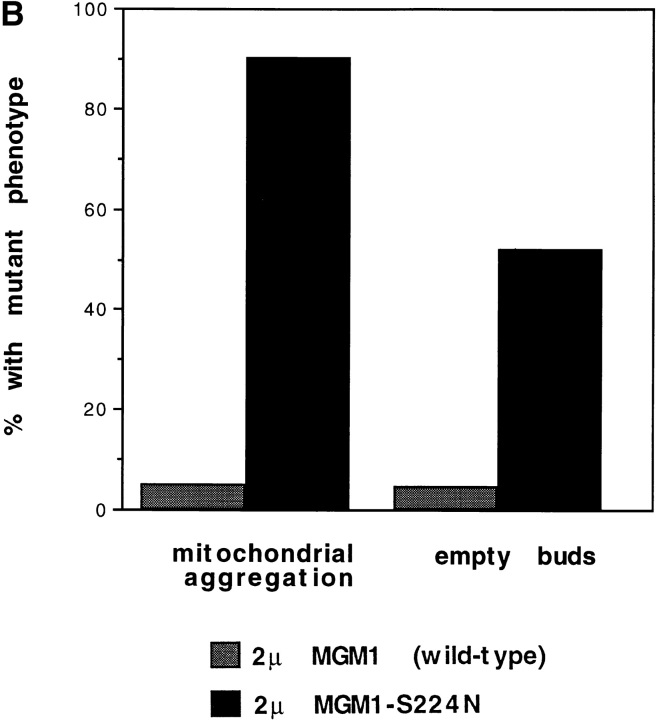
Overexpression of dynamin family members mutated in critical residues of the GTP-binding site was previously found to cause dominant-negative effects in wild-type cells (Warnock and Schmid, 1996). To test whether this is also true for MGM1, high-copy plasmids encoding either wild-type MGM1 or mgm1-S224N were transformed into wild-type cells. Only a small number of transformants harboring the mutant gene was recovered, and Western blot analysis indicated that the mutant protein was not overexpressed in these cells (perhaps reflecting a regulatory or suppression phenomenon). However, both wild-type and mutant proteins were overexpressed in mgm1-null cells harboring the respective plasmids (data not shown). These transformed cells were mated to wild-type cells, and the effect of overexpression in the resulting zygotes was analyzed by fluorescence microscopy. High levels of wild-type MGM1 had no apparent effect on mitochondrial distribution or morphology (Fig. 8 A), but zygotes with elevated levels of the mutant protein displayed defects very similar to those caused by the original mdm17 mutation (Fig. 8 A). In particular, 90% of the latter zygotes displayed aggregated mitochondria (Fig. 8 B), and 52% of the buds produced by these zygotes contained no mitochondria (Fig. 8 B). These results demonstrate that the mgm1-S224N mutant gene product confers dominant-negative effects on mitochondrial distribution and morphology.
Figure 8.
Dominant-negative effects of Mgm1p mutated in the GTP-binding site. Haploid cells with an mgm1-null mutation harboring multicopy plasmids encoding either wild-type (p423-MGM1) or S224N mutant (p423-MGM1-S224N) forms of MGM1 were crossed to wild-type haploid cells. Mitochondria in zygotes were visualized by staining with DASPMI and fluorescence microscopy. (A) Phase-contrast and fluorescence images of two representative budded zygotes harboring wild-type (upper panels) or mutant (lower panels) versions of MGM1. Bar, 2 μm. (B) Quantitation of zygote phenotypes. Stained zygotes (budded and unbudded) were scored for mitochondrial aggregation, and budded zygotes were scored for the presence of stained mitochondria in the buds. At least 300 zygotes were counted for each sample.
Discussion
We have identified a role for Mgm1p in mitochondrial inheritance through the analysis of cells harboring mdm17, a mutation causing temperature-sensitive growth and alterations in mitochondrial distribution and morphology. Genetic analysis revealed that mdm17 is a mutant allele of MGM1, a nuclear gene originally identified by its role in maintenance of the mitochondrial genome (Jones and Fangman, 1992). Two key observations indicate that the primary functions of Mgm1p are to facilitate the maintenance of mitochondrial morphology and to mediate mitochondrial inheritance and that mitochondrial genome maintenance is a secondary role of the protein. First, analysis of mdm17 cells revealed that alterations in mitochondrial morphology and distribution appeared very quickly after a shift to elevated temperature, whereas loss of respiratory competence and the mitochondrial genome occurred only much later. Second, the location of Mgm1p on the mitochondrial surface is consistent with a primary function of the protein in morphology and distribution, but not with a direct role in maintenance of the matrix-localized mitochondrial DNA. In addition, several other mutations that cause aberrant mitochondrial morphology also cause increased loss of mitochondrial DNA (Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997). This loss appears to be a secondary effect of the morphological changes.
Mgm1p is one of a small group of outer membrane proteins that have been shown to be required for mitochondrial inheritance. Mdm10p, Mdm12p, and Mmm1p are also integral proteins of the outer membrane whose loss leads to dramatic alterations in mitochondrial morphology and defects in transmission of mitochondria into developing daughter buds (Burgess et al., 1994; Sogo and Yaffe, 1994; Berger et al., 1997). Although the specific molecular activities of these other components are unknown, their location in the mitochondrial outer membrane and their exposed cytoplasmic domains suggest two potential models of function. One possibility is that these proteins serve as binding sites or anchor points for interaction with cytoskeletal structures or molecular motors. Such interactions might pull mitochondria into tubular structures and mediate mitochondrial movement into buds. A second possibility is that the outer membrane components regulate structural properties of the outer membrane, such as membrane fluidity, which are themselves essential for normal mitochondrial morphology and inheritance. Additionally, mitochondrial inheritance may depend on a tubular morphology that is maintained by these outer membrane proteins. Mgm1p may function in concert with the other outer membrane proteins, or it may fulfill a unique role. This latter possibility is suggested by the observation that phenotypes caused by mdm10, mdm12, and mmm1 are suppressed by the SOT1 mutation (Berger et al., 1997) but defects caused by mutations in mgm1 are not (data not shown).
Mgm1p is a member of the dynamin family of large GTP-binding proteins. The best characterized member of this family is dynamin itself, which plays an essential role in clathrin-mediated endocytosis in animal cells (Herskovits et al., 1993; van der Bleik et al., 1993). Although S. cerevisiae does not have a true dynamin orthologue, this yeast possesses two other dynamin-like proteins, Vps1p (Vater et al., 1992) and Dnm1p (Gammie et al., 1995). Vps1p is localized to the Golgi apparatus and participates in the vesicular transport of proteins to the vacuole (Vater et al., 1992). Dnm1p may function in the endosomal pathway (Gammie et al., 1995), but was recently found to play an important role in mitochondrial distribution and morphology (Otsuga et al., 1998). In particular, cells deleted for Dnm1p displayed mitochondrial tubules collapsed along one side of the cell and extending from the mother portion of the cell into the daughter bud. A related phenotype, the collapse of mitochondrial tubules into perinuclear aggregates, was observed in an independent study in which a mutant form of the human dynamin-like protein, Drp1, was expressed in cultured mammalian cells (Smirnova et al., 1998). These observations suggest that Dnm1p and its mammalian homologue Drp1 may mediate the lateral distribution or branching of mitochondrial tubules.
Many molecular details of dynamin's function in endocytosis remain to be clarified, but key features of the protein's activity include GTP hydrolysis and assembly of dynamin into oligomeric structures (Herskovits et al., 1993; Hinshaw and Schmid, 1995). Dynamin has been proposed to act as a mechanical constrictor for the neck of an invaginating coated pit (Warnock and Schmid, 1996), or as a regulator or recruiter of the constriction machinery (Roos and Kelly, 1997). Although it is not apparent that a constricting activity is necessary for the maintenance of mitochondrial morphology and distribution, our results suggest that Mgm1p shares two key functional features with dynamin. First, mutational analysis revealed that the GTP-binding site is essential for Mgm1p function, suggesting the importance of GTP hydrolysis. Second, the dominant-negative effect of the S224N mutation and the wild-type protein's gel filtration profile suggest that Mgm1p functions as part of an oligomeric complex. Mgm1p might play a structural role in locally influencing the properties of the outer membrane. For example, Mgm1p could assemble into a structure that promotes a tubular mitochondrial morphology. Alternatively, Mgm1p might fulfill a regulatory function in controlling the activity of other membrane components. More details of the protein's function should emerge from identification of interacting proteins.
Acknowledgments
This work was supported by grant GM44614 from the National Institutes of Health.
Abbreviations used in this paper
- DASPMI
2-(4-dimethylaminostryl)- 1-methylpyridinium iodide
- mdm
mitochondrial distribution and morphology mutant
- YPD
yeast extract/peptone/glucose
- YPG
yeast extract/ peptone/glycerol
Footnotes
Address correspondence to Dr. Michael Yaffe, Department of Biology, 0347, University of California, San Diego, La Jolla, CA 92093-0347. Tel.: 619-534-4769. Fax: 619-534-4403. E-mail: myaffe@ucsd.edu
This paper is dedicated to the memory of Segall Livneh (1973–1991), who generated the collection of temperature-sensitive strains from which the mdm17 mutant was isolated. We are grateful to Randy Hampton, Karen Berger, and Peter Fekkes for valuable comments on the manuscript and members of the Yaffe lab for their insightful advice and helpful suggestions. We thank Rick Roberts for purification of membrane fractions, Jim Kadonaga for use of the FPLC apparatus, and Alan Kutach for his invaluable help in FPLC analysis.
References
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. . Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Yaffe MP. Mitochondrial distribution and inheritance. Experientia (Basel) 1996;52:1111–1116. doi: 10.1007/BF01952109. [DOI] [PubMed] [Google Scholar]
- Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE. MMM1encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Feig LA, Cooper GM. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment. Application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie AE, Kurihara LJ, Vallee RB, Rose MD. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratzer S, Lithgow T, Bauer RE, Lamping E, Paltauf F, Kohlwein SD, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Farh L, Marshall TK, Deschenes RJ. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr Genet. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Shaw JM. Mitochondrial dynamics in yeast. Annu Rev Cell Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits JS, Burgess CC, Obar RA, Vallee RB. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Horton RM. PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol Biotechnol. 1995;3:93–99. doi: 10.1007/BF02789105. [DOI] [PubMed] [Google Scholar]
- Jones BA, Fangman WL. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Yaffe MP. Nuclear and mitochondrial inheritance in yeast depends on novel cytoplasmic structures defined by the MDM1 protein. J Cell Biol. 1992;118:385–395. doi: 10.1083/jcb.118.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SJ, Yaffe MP. Intermediate filament formation by a yeast protein essential for organelle inheritance. Science. 1993;260:687–689. doi: 10.1126/science.8480179. [DOI] [PubMed] [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP. Temperature-sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol. 1990;111:967–976. doi: 10.1083/jcb.111.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. . J Biol Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Roos J, Kelly RB. Is dynamin really a ‘pinchase'? . Trends Cell Biol. 1997;7:257–259. doi: 10.1016/S0962-8924(97)01068-4. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer I, Emr S, Gross C, Schekman RW. Invertase signal and mature sequence substitutions that delay intercompartmental transport of active enzyme. J Cell Biol. 1985;100:1664–1675. doi: 10.1083/jcb.100.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BJ, Yaffe MP. A mutation in the yeast heat-shock factor gene causes temperature-sensitive defects in both mitochondrial protein import and the cell cycle. Mol Cell Biol. 1991;11:2647–2655. doi: 10.1128/mcb.11.5.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Stinchcomb DT, Scherer S, Davis RW. High frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an early stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock DE, Schmid SL. Dynamin GTPase, a force-generating molecular switch. Bioessays. 1996;18:885–893. doi: 10.1002/bies.950181107. [DOI] [PubMed] [Google Scholar]
- Yaffe MP. Analysis of mitochondrial function and assembly. Methods Enzymol. 1991;194:627–643. doi: 10.1016/0076-6879(91)94046-f. [DOI] [PubMed] [Google Scholar]
- Yaffe MP. Isolation and analysis of mitochondrial inheritance mutants from Saccharomyces cerevisiae. . Methods Enzymol. 1995;260:447–453. doi: 10.1016/0076-6879(95)60157-0. [DOI] [PubMed] [Google Scholar]