Abstract
Members of the eukaryotic heat shock protein 70 family (Hsp70s) are regulated by protein cofactors that contain domains homologous to bacterial DnaJ. Of the three DnaJ homologues in the yeast rough endoplasmic reticulum (RER; Scj1p, Sec63p, and Jem1p), Scj1p is most closely related to DnaJ, hence it is a probable cofactor for Kar2p, the major Hsp70 in the yeast RER. However, the physiological role of Scj1p has remained obscure due to the lack of an obvious defect in Kar2p-mediated pathways in scj1 null mutants. Here, we show that the Δscj1 mutant is hypersensitive to tunicamycin or mutations that reduce N-linked glycosylation of proteins. Although maturation of glycosylated carboxypeptidase Y occurs with wild-type kinetics in Δscj1 cells, the transport rate for an unglycosylated mutant carboxypeptidase Y (CPY) is markedly reduced. Loss of Scj1p induces the unfolded protein response pathway, and results in a cell wall defect when combined with an oligosaccharyltransferase mutation. The combined loss of both Scj1p and Jem1p exaggerates the sensitivity to hypoglycosylation stress, leads to further induction of the unfolded protein response pathway, and drastically delays maturation of an unglycosylated reporter protein in the RER. We propose that the major role for Scj1p is to cooperate with Kar2p to mediate maturation of proteins in the RER lumen.
Keywords: glycosylation, endoplasmic reticulum, chaperones, heat-shock protein 70, oligosaccharides
Kar2p and Lhs1p (Cer1p/Ssi1p), members of the heat shock protein 70 family (Hsp70s)1 perform several diverse functions in the lumen of the rough endoplasmic reticulum of Saccharomyces cerevisiae. Kar2p, which is the yeast homologue of mammalian BiP, acts as a chaperone to mediate protein folding in the ER lumen (te Heesen and Aebi, 1994; Simons et al., 1995). BiP/Kar2p prevents inappropriate interactions between hydrophobic segments of nascent polypeptides via ATP hydrolysis-dependent cycles of peptide binding and release (Flynn et al., 1989; Gaut and Hendershot, 1993). Reducing agents, glycosylation inhibitors, and other agents that interfere with protein folding in the RER induce the unfolded protein response (UPR) pathway leading to enhanced expression of Kar2p and other RER chaperones including protein disulfide isomerase (Kozutsumi et al., 1988; Cox et al., 1993). Mutations in the essential KAR2 gene were first isolated in a screen for yeast that have unilateral karyogamy defects (Rose et al., 1989). Although the precise role of Kar2p in nuclear fusion during karyogamy is not fully understood, kar2 mutants are also defective in an in vitro assay that monitors homotypic fusion of ER derived vesicles (Latterich and Schekman, 1994). Class I kar2 mutants are also defective in translocation of proteins across the endoplasmic reticulum (Vogel et al., 1990). Loss of Lhs1p, a nonessential and presumably less abundant RER Hsp70 protein, causes cytoplasmic accumulation of a subset of yeast secretory precursors suggesting a partial overlap in function between Kar2p and Lhs1p in nascent chain translocation (Baxter et al., 1996; Craven et al., 1996; Hamilton and Flynn, 1996). Disruption of the LHS1 gene reduces the ability of yeast to refold heat-induced protein aggregates in the endoplasmic reticulum (Saris et al., 1997). Transcription of the LHS1 gene is induced by the UPR pathway but not by elevated temperatures (Baxter et al., 1996; Craven et al., 1996).
The ATP hydrolysis cycle of Hsp70 proteins is regulated by cofactors that are members of the DnaJ protein family (as reviewed by Bukau and Horwich, 1998). DnaJ proteins interact with Hsp70 via a conserved ∼80-residue J domain. The yeast endoplasmic reticulum contains three members of the DnaJ protein family (Sec63p, Scj1p and Jem1p) that likely cooperate with Kar2p and Lhs1p (Sadler et al., 1989; Schlenstedt et al., 1995; Nishikawa and Endo, 1997). Sec63p, an integral component of the yeast translocation complex (Deshaies et al., 1991; Brodsky and Schekman, 1993; Panzner et al., 1995), has a J domain located within a lumenally disposed loop (Sadler et al., 1989). Synthetic interactions between kar2 mutants and sec63-1 provide genetic support for a direct interaction between Kar2p and Sec63p (Scidmore et al., 1993). An amino acid substitution in the J domain of Sec63p (sec63-1 allele) confers a temperature sensitive defect in protein translocation due to a reduced interaction with Kar2p (Brodsky and Schekman, 1993) resulting in precursor stalling within the Sec61 translocation pore (Lyman and Schekman, 1995). Hydrolysis assays have shown that the J domain of wild-type Sec63p, but not the sec63-1 mutant, stimulates ATP hydrolysis by Kar2p (Corsi and Schekman, 1997). Posttranslational translocation of proteins across the yeast RER is ATP dependent (Brodsky and Schekman, 1993; Panzner et al., 1995) leading to the hypothesis that the interaction between Kar2p and Sec63p promotes nascent chain transport through the Sec61 complex by iterative cycles of nascent chain binding and ATP hydrolysis–dependent release (Brodsky and Schekman, 1993; Lyman and Schekman, 1995).
The roles of Scj1p and Jem1p in the RER are less well defined. The growth rate of yeast is not altered by disruption of either the SCJ1 or the JEM1 gene (Schlenstedt et al., 1995; Nishikawa and Endo, 1997); simultaneous disruption of both genes yields a strain that is inviable at 37°C (Nishikawa and Endo, 1997). Synthesis of Jem1p and Scj1p, unlike Sec63p, is induced by treatments that activate the UPR pathway (Schlenstedt et al., 1995; Nishikawa and Endo, 1997). The COOH-terminal J domain of Jem1p is believed to interact with Kar2p, as the nuclear fusion defect that occurs when Δjem1 strains are mated also occurs when the J domain of Jem1p bears a point mutation identical to the sec63-1 allele that interferes with the Sec63p-Kar2p interaction (Nishikawa and Endo, 1997). Several lines of evidence suggest that the lumenal Scj1 protein can interact with Kar2p in the ER via the conserved NH2-terminal J domain (Schlenstedt et al., 1995). The J domain from Scj1p, but not the J domains from DnaJ homologues in the yeast cytoplasm (Sis1p) or mitochondria (Mdj1p), can replace the essential J domain in Sec63p to promote translocation of proteins across the RER (Schlenstedt et al., 1995). Synthetic genetic interactions between Δscj1 and certain kar2 alleles strongly suggest that Scj1p serves as a cofactor for Kar2p (Schlenstedt et al., 1995). Despite this genetic evidence for a functional interaction between Scj1p and Kar2p, cells lacking Scj1p do not have detectable defects in protein translocation across the RER (Blumberg, H., and P.A. Silver, unpublished results) or in nuclear membrane fusion during mating (Nishikawa and Endo, 1997). Consequently, a functional role for Scj1p in cellular processes that require Kar2p remains to be elucidated.
The oligosaccharyltransferase (OST) catalyzes the transfer of a high-mannose oligosaccharide onto asparagine residues of nascent polypeptides entering the lumen of the rough endoplasmic reticulum. Mutagenesis of the N-X-T/S sites in acid phosphatase (AP) and carboxypeptidase Y (CPY) has shown that hypoglycosylated proteins either fail to fold and are retained in the ER (AP), or exit the ER at reduced rates (CPY; Riederer and Hinnen, 1991; Winther et al., 1991) indicating that N-linked oligosaccharides are important for protein folding in yeast. Mutations in each of the eight genes that encode the subunits of the yeast OST result in reductions in the in vivo and in vitro OST activity (for a review see Silberstein and Gilmore, 1996). Isolation of the OST mutants indicates that significant underglycosylation of glycoproteins is tolerated by yeast with a functional UPR pathway (Cox et al., 1993).
In an effort to obtain insight into the role of the Scj1 protein in the endoplasmic reticulum, yeast mutants were sought that were inviable when combined with a Δscj1 mutation. The synthetic lethal screen yielded a mutant that could be complemented by the OST1 gene. We found that mild reductions in N-linked glycosylation are poorly tolerated in the absence of Scj1p, as a Δost3Δscj1 mutant has a severe growth defect at 37°C compared with Δost3 mutants that grow at wild-type rates. Loss of Scj1p induces the UPR pathway, without yielding a detectable defect in cell wall biosynthesis. However, Δscj1 cells are hypersensitive to reducing agents and tunicamycin, and show a delay in the transport of nonglycosylated CPY from the RER to the Golgi. Jem1p and Scj1p appear to have partially overlapping functions as cofactors for Kar2p. Although the mild reduction in growth rate of the Δost3Δjem1 mutant at 37°C appears to argue against an important role for Jem1p in protein folding, overexpression of Jem1p can suppress the slow growth phenotype of the Δost3Δscj1 mutant at 37°C. Our results suggest that Scj1p cooperates with Kar2p to mediate folding of proteins under conditions of stress induced by hypoglycosylation.
Materials and Methods
Growth Media
Yeast were grown in YPDA medium (2% bacto-peptone, 1% yeast extract, 2% glucose, 0.003% adenine sulfate) to avoid accumulation of the red pigment in ade2 cells, or in synthetic minimal media (0.67% yeast nitrogen base, 2% glucose) supplemented with adenine and the appropriate amino acids or uracil. Growth media for drug sensitivity assays was prepared by adding aliquots from stock solutions of tunicamycin (10 mg/ml in 0.1 M NaOH), Calcofluor White (10 mg/ml in 1:1 ethanol-water), hygromycin B (10 mg/ml in water), or β-mercaptoethanol to YPDA-2% agar immediately before pouring the plates.
Isolation of Mutants Synthetically Lethal with Δscj1
The strain for the synthetic lethality screen was constructed in several steps. PSY586 was obtained as a segregant from a cross of MS4 and MS20 (provided by Mark Rose, Princeton University, Princeton, NJ). The plasmid pPS753 was constructed to precisely disrupt the SCJ1 gene. Plasmid pPS176, which contains the SCJ1 gene (Blumberg and Silver, 1991) as a 4-kb KpnI fragment in pBluescript, was digested with NsiI and BalI to excise the SCJ1 gene plus 100 bp of 5′ and 29 bp of 3′ flanking sequence. A 958-bp SmaI-PstI fragment from pJJ248 (Jones and Prakash, 1990) containing the TRP1 gene was cloned into pPS176 digested with NsiI and BalI to obtain pPS753. KpnI-digested pPS753 was used to transform PSY586 to tryptophan prototrophy to obtain the strain designated PSY729. Southern blotting confirmed the disruption of the SCJ1 gene in PSY729. PSY586 was crossed to CH1305 (Kranz and Holm, 1990) to introduce the ade3 mutation; after sporulation and tetrad dissection white colonies (ade2ade3) were selected to obtain PSY7-10D. The strains for the synthetic lethality screen (PSY759 and PSY761) were segregants from the diploid obtained by crossing PSY7-10D with PSY729. To construct the URA3 ADE3 SCJ1 plasmid (pPS720) for the colony sectoring assay, a 3,673-bp fragment containing the ADE3 gene was cloned into BamHI-SpeI digested pRS426, a URA3 marked 2μ vector (Christianson et al., 1992) to obtain the plasmid pPS719. A 1.7 kb fragment containing the SCJ1 gene was excised from YEpSCJ1 (Schlenstedt et al., 1995) by digestion with BamHI and SalI, and ligated to BamHI-SalI digested pPS719. The strain PSY759 was transformed to uracil prototrophy with pPS720. The resulting strain was grown to a density of 5 × 107 cells/ml at 30°C in synthetic minimal media lacking uracil and mutagenized by UV-exposure to 50% survival as described (Lawrence, 1991). Nonsectoring mutants were selected and isolated by restreaking three times on YPD plates. Nonsectoring mutants did not grow on plates containing 5-fluoro-orotic acid (5-FOA) unless they were first transformed with a LEU2 marked SCJ1 plasmid, thereby showing that the synthetic phenotype of the mutant strain is due to a requirement for the SCJ1 gene, not the ADE3 or URA3 genes. Nonsectoring mutants were transformed with a yeast genomic library on a 2μ LEU2-marked plasmid (a gift from Richard Kolodner, Harvard Medical School, Boston, MA) to identify genes that could suppress the synthetic lethal phenotype.
Strain Constructions
To minimize the effects of strain background, PSY771 was crossed with RGY132, an α-haploid derived by sporulation of YPH274 (Sikorski and Hieter, 1989). A temperature-sensitive, trp− his− lys− haploid (ost1-6 SCJ1) derived from the initial cross was backcrossed with RGY132 and sporulated to obtain RGY140, and a nontemperature-sensitive, trp+ his− lys− haploid (OST1 Δscj1::TRP1) from the initial cross was backcrossed to RGY132 and sporulated to obtain trp− (RGY141) and trp+ (RGY142) haploids. RGY324 (derived from the diploid RGY302; Karaoglu et al., 1995a ) was mated with RGY142. The resulting diploid was sporulated, and asci dissected to obtain strains RGY143A-D that are the haploid progeny from a tetratype ascus. To obtain RGY144, the SCJ1 gene was disrupted by transforming RGY324 to uracil prototrophy with the KpnI-linearized URA3 marked plasmid pΔSCJ1 (Blumberg and Silver, 1991). Correct integration of the plasmid into the SCJ1 gene was confirmed by PCR. The strain RGY145 was obtained by sporulation of the diploid obtained by crossing RGY144 with RGY131.
A one-step PCR-based gene disruption method (Wach et al., 1994; Wach, 1996) was used to disrupt the JEM1 gene to obtain RGY146. PCR was used to generate a DNA fragment containing a HIS5 gene from S. pombe flanked by 5′ (nucleotides −37 to 5) and 3′ (nucleotides 2015 to 2055) regions from the JEM1 gene (Wach, 1996). The pFA6a-HIS3MX6 template for the PCR was kindly provided by Peter Philippsen (University of Basel, Switzerland). After transformation of RGY324 with the disruption cassette, histidine prototrophs were selected and integration of the cassette into the JEM1 gene was confirmed by PCR. RGY146 was crossed with RGY142 to obtain a diploid, which after sporulation and dissection yielded RGY147 and RGY148.
Radiolabeling and Immunoprecipitation of Proteins
To obtain strains expressing unglycosylated CPY (ug-CPY), yeast were transformed with the URA3 marked centromeric plasmid pJW373, that bears a modified PRC1 gene that lacks all four N-glycosylation sites (Winther et al., 1991). The plasmid pJW373 was a generous gift from Jakob R. Winther (Carlsberg Laboratory, Denmark).
Yeast were grown at 25°C in synthetic minimal media supplemented with the appropriate amino acids until mid-log phase (0.5–0.8 OD at 600 nm). Cells collected by centrifugation were resuspended at a density of 6 A600/ml of media and radiolabeled for 10 min at 25°C with 50 μCi of Tran- 35S-label (ICN Biomedicals Inc., Costa Mesa, CA) per A600 unit of cells. In pulse–chase experiments, 100 μl of a fresh solution of unlabeled methionine and cysteine (each at 5 mg/ml) was added after 10 min to initiate the chase incubation that continued at the same temperature. Aliquots of the radiolabeled cultures were taken at the indicated times, added to an equal volume of 20 mM sodium azide, frozen in liquid nitrogen and stored at −80°C. Rapid lysis of cells with glass beads and immunoprecipitation of carboxypeptidase Y (CPY) was done as described (Rothblatt and Schekman, 1989). Immunoprecipitation of Kar2p was performed as described for CPY using 5 μl of antiserum against Kar2p (a gift from Peter Walter, UCSF) per 1.5 A600 of labeled yeast cells. Coimmunoprecipitation of unfolded protein substrates with Kar2p was performed as described (Simons et al., 1995). In brief, Kar2p was immunoprecipitated from radiolabeled yeast cells that were lysed with glass beads in nondenaturing lysis buffer (10 mM Tris-Cl [pH 8.0], 0.3M NaCl) containing 2% CHAPS and 30 U/ml potato apyrase. The Kar2p immunoprecipitates were washed once with nondenaturing lysis buffer containing 0.5% CHAPS and boiled in the presence of 1% SDS. CPY was immunoprecipitated from denatured Kar2p immunocomplexes as described (Simons et al., 1995).
RNA Extraction and Northern Blot Analysis
Yeast cells were grown in YPDA at 25°C until mid-log phase. As indicated, tunicamycin was added 2 h before cells were collected at a concentration of 1 μg/ml. Whole-cell RNA was prepared using the hot phenol procedure (Kohrer and Domdey, 1991). Samples of RNA (20 μg) were resolved by 1% agarose/formaldehyde gel electrophoresis and transferred to Hybond-N+ membranes (Amersham Corp.) in 20× SSC for 20 h (Sambrook et al., 1989). The membranes were prehybridized for 2 h and hybridized overnight in 35% formamide at 42°C as described (Sambrook et al., 1989) with probes specific for yeast KAR2 (nucleotides 1088 to 2376) or JEM1 (nucleotides 156 to 1293) generated by PCR. Membranes were washed twice with 1× SSC, 0.1% SDS at 55°C and exposed. The Hybond membranes were stripped according to manufacturer's recommendations before reprobing with a 1.5-kb EcoRI/ScaI fragment of the ACT1 gene. pDH6, a construct bearing the ACT1 was provided by Allan Jacobson, UMMC. Prehybridization, hybridization, and washes were performed as described above for the KAR2 probe. Hybridization probes were 32P-labeled using the Oligolabeling Kit (Pharmacia Biotech, Inc., Piscataway, NJ). Radioactive bands were quantified with a PhosphorImager (Molecular Dynamics, Inc., Sunnyvale, CA) and visualized by autoradiography.
Results
Identification of ost1-6 as a Temperature-sensitive Allele of the OST1 Gene
Previous studies have established that expression of Scj1p is not essential for viability of yeast, yet the homology between Scj1p and the E. coli DnaJ protein strongly suggests that Scj1p functions as a cofactor for Kar2p and/or Lhs1p, DnaK homologues located within the yeast endoplasmic reticulum. To further explore the role of Scj1p in the yeast RER, the ade2ade3 colony-sectoring screen (Koshland et al., 1985; Bender and Pringle, 1991) was used to identify mutations that were incompatible with a null allele of the SCJ1 gene. Two Δscj1ade2ade3 strains that differed with respect to mating type were transformed with a plasmid bearing the URA3, ADE3 and SCJ1 genes. After UV mutagenesis, yeast mutants were isolated that were unable to lose the plasmid by their failure to form sectored red and white colonies. Potential synthetic lethal strains were then tested for the ability to grow on plates containing 5-FOA. From among 45,000 colonies that were screened, a single mutant strain (PSY771) was unable to lose the plasmid bearing the SCJ1 gene.
PSY771 was inviable at 37°C in the presence of the SCJ1 plasmid. Mating with a parental strain (PSY761) showed that the synthetic lethal and temperature sensitive phenotypes were both recessive, and furthermore these two phenotypes cosegregated after sporulation and tetrad dissection. To identify the mutant gene responsible for the synthetic lethal phenotype, PSY771 was transformed with a high copy yeast genomic plasmid bank. Plasmids that were subsequently isolated from sectoring transformants were found to contain the OST1 gene encoding the α subunit of the yeast OST complex (Silberstein et al., 1995). Crosses between PSY771 and two previously characterized temperature sensitive ost1 mutants (RGY121 and RGY122; Silberstein et al., 1995) yielded diploids that were temperature sensitive, indicating that PSY771 bears a mutant allele of ost1 which we designate as ost1-6. Finally, we noted that PSY771 could lose the SCJ1 plasmid at 25°C, provided that the growth media contained 2 M glycerol, suggesting that the Δscj1ost1-6 strain is osmotically fragile in standard growth media.
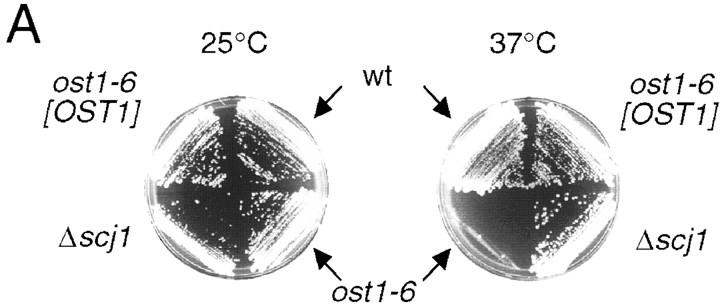
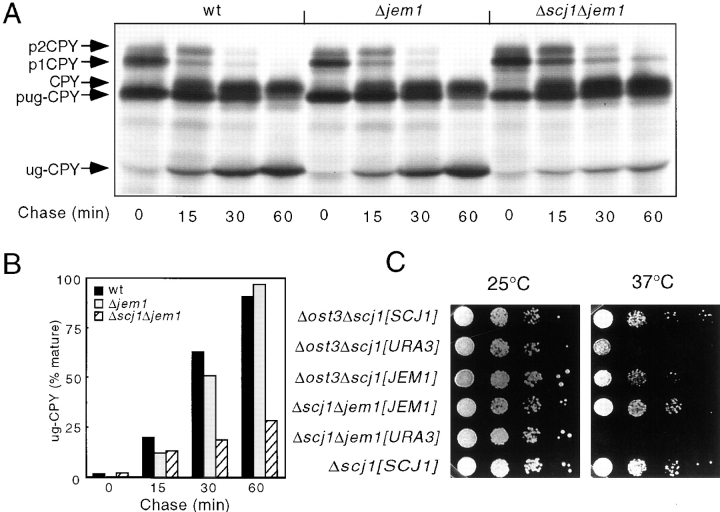
The initial ost1-6 strain was backcrossed several times with RGY131 to minimize the effect of strain background on the subsequent analysis, and to separate the Δscj1 and ost1-6 mutations. In this new genetic background, the resulting ost1-6 strain displays a temperature-sensitive phenotype (Fig. 1 A). Previously isolated ost1 mutants have reduced OST activity as detected by pleiotropic hypoglycosylation of N-linked glycoproteins synthesized in vivo at both the permissive (25°C) and restrictive (37°C) temperatures (Silberstein et al., 1995). Biosynthesis of the soluble vacuolar glycoprotein CPY by wild-type, Δscj1, ost1-6, and ost1-6 cells complemented with pRS316-OST1 was analyzed to determine whether the ost1-6 mutant has reduced OST activity at the permissive temperature. CPY is synthesized as a proenzyme that acquires four N-linked oligosaccharides upon translocation into the lumen of the endoplasmic reticulum. The 67-kD ER form of proCPY (p1CPY) is transported to the Golgi complex, where the core oligosaccharides are elongated by the addition of mannose residues to yield the 69-kD Golgi form of proCPY (p2CPY). Upon transport to the vacuole, proteolytic removal of an 8-kD propeptide generates the mature 61-kD vacuolar form of CPY. CPY was immunoprecipitated from yeast cultures that were radiolabeled with 35S methionine for 1 h at 25°C (Fig. 1 B). As expected, the predominant form of CPY synthesized by the wild-type and Δscj1 mutant has four N-linked oligosaccharides. In addition to fully glycosylated CPY, the ost1-6 mutant strain synthesized hypoglycosylated variants of CPY that lack between one and three oligosaccharides (Fig. 1 B, −1, −2, and −3 forms of CPY). Notably, the synthetic lethal phenotype, the temperature sensitive growth defect (Fig. 1 A), and the glycosylation defect of the ost1-6 mutant (Fig. 1 B) are corrected by expression of Ost1p from a centromeric plasmid. From the preceding results, we conclude that the temperature-sensitive phenotype of the ost1-6 allele is due to reduced levels of N-glycosylation caused by a mutation in OST1, the α subunit of the OST complex.
Figure 1.
Characterization of ost1-6, a temperature-sensitive allele of the OST1 gene. (A) Growth phenotype of the ost1-6 mutant. Yeast strains (RGY140, RGY140 [pRS316-OST1], RGY141, and RGY142) were streaked on YPDA-agar and incubated for 2 d at the indicated temperatures. (B) Hypoglycosylation of CPY by the ost1-6 mutant. CPY immunoprecipitates from glass bead extracts of yeast strains radiolabeled for 1 h at 25°C were resolved by PAGE in SDS. Fully glycosylated vacuolar CPY and hypoglycosylated variants of CPY lacking between 1 and 4 N-linked oligosaccharides are indicated by the labeled arrows designated −1 through −4.
A Synthetic Growth Defect Results when Δost3 and Δscj1 Are Combined
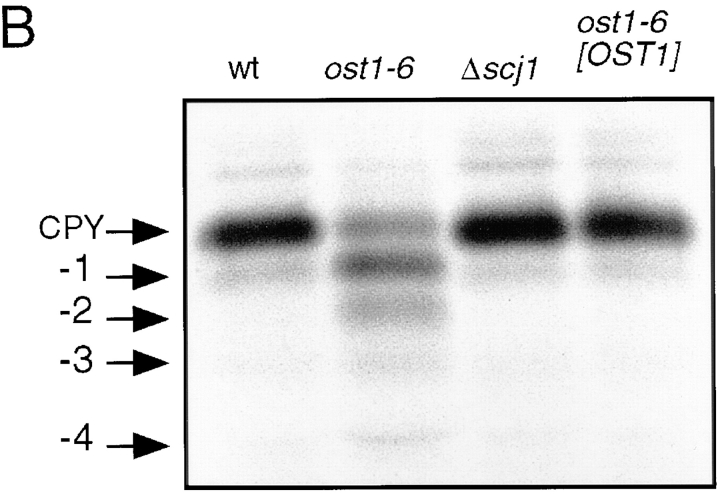
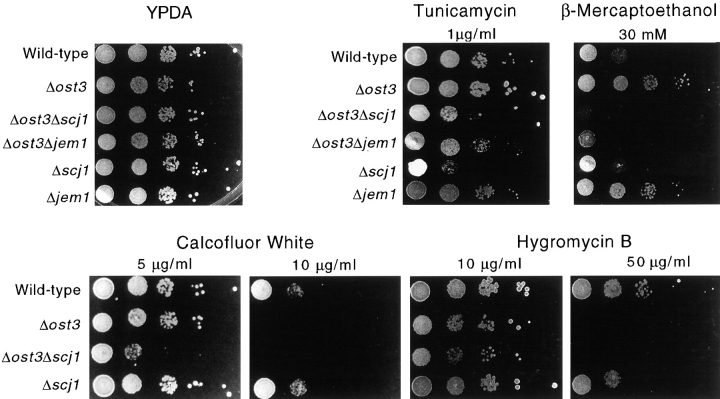
We postulated that the synthetic lethal phenotype produced by the combination of the Δscj1 and ost1-6 mutations was not caused by an exaggeration of the glycosylation defect inherent to ost1-6, as glycosylation of CPY in the Δscj1 strain is indistinguishable from that of the wild-type strain. However, given that Scj1p is proposed to cooperate with the ER chaperone Kar2p (Schlenstedt et al., 1995), it is formally possible that folding or oligomeric assembly of the mutant form of Ost1p is impaired in a Δscj1 strain thereby causing an indirect reduction in OST activity. To address this possibility, the Δscj1 strain was crossed to a milder OST mutant caused by disruption of the nonessential OST3 gene encoding the 34-kD subunit of the OST. We selected the Δost3 mutant for this analysis because the strain grows at a wild-type rate at 25, 30, and 37°C (Karaoglu et al., 1995a ). Moreover, immunoprecipitation experiments from strains that express an epitope-tagged OST subunit (Stt3p) have revealed that the integrity and stability of the OST complex is not compromised in yeast cells that do not express Ost3p (Karaoglu et al., 1997). After sporulation of the Δost3Δscj1 diploid, dissection of tetrads yielded four viable colonies at 25°C from tetratype asci. As reported previously (Blumberg and Silver, 1991; Karaoglu et al., 1995a ), the Δost3 and Δscj1 mutants grow at wild-type rates on YPD plates incubated at 25°C or 37°C. In contrast, the Δost3Δscj1 double mutant has a severe growth defect at 37°C, but not at 25°C (Fig. 2 A).
Figure 2.
Growth and N-linked glycosylation of the Δost3Δscj1 mutant. (A) Severe growth defect of the Δost3Δscj1 mutant at 37°C. Yeast strains (RGY143A, RGY143B, RGY143C, and RGY143D) were grown in liquid YPDA medium at 25°C and diluted to a density of 106 cells/ml. 5-μl aliquots of 10-fold serial dilutions of the cultures were plated on YPD-agar and incubated at 25 and 37°C for 2 d. (B) Glycosylation of CPY in vivo. CPY was immunoprecipitated from glass bead extracts of yeast cultures radiolabeled for 10 min at 25°C and then chased for 0 or 15 min. The ER form (p1CPY), the Golgi form (p2CPY), and fully glycosylated vacuolar CPY were resolved by PAGE in SDS and are designated by labeled arrows. The migration positions corresponding to hypoglycosylated variants of CPY lacking between 1 and 4 N-linked oligosaccharides are indicated.
CPY was immunoprecipitated from yeast cultures that were pulse labeled at 25°C for 10 min and then chased for 0 or 15 min to determine whether the loss of Scj1p exaggerates the glycosylation defect of a Δost3 mutant (Fig. 2 B). Nontranslocated prepro-CPY is not detected in the immunoprecipitates from Δscj1 cells (Fig. 2 B), confirming the previous observation that Scj1p does not have a role in protein translocation across the ER (Blumberg, H., and P.A. Silver, unpublished observations). The 67-kD p1 form of CPY was the predominant product after the 10-min pulse of wild-type or Δscj1 cells. CPY immunoprecipitates from the Δost3 mutant contain p1CPY as well as more rapidly migrating variants of p1CPY that lack one or two oligosaccharides. Notably, the same glycoforms of p1CPY were seen in the Δost3Δscj1 mutant after the 10-min pulse label. After a 15 min chase, mature vacuolar CPY is the major product detected in the wild-type and mutant strains, together with residual p1CPY and p2CPY. Additional polypeptides corresponding to CPY glycoforms lacking one, and in some cases, two core oligosaccharides are obtained from both the Δost3 and the Δost3Δscj1 strains. Since the absence of Scj1p does not cause a defect in N-linked glycosylation, the Δost3 and Δost3Δscj1 strains synthesized similar CPY glycoforms. We conclude that the synthetic lethal phenotype of the Δscj1ost1-6 mutant is not explained by a further reduction in OST activity in the mutant strain, but instead indicates that Δscj1 cells do not tolerate hypoglycosylation of proteins.
As Scj1p is proposed to interact functionally with Kar2p, one would predict a synthetic growth defect for an ost1-6kar2 double mutant. To test this prediction, we crossed the ost1-6 mutant with the temperature-sensitive (ts) kar2-159 mutant. After sporulation and dissection of 27 asci at 25°C on YPD plates, we obtained 21 parental ditype tetrads (4 viable ts colonies), one nonparental ditype tetrad (2 viable non-ts colonies) and 5 tetratype tetrads (3 viable colonies, 2 of which were ts). This distribution of tetrads is consistent with the linkage of both KAR2 (YJL034W) and OST1 (YJL002C) to the centromere of chromosome X. Spores corresponding to the ost1-6kar2-159 double mutant germinated at 25°C and gave rise to microcolonies containing 2–4 cells, indicating a synthetic lethal interaction between the kar2-159 and ost1-6 mutations.
The Maturation of Unglycosylated CPY Is Delayed in Δscj1 Strains
A reduction in N-linked glycosylation due to a mutation in the OST would be predicted to interfere with the glycoprotein folding in cells that lack a component of the ER chaperone machinery. Although the results obtained by pulse-chase labeling of the Δost3Δscj1 mutant (e.g., Fig. 2 B) suggest a possible delay in ER-exit of the hypoglycosylated variants of CPY, the presence of multiple CPY glycoforms prevented accurate quantitation of CPY transport to the vacuole. To determine whether Scj1p is involved in folding and ER exit of glycoproteins, we asked whether processing and transport of CPY and a nonglycosylated mutant form of CPY is altered in the Δscj1 mutant. We constructed wild-type and Δscj1 mutant strains that have the chromosomal CPY gene (PRC1) as well as a CEN plasmid-borne CPY gene that contains point mutations in each of the four consensus N-X-T/S sites for N-linked glycosylation. The unglycosylated CPY mutant has been used as a reporter protein in previous studies since the expression, stability and activity of the CPY mutant are not affected by elimination of the glycosylation sites (Winther et al., 1991). However, transport of the unglycosylated CPY mutant from the RER to the vacuole occurs at a reduced rate in wild-type cells (Winther et al., 1991), and is further slowed in kar2 mutants (te Heesen and Aebi, 1994).
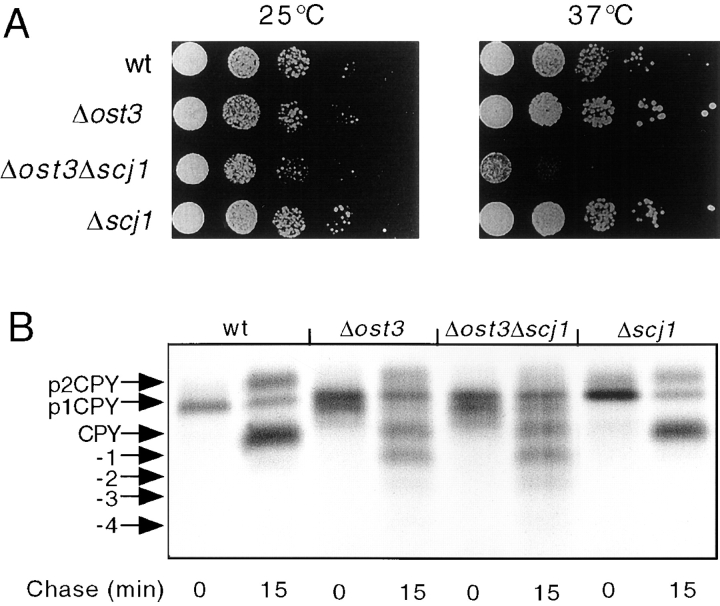
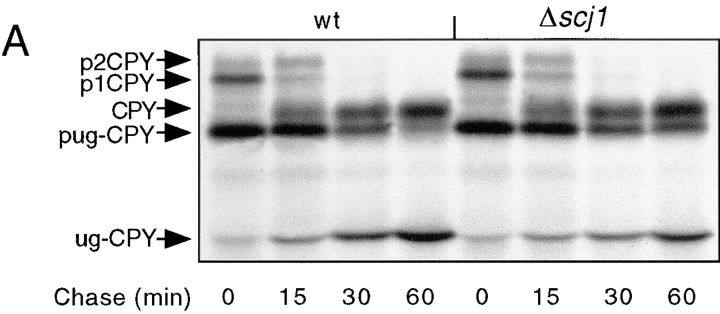
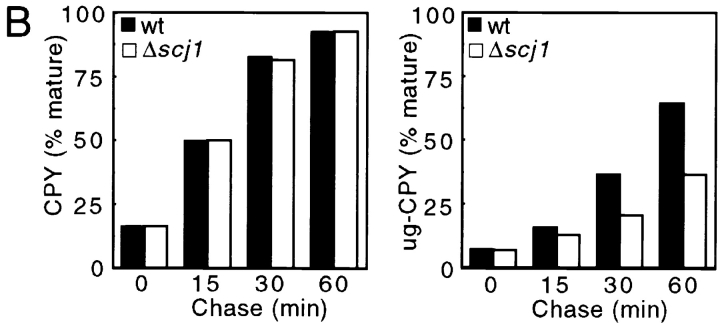
CPY was immunoprecipitated from yeast cultures that were pulse-labeled for 10 min at 25°C, and then chased for the indicated times (Fig. 3 A). The ER and Golgi precursor forms of wild-type CPY (p1CPY, p2CPY) and of the mutant CPY (pug-CPY) can be readily distinguished from the mature vacuolar forms of wild-type CPY and mutant ug-CPY by their different mobilities after PAGE in SDS. After a 10-min pulse label, the ER and Golgi precursors are the predominant forms of CPY and ug-CPY detected in both yeast strains. Conversion of the precursors to the more rapidly migrating mature forms during the chase period can be used to estimate the rate at which folded CPY exits the ER. The effect of the Scj1 protein on transport was quantified by determining the percentage of mature CPY (Fig. 3 B, left) and mature ug-CPY (Fig. 3 B, right) during the 1-h chase incubation. During the chase period, similar percentages of mature CPY were obtained from the wild-type and Δscj1 strains, suggesting that Scj1p is not required for a rate-limiting step in transport of fully glycosylated CPY from the ER to the vacuole (Fig. 3 B). Consistent with previous reports (Winther et al., 1991), transport of unglycosylated CPY from the ER to the vacuole was slightly delayed compared with transport of wild-type CPY (Fig. 3 B, compare solid bars in left and right panels). In contrast to what was observed for wild-type CPY, the Δscj1 yeast cells were found to transport unglycosylated CPY to the vacuole 1.7-fold slower than the wild-type strain (Fig. 3 B, right). Loss of Scj1p delays the folding of hypoglycosylated forms of CPY, albeit to a lesser extent than reported previously for several kar2 alleles (te Heesen and Aebi, 1994).
Figure 3.
Transport of unglycosylated CPY from the ER to the vacuole is delayed in a Δscj1 mutant. (A) Wild-type (RGY141) and Δscj1 (RGY142) strains bearing pJW373 (encoding ug-CPY) were radiolabeled for 10 min at 25°C and chased for the indicated times. CPY immunoprecipitates were resolved by PAGE in SDS. The ER, Golgi and vacuolar forms of CPY, expressed from the genomic PRC1 gene, are designated as p1CPY, p2CPY, and CPY, respectively. The precursor and vacuolar forms of unglycosylated CPY encoded by pJW373 are designated as pug-CPY and ug-CPY. (B) The radiolabeled bands from the gel in A were quantified, normalized to the total amount of CPY (precursors plus mature) present in a sample, and expressed as the percentage of mature CPY (left) or mature ug-CPY (right) for each time point of the chase.
The Folding Defect of Δscj1 Strains Is Aggravated in the Absence of Jem1p
Expression of two of the ER DnaJ homologues (Scj1p and Jem1p) is induced by tunicamycin (Schlenstedt et al., 1995; Nishikawa and Endo, 1997); hence the UPR pathway regulates expression of both proteins. A partial overlap of function for Jem1p and Scj1p is suggested by the observation that the Δscj1Δjem1 double mutant is viable at 25°C but not at 37°C (Nishikawa and Endo, 1997). We constructed a series of strains lacking the nonessential JEM1 gene to determine whether Jem1p participates in processes that mediate folding of hypoglycosylated glycoproteins in the ER. Growth on YDP plates revealed a much weaker growth defect for the Δost3Δjem1 mutant than for the Δost3Δscj1 mutant at 37°C (not shown). However, the Δost3Δscj1Δjem1 triple mutants were inviable at 25°C (data not shown). These results reinforce the hypothesis that Jem1p and Scj1p perform related but nonidentical functions in the RER.
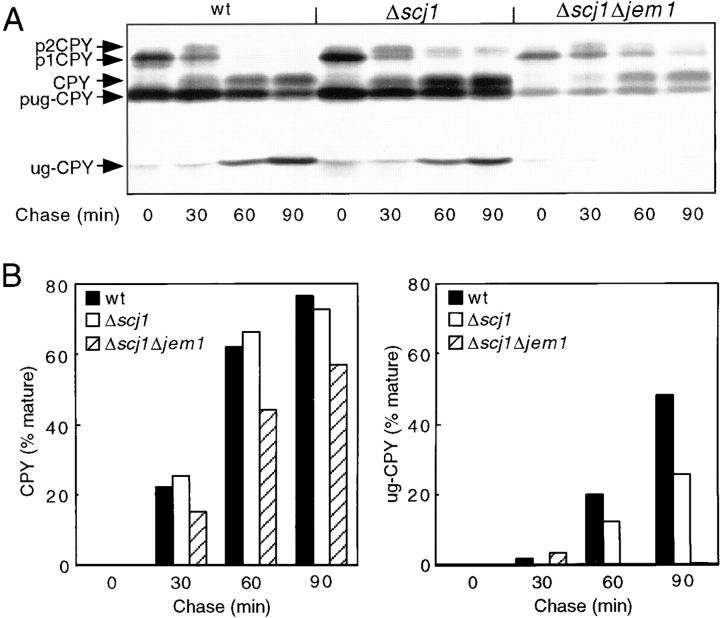
The intracellular transport of glycosylated and unglycosylated CPY was analyzed in the wild-type, Δjem1 and Δscj1Δjem1 mutant strains to determine whether Jem1p is also involved in protein folding (Fig. 4 A). The time course for conversion of pug-CPY to mature unglycosylated CPY was remarkably similar in the wild-type and Δjem1 strains (Fig. 4 B). However, the transport of unglycosylated CPY to the vacuole was drastically delayed in the Δscj1Δjem1 mutant; after a 60-min chase period, <30% of the total ug-CPY precursor had been converted to the mature form (Fig. 4 B). The 3.2-fold slower transport of ug-CPY in the Δscj1Δjem1 mutant relative to wild-type cells is similar to that observed for several kar2 alleles (te Heesen and Aebi, 1994), and is more severe than that observed for the Δscj1 mutant (Fig. 3 B). Interestingly, the maturation of CPY expressed from the chromosomal PRC1 gene was also delayed in the strain that lacks both DnaJ homologues, since the ER precursor (p1CPY) can still be detected after a 60-min chase in the immunoprecipitates from the Δscj1Δjem1 strain (Fig. 4 A). This delay in transport of wild-type CPY from the ER to the vacuole in the Δscj1Δjem1 mutant was dependent upon the simultaneous expression of the unglycosylated CPY mutant (data not shown). Nontranslocated precursor forms of Kar2p were not detected when Kar2p was immunoprecipitated from pulse-labeled Δscj1Δjem1 cells (not shown), indicating that protein translocation is not perturbed when two of the three DnaJ homologues are absent.
Figure 4.
Loss of Jem1p aggravates the folding defect of the Δscj1 mutant. (A) Wild-type (RGY131), Δjem1 (RGY147), and Δscj1Δjem1 (RGY148) strains bearing pJW373 were radiolabeled as described in Fig. 3. CPY was immunoprecipitated from glass bead extracts of radiolabeled cells and resolved by PAGE in SDS. (B) The radioactive bands corresponding to pug-CPY and ug-CPY in the gel shown in A were quantified, and the amount of mature ug-CPY present in each sample was expressed as a percentage of the total amount of pug-CPY and ug-CPY present at each time point during the chase. (C) Δscj1 (RGY142), Δost3Δscj1 (RGY144) and Δscj1Δjem1 (RGY148) strains were transformed with URA3 marked 2μ plasmids that either lack an insert (yEp352), contain the SCJ1 gene (pPS720) or the JEM1 gene (pMR3270). Yeast strains were grown in liquid minimal media (−uracil) at 25°C and diluted to a cell density of 106 cells/ml. 5-μl aliquots of tenfold serial dilutions were plated on YPDA plates and incubated at 25 or 37°C for 2 d.
Next, we tested whether overexpression of Jem1p could suppress the temperature sensitive growth defect of the Δost3Δscj1 mutant. The double mutant was transformed with URA3 marked high copy plasmids that either lack a DNA insert, or contained the SCJ1 gene or the JEM1 gene as inserts. As additional controls, the Δscj1Δjem1 mutant was transformed with the empty vector, or the JEM1 plasmid. As expected, all the strains grew at similar rates at 25°C (Fig. 4 C). The temperature sensitive lethal phenotype of the Δscj1Δjem1 mutant was rescued by the plasmid that contained the JEM1 insert, but not by the empty vector. The severe growth defect of the Δost3Δscj1 mutant was also suppressed by high copy plasmids that contain either the SCJ1 gene or the JEM1 gene. Suppression of the growth defects for the Δscj1Δjem1 and Δost3Δscj1 strains was plasmid dependent as neither strain grew at a wild-type rate at 37°C on minimal media plates that contain 5-FOA (not shown).
Prolonged Binding of Kar2p to Unglycosylated CPY in the Δscj1 and Δscj1Δjem1 Mutants
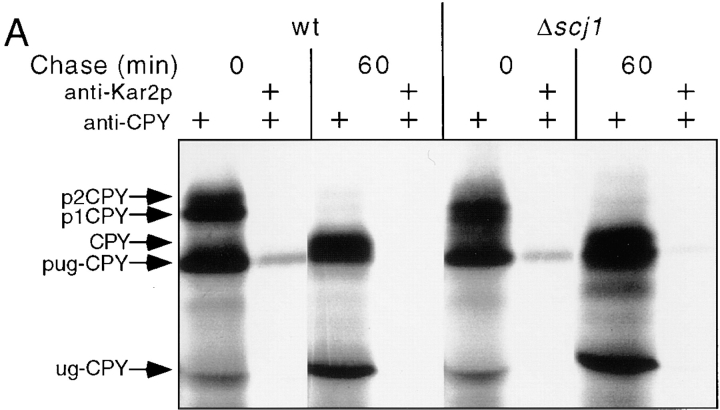
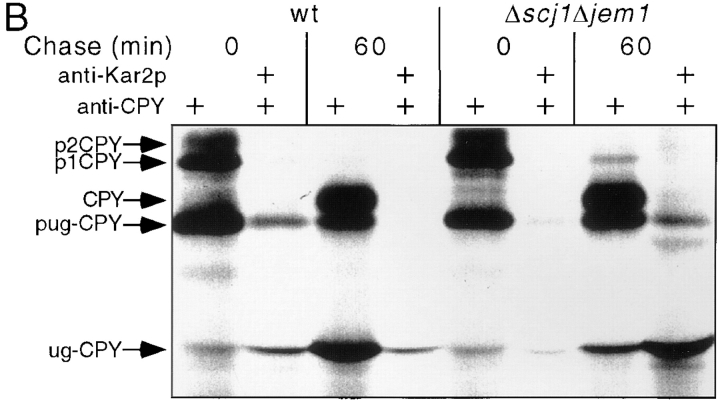
Loss of the Scj1p selectively delays export of unglycosylated CPY from the ER. Presumably, the ability of Kar2p to mediate pug-CPY folding is impaired when one or more of the DnaJ homologues are not expressed. Based upon previous studies using conditional kar2 mutants (te Heesen et al., 1994; Simons et al., 1995), we would predict that the unfolded pug-CPY remains associated with Kar2p for an extended period in Δscj1 and Δscj1Δjem1 cells. To test this prediction, antibodies specific for Kar2p were used to immunoprecipitate the chaperone under nondenaturing conditions together with any unfolded protein substrates. A subsequent immunoprecipitation under denaturing conditions using antibodies to CPY allowed an estimation of the fraction of p1CPY and pug-CPY that were associated with Kar2p either 0 or 60 min after pulse labeling (Fig. 5). Although a faint p1CPY band could be detected in the Kar2p immunoprecipitates from the 0-min chase, quantitation of the data revealed that <5% of the total p1CPY was associated with Kar2p in wild-type, Δscj1 and Δscj1Δjem1 cells. A greater fraction of pug-CPY was associated with Kar2p in wild-type cells immediately after pulse labeling (Fig. 5 A [8%] and B [13%]). After a 1-h chase, neither p1CPY nor pug-CPY remained bound to Kar2p in wild-type cells, nor did we observe artifactual coimmunoprecipitation of p2-CPY or mature CPY with Kar2p in 0 min or 60 min samples. Typically, a slightly greater fraction of the pug-CPY was associated with Kar2p in the Δscj1 (Fig. 5 A, 11%) and Δscj1Δjem1 (Fig. 5 B, 12%) mutants than in the wild-type strain immediately after pulse labeling. In contrast to what we observe in wild-type cells, complexes between Kar2p and pug-CPY persist in the Δscj1 and Δscj1Δjem1 mutants. Quantification of the data in Fig. 5 B indicated that 21% of the remaining pug-CPY was bound to Kar2p in the Δscj1Δjem1 mutant after a 1-h chase. The native coimmunoprecipitation procedure may underestimate the fraction of unfolded proteins that are bound to Kar2p, given that chaperone–substrate complexes may dissociate during the time required for the immunoprecipitation.
Figure 5.
Complexes between Kar2p and ER precursor form of unglycosylated CPY. Cultures of the wild-type (A, RGY141; B, RGY131), Δscj1 (A, RGY142) and Δscj1Δjem1 (B, RGY148) strains bearing pJW373 were radiolabeled for 10 min at 25°C, and chased for 0 or 60 min. Glass bead extracts of the labeled cultures were treated with apyrase to deplete ATP. One aliquot from each sample was immunoprecipitated in nondenaturing buffers using antibodies to Kar2p, while a second aliquot was immunoprecipitated with antibodies to CPY. The Kar2p immunoprecipitates were denatured and precipitated with antibodies to CPY. The CPY immunoprecipitates were resolved by PAGE in SDS. The radioactive bands corresponding to pug-CPY was quantified to determine the percentage of p1CPY and pug-CPY that was associated with Kar2p. The radioactive band that migrates closely to ug-CPY (e.g., see 0 min of chase) is an artifact caused by compression of the background by the 50-kD IgG heavy chain.
ER DnaJ Proteins Help Protect Yeast from Stress-inducing Agents
The synthetic growth defect produced by the combination of a reduction in OST activity and a null allele of Scj1p is most readily explained by a protein folding defect that is ultimately responsible for reduced exit of hypoglycosylated proteins from the ER. To determine whether loss of Scj1p or Jem1p makes yeast cells more sensitive to the stress caused by accumulation of unfolded proteins in the ER, the growth of wild-type and mutant strains was compared on plates that contained sublethal concentrations of tunicamycin or β-mercaptoethanol (Fig. 6). All the strains analyzed grew at wild-type rates on YPDA plates incubated at 25°C. The Δscj1 and the Δost3Δscj1 mutant strains showed reduced growth rates compared with the wild-type strain when the YPDA plates contained tunicamycin at a concentration of 1 μg/ml. A less-pronounced sensitivity to tunicamycin, as detected by a slight reduction in colony size, was observed for the Δost3Δjem1 strain relative to the Δost3 strain. The Δost3 strain is slightly more resistant to tunicamycin than the wild-type strain. Resistance to low concentrations of tunicamycin is also a property of the ost1 and ost2 mutants; the degree of resistance is roughly proportional to the severity of the glycosylation defect (Silberstein and Gilmore, unpublished observation). The Δost3 and the Δjem1 mutants were also less sensitive to β-mercaptoethanol than the wild-type strain or the other mutants. Notably, RER proteins involved in disulfide bond formation including protein disulfide isomerase and Ero1p are regulated by the UPR pathway (Frand and Kaiser, 1998). Whereas mutations in the OST result in increased, rather than decreased, resistance to agents that promote protein folding stress, we consider it unlikely that the OST has an intrinsic role as a chaperone. In contrast, the Δost3 Δscj1 and Δost3Δjem1 double mutant strains showed a more pronounced growth defect than the wild-type and Δscj1 strains in plates containing β-mercaptoethanol. Apparently, the combination of hypoglycosylation stress and redox stress is not tolerated in cells that are deficient in Scj1p or Jem1p.
Figure 6.
Growth of wild-type (RGY131) and mutant strains (RGY324, RGY144, RGY145, RGY147, RGY146) on media containing tunicamycin, β-mercaptoethanol, Calcofluor White, or hygromycin B. Yeast strains were grown in liquid YPDA medium at 25°C and diluted to a cell density of 106 cells/ml. 5-μl Aliquots of 10-fold serial dilutions were plated on YPDA-agar or on plates with the same medium containing drugs at the indicated concentrations. YPDA plates without drug were incubated at 25°C for 2 d. Plates containing tunicamycin, β-mercaptoethanol, Calcofluor White and hygromycin B were incubated in the dark for 5–6 d at 25°C.
Assembly of the yeast cell wall requires the biosynthesis and transport of glycoproteins, GPI anchored proteins, and β-1,6 and β-1,3 glucans (Stratford, 1994). Yeast with mutations that block different stages in the synthesis of N-linked oligosaccharides were found to exhibit increased sensitivity to aminoglycoside antibiotics like hygromycin B (Dean, 1995) or compounds that interfere with cell wall assembly such as Calcofluor White (Ram et al., 1994; Lussier et al., 1997). To determine whether the absence of Scj1p results in a defect in cell wall biosynthesis, mutant and wild-type strains were plated on YPD plates that had various concentrations of Calcofluor White or hygromycin B (Fig. 6). As expected for an OST mutant, the Δost3 strain was hypersensitive to hygromycin B and Calcofluor White. It should be noted that yeast with more severe defects in the OST (e.g., ost1-3 or ost1-4) show little or no growth on plates that contain 10 μg/ml of hygromycin B (Karaoglu et al., 1995b ). The Δscj1 strain did not show a significant increase in sensitivity to either Calcofluor White or hygromycin B. In contrast, the loss of Scj1p aggravates the cell wall defect of the Δost3 mutant as shown by a further decrease in colony size on plates that contain Calcofluor White.
The UPR Pathway Is Induced in Δscj1, Δost3 and Δjem1 Strains
The accumulation of unfolded proteins in the ER is a signal for increased synthesis of ER chaperones that are regulated by the UPR pathway (Kozutsumi et al., 1988). Increased transcription of the KAR2 gene (Normington et al., 1989; Rose et al., 1989), the promoter of which contains a 22-bp UPR element (UPRE; Mori et al., 1992), is indicative of an increased content of unfolded proteins in the yeast ER. The SCJ1 and JEM1 genes contain UPRE (Blumberg and Silver, 1991), and transcription of both genes is induced by agents that cause protein folding stress (Schlenstedt et al., 1995; Nishikawa and Endo, 1997). Transcription of KAR2 and JEM1 mRNAs was quantified by probing Northern blots of total RNA prepared from yeast cultures grown at 25°C (Fig. 7, A and B). Incubation of wild-type cells for 2 h in the presence of sufficient tunicamycin to completely block assembly of the dolichol-linked oligosaccharide donor for N-linked glycosylation results in a 4.9-fold increase in KAR2 mRNA expression and a 3.5-fold increase in JEM1 mRNA expression. Previous investigators have reported similar extents of KAR2 mRNA induction by tunicamycin treatment (Cox et al., 1993; Baxter et al., 1996). Disruption of the OST3 gene or the SCJ1 gene results in a 2- and a 2.7-fold increase in KAR2 mRNA relative to the wild-type strain, respectively. The combined loss of both Ost3p and Scj1p results in a 3.5-fold induction of KAR2 mRNA, consistent with the view that loss of Scj1p aggravates the protein folding stress caused by hypoglycosylation. In each strain, induction of JEM1 mRNA was slightly lower than induction of KAR2 mRNA (Fig. 7 B). Similar results were obtained for the Δost3, Δost3Δscj1 and Δscj1 strains when induction of the UPR pathway was analyzed by comparing the incorporation of 35S methionine into Kar2p in the wild-type and mutant strains (Fig. 7 C and not shown). Induction of Kar2p was also evaluated in strains in which the JEM1 gene was disrupted (Fig. 7 C). We found that loss of Jem1p has less effect on Kar2p expression than loss of Scj1p. As observed for the Δost3Δscj1 double mutant (Fig. 7 A), disruption of the JEM1 gene in the context of a Δost3 strain or a Δscj1 strain leads to a further increase in Kar2p expression (Fig. 7 C).
Figure 7.
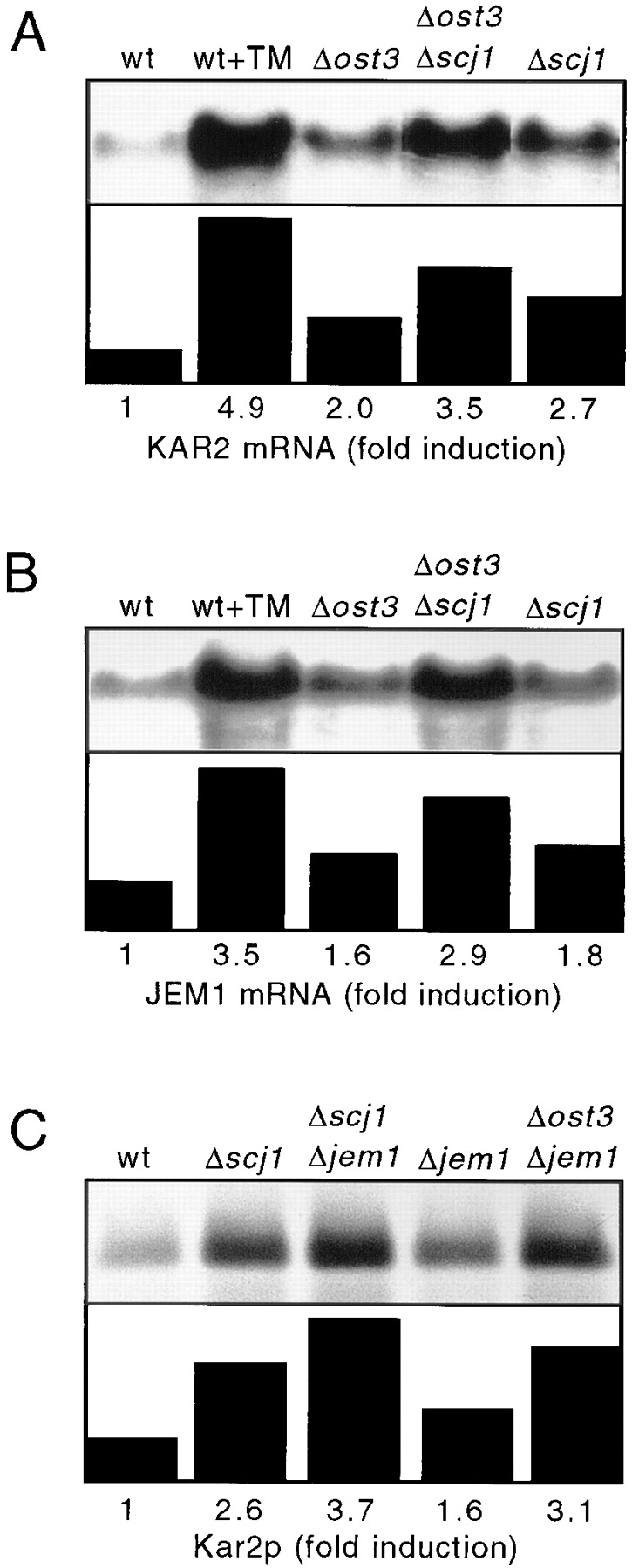
Induction of the unfolded protein response pathway. (A and B) Wild-type (RGY131) and mutant (RGY144, RGY145, RGY324) yeast strains were grown at 25°C in the absence or presence of 10 μg/ml of tunicamycin (TM). Northern blot analysis of RNA preparations were performed as described in Materials and Methods. (A) KAR2 mRNA, normalized to ACT1 mRNA probed on the same blot, is expressed as bars corresponding to the fold-induction of KAR2 mRNA relative to that expressed by the wild-type strain. (B) JEM1 mRNA, normalized to ethidium bromide stained rRNAs, is expressed as bars corresponding to the fold-induction of JEM1 mRNA relative to that expressed by the wild-type strain. (C) Kar2p immunoprecipitates from wild-type (RGY131) and mutant strains (RGY145, RGY146, RGY147, and RGY148) that were radiolabeled for 10 min at 25°C were resolved by PAGE in SDS. The radioactive band corresponding to Kar2p was quantified and is expressed as fold-induction of Kar2p relative to that expressed by the wild-type strain at 25°C.
Redox Stress Aggravates the Protein Folding Defect in the Δscj1Δjem1 Mutant
The presence of a reducing agent (e.g., DTT) in yeast or mammalian cell culture media arrests the folding of proteins that contain disulfide bonds in the ER (Braakman et al., 1992; Simons et al., 1995). The arrest in folding is reversible; dilution of the reducing agent restores the normal redox potential of the ER, at which point folding of disulfide bond containing proteins resumes (Braakman et al., 1992). Studies in yeast have shown that unfolded p1CPY accumulates when yeast are pulse labeled after DTT treatment (Jämsä et al., 1994). After removal of DTT, the subsequent folding and transport of p1CPY to the Golgi is blocked in yeast strains that have conditional kar2 mutations (Simons et al., 1995).
To determine whether Scj1p and Jem1p were also required for the conformational maturation of DTT-reduced p1CPY and pug-CPY, we compared transport of CPY and ug-CPY to the vacuole in wild-type, Δscj1 and Δscj1Δjem1 mutant cells that were radiolabeled in the presence of DTT. In agreement with previous reports (Jämsä et al., 1994; Simons et al., 1995), only the reduced p1 precursor forms of CPY or pug-CPY accumulate when cells are pulse labeled for 30 min in the presence of DTT (Fig. 8 A). After dilution of the reducing agent, p1CPY and pug-CPY are oxidized and fold into transport competent forms, as judged by the disappearance of p1CPY and pug-CPY. Transport of fully glycosylated CPY occurred with very similar kinetics in wild-type and Δscj1 mutant cells (Fig. 8 B, left). We conclude that Scj1p is not required for a rate-limiting step in the folding of DTT-reduced CPY in the endoplasmic reticulum. Exit of p1CPY from the ER was delayed, but not prevented, in theΔscj1Δjem1 mutant relative to the wild-type strain (Fig. 8 B, right). After dilution of the reducing agent, transport of unglycosylated CPY to the vacuole was delayed roughly twofold in the Δscj1 mutant relative to the wild-type strain, just as we had observed in the absence of DTT treatment. The loss of both DnaJ homologues had a much more profound effect upon folding and ER-exit of pug-CPY. We did not detect conversion of pug-CPY to ug-CPY during the 90 minute incubation after dilution of the DTT in the Δscj1Δjem1 mutant (Fig. 8 B, right), suggesting that the DTT-reduced unglycosylated CPY cannot be rescued when the folding machinery is compromised by loss of both Scj1p and Jem1p.
Figure 8.
Intracellular transport of CPY and unglycosylated CPY after removal of DTT. Wild-type (RGY141), Δscj1 (RGY142) and Δscj1Δjem1 (RGY148) strains that were incubated for 15 min at 25°C in the presence of 5 mM DTT, were pulse labeled for 10 min with Tran-35S-label. The cultures were then diluted 10-fold with DTT-free medium containing an excess of cold methionine and cysteine, and aliquots were taken at the indicated times. CPY was immunoprecipitated from glass bead extracts of the cells and resolved by PAGE in SDS. The ER, Golgi, and vacuolar forms of CPY, expressed from the genomic PRC1 gene, are designated as p1CPY, p2CPY, and CPY, respectively. The radioactive band that migrates closely to ug-CPY (e.g., see 0 min of chase) is an artifact caused by compression of the background by the 50-kD IgG heavy chain. The precursor and vacuolar forms of unglycosylated CPY encoded by pJW373 are designated as pug-CPY and ug-CPY. (B) The radiolabeled bands from the gel in A were quantified, normalized to the total amount of CPY (precursors plus mature) present in a sample, and expressed as the percentage of mature CPY (left) or mature ug-CPY (right) for each time point of the chase.
Discussion
Here, we have analyzed the genetic interactions between mutations in genes encoding subunits of the OST and null alleles of the SCJ1 and JEM1 genes encoding homologues of the bacterial chaperone DnaJ. We observed that the protein folding stress induced by hypoglycosylation causes a synthetic growth defect in cells that do not express Scj1p. Cells that lack both Scj1p and Jem1p are extremely sensitive to hypoglycosylation. Our data provide the first direct evidence that Scj1p and Jem1p perform a role in protein folding in the lumen of the yeast ER.
Insight into the role of Scj1p as a regulatory cofactor for Kar2p in the ER lumen is provided by considering the ATP hydrolysis-dependent reaction cycle of their prokaryotic homologues DnaJ and DnaK. The affinity of the peptide binding domain of DnaK for substrate polypeptides is controlled by the ATPase cycle of DnaK, which is in turn regulated by DnaJ and GrpE which stimulate ATP hydrolysis and ribonucleotide exchange, respectively (for recent reviews see Hartl, 1996; Bukau and Horwich, 1998). The ATP-bound conformation of DnaK binds and releases polypeptide substrates rapidly, whereas the ADP-bound conformation of DnaK binds peptides with higher affinity due to a reduced rate of peptide dissociation. Eukaryotic DnaJ homologues have been shown to stimulate the intrinsic ATPase activity of Hsp70 proteins (Cyr et al., 1992; Braun et al., 1996; Schumacker et al., 1996); hence it is likely that Scj1p stimulates the ATPase activity of Kar2p. DnaJ is able to bind unfolded polypeptides and deliver these substrates to the ATP-bound conformation of DnaK (Szabo et al., 1994). The substrate binding domain of DnaJ has been mapped to a zinc finger-like domain that is conserved in many (Scj1p, Mdj1p and Ydj1p), but not all (Sec63p and Jem1p) eukaryotic DnaJ homologues (Szabo et al., 1996). The presence in Scj1p of the cysteine-rich zinc finger-like domain argues strongly that Scj1p recognizes unfolded polypeptides in the ER lumen, and initiates folding reactions by cooperation with Kar2p. After ATP hydrolysis and GrpE-mediated dissociation of ADP the substrate polypeptide dissociates from DnaK and proceeds along a folding pathway or is instead rebound by DnaJ for another cyclic interaction with DnaK
Genetic Interactions between OST Mutants and ER Chaperone Mutants
The ost1-6 allele that was identified in the synthetic lethality screen is phenotypically similar to other previously described ost1 mutants (Silberstein et al., 1995). The mutation in Ost1p confers a temperature sensitive growth phenotype even though nascent glycoproteins are hypoglycosylated at both the permissive and restrictive temperatures. As reported previously for other ost1 mutants (Silberstein et al., 1995), expression of Kar2p is induced in ost1-6 cells at the permissive temperature (Silberstein, S., and R. Gilmore, unpublished observation). Given the role for N-linked oligosaccharides in protein folding, a synthetic genetic interaction is anticipated for yeast strains that have defects in an OST subunit and the Kar2 protein. Indeed, a slow-growth phenotype is observed in each case for double mutant strains that combine a wbp1 mutation (wbp1-1 or wbp1-2) with a kar2 mutation (kar2-1, kar-159, or kar2-203; te Heesen and Aebi, 1994). Here, we observed a synthetic lethal phenotype for the ost1-6kar2-159 double mutant.
The glycosylation defect caused by ost1-6 does not severely compromise growth of the mutant strain at 25°C. However, loss of Scj1p was not tolerated in an ost1-6 background at any temperature. Unlike the kar2 mutants that are defective in multiple RER-associated pathways, Δscj1 mutants have no previously reported defects in protein translocation, nuclear membrane fusion during karyogamy, protein folding, or resistance to severe heat shock. Consequently, several different explanations were considered for the inviability of the Δscj1ost1-6 mutant. The discovery that the Δscj1ost1-6 mutant is viable on plates that contain an osmotic stabilizing agent argues strongly against the catastrophic loss of fully assembled OST complexes in the Δscj1ost1-6 mutant. As Δscj1 mutants are wild-type with respect to N-linked glycosylation of proteins, the lethal phenotype of the double mutant cannot be easily explained by the additive effect of two lesions in oligosaccharide addition. Synthetic lethal interactions of the latter type do occur when an OST mutant (e.g., wbp1-2) is combined with an alg (asparagine linked glycosylation) mutant that blocks assembly of the full-sized dolichol-linked oligosaccharide donor for glycosylation (Stagljar et al., 1994).
To investigate the effect of deleting Scj1p in a viable strain with defects in N-glycosylation, we constructed the Δost3Δscj1 mutant. The Δost3Δscj1 mutant shows a synthetic growth defect, without showing a concomitant reduction in N-glycosylation of proteins, indicating that Scj1p does not have a direct or indirect effect upon the efficiency of the glycosylation reaction. The more restrictive phenotype of the Δscj1ost1-6 mutant relative of the Δost3Δscj1 mutant suggests that the requirement for Scj1p becomes greater as the severity of hypoglycosylation increases.
A Role for Scj1p in Protein Folding
For many proteins that enter the secretory pathway, the addition of N-linked oligosaccharides is an important modification that precedes the successful acquisition of tertiary structure. Until proteins fold, they are not packaged into vesicles for intracellular transport, but are instead retained in the endoplasmic reticulum. Here, we observed that the rate of ER-export for a mutant form of CPY that lacks N-linked oligosaccharides was reduced twofold in cells that lack Scj1p. The reduced transport rate for unglycosylated-CPY was in marked contrast to the wild-type transport rate for fully-glycosylated CPY in cells that lack Scj1p. Folding-deficient proteins are retained in the ER via prolonged interactions with the Hsp70 chaperone BiP or Kar2p (De Silva et al., 1990). The slow maturation of unglycosylated CPY, but not fully glycosylated CPY, was previously observed in kar2 mutants (te Heesen and Aebi, 1994). The unglycosylated precursor (pug-CPY) was shown to be present in a complex with Kar2p mutant proteins that had lesions in the ATPase domain (i.e., kar2-159 and kar2-203; te Heesen and Aebi, 1994). Here, we observed a prolonged association between Kar2p and the unglycosylated CPY in the endoplasmic reticulum. Our results indicate that in addition to Kar2p, efficient folding of unglycosylated CPY requires the DnaJ homologue Scj1p. The major fate for soluble proteins that fail to fold in the yeast RER is dislocation back to the cytosol for subsequent degradation by the proteasome (Hiller et al., 1996). As reported previously, the degradative pathway does not appear to be taken by pug-CPY in wild-type cells (Winther et al., 1991). Here, we did not observe a selective loss of pug-CPY during the 1-h chase incubation in mutant strains that lack Scj1p or Jem1p. An in vitro assay for the proteasome-mediated degradation of unglycosylated pro-α-factor did not disclose an important role for Scj1p in degradation of this substrate (McCracken and Brodsky, 1996).
Delayed transport of folding-impaired hypoglycosylated proteins to the cell wall provides the most likely explanation for the synthetic phenotypes that arise when Δscj1 is combined with mutations in the OST. Support for the latter conclusion is provided by the observation that Δscj1 cells are hypersensitive to tunicamycin. In the absence of an OST mutation, Δscj1 mutants lack an obvious cell wall defect. However, the cell wall defect of the Δost3 mutant was greatly exaggerated in a yeast strain that lacks Scj1p. Folding of DTT-reduced CPY has been found to require Kar2p, as strains carrying kar2 alleles with mutations that map in the ATPase domain (e.g., kar2-159) are unable to form native disulfide bonds in p1CPY after removal of DTT resulting in the formation of high molecular mass aggregates of Kar2p and unfolded p1CPY (Simons et al., 1995). We did not obtain evidence that Scj1p was required for CPY maturation after redox stress, suggesting that DTT-reduced CPY may exist as a compact folding intermediate that rapidly acquires a native conformation after removal of reductant without intervention of Scj1p. However, unglycosylated CPY did not fold correctly in the Δscj1Δjem1 mutant after redox stress, but instead was retained in the endoplasmic reticulum. Interestingly, we did observe that Δost3Δscj1 cells are hypersensitive to DTT, suggesting that Scj1p does not act exclusively during glycoprotein folding, but instead has a broader role in the maturation of folding impaired proteins in the RER.
Survival of Yeast without Scj1p
How can we resolve the apparent paradox between the nonessential status of the SCJ1 gene and the postulated role for Scj1p as a regulatory cofactor for the essential Kar2 protein during protein folding in the ER lumen. The UPR pathway induces enhanced expression of RER chaperones and protein folding enzymes to permit survival under conditions that interfere with protein folding in the RER. Interestingly, the UPR pathway is dispensable for cell viability under normal growth conditions (Cox et al., 1993). One possible explanation for the nonessential nature of Scj1p would be that Scj1p only acts as a chaperone when unfolded proteins accumulate in the RER. However, we found that strains lacking Scj1p express elevated levels of Kar2p and Jem1p in the absence of other perturbants, indicating that loss of Scj1p causes protein folding stress in the RER. We propose that Scj1p is the primary cochaperone for Kar2p in protein folding reactions under normal physiological conditions.
A second explanation for the nonessential nature of Scj1p is suggested by the presence of one essential and two nonessential DnaJ homologues in the same cellular compartment raising the possibility that these proteins perform partially redundant functions. A complete functional redundancy of Scj1p, Sec63p and Jem1p can be discounted for several reasons. Of the three DnaJ homologues, only Sec63p has the correct localization as a component of the large Sec complex (Deshaies et al., 1991; Panzner et al., 1995) to recruit Kar2p to the protein translocation channel to interact with translocation substrates (Brodsky and Schekman, 1993; Corsi and Schekman, 1997). Before this report, a partial overlap of function for Scj1p and Jem1p was suggested by the temperature sensitive growth phenotype of the Δscj1Δjem1 mutant and by the observation that expression of both proteins is controlled by the UPR pathway (Schlenstedt et al., 1995; Nishikawa and Endo, 1997).
The wild-type growth phenotype of the Δscj1 mutant is most readily explained by the induction of the UPR pathway leading to enhanced expression of RER chaperones including Kar2p, Lhs1p and Jem1p. Kar2p binds nascent polypeptides as they enter the ER lumen via the Sec61 channel (Sanders et al., 1992), hence the initial interaction between an unfolded protein and Kar2p is mediated by Sec63p (Corsi and Schekman, 1997). For wild-type proteins that fold rapidly, this initial Scj1p-independent interaction between Kar2p and the hydrophobic segments of the nascent polypeptide may be sufficient to initiate productive folding in the RER lumen. In the absence of mutations or perturbants that interfere with protein folding in the RER, enhanced expression of Kar2p and Jem1p may compensate for loss of Scj1p. Conceivably, a higher lumenal concentration of Kar2p may result in Scj1p-independent binding of Kar2p to slowly folding proteins in the RER lumen. Furthermore, suppression of the temperature-sensitive growth phenotype of the Δost3Δscj1 strain by the high copy JEM1 plasmid argues strongly that UPR-mediated induction of Jem1p expression will help compensate for loss of Scj1p.
Overlapping, but Nonidentical Roles for Jem1p and Scj1p
Induction of Kar2p expression was greater in the Δscj1 mutant than in the Δjem1 mutant. Consistent with the relatively minor reduction in growth rate at 37°C for the Δost3Δjem1 mutant, we observed that Δjem1 yeast cells are not hypersensitive to tunicamycin and are able to transport unglycosylated CPY to the vacuole at a wild-type rate. We conclude that the viability of Δscj1 cells cannot be adequately explained by a complete redundancy of Scj1p and Jem1p. Nonetheless, several observations reported here suggest a role for Jem1p in protein folding in the RER. In comparison to the Δjem1 mutant, the Δscj1Δjem1 double mutant shows a further elevation in Kar2p expression, and a more severe delay in transport of ug-CPY to the vacuole. Furthermore, expression of a folding impaired protein (i.e., ug-CPY) in the Δscj1Δjem1 double mutant interferes with ER exit of fully-glycosylated wild-type CPY suggesting that the protein folding capacity of the ER has been exceeded. Surprisingly, expression of Jem1p from a high copy vector largely alleviates the temperature sensitive growth defect of the Δscj1Δost3 mutant. Overexpression of Jem1p may be able to compensate for loss of Scj1p, even though Jem1p lacks the cysteine rich domain that has been implicated in substrate recognition. These data, together with the inviability of a Δost3Δscj1 Δjem1 strain, are consistent with Jem1p acting as a second regulatory chaperone for Kar2p-mediated protein folding in the RER. Although Jem1p was initially believed to be a type II integral membrane protein (Nishikawa and Endo, 1997), a more recent study has established that the initiation codon for Jem1p was incorrectly assigned, and that Jem1p is a peripheral membrane protein associated with the lumenal face of the yeast RER (Nishikawa and Endo, 1998). Conceivably, the primary role of Jem1p as a chaperone may be to recruit Kar2p (or Lhs1p) to the membrane to interact with unfolded membrane proteins. Consequently, a soluble substrate like unglycosylated proCPY may not be the optimal substrate to reveal a folding delay caused by loss of Jem1p.
Acknowledgments
We thank Mark Rose for yeast strains (kar2-159, MS4, and MS20) and for the 2μ JEM1 plasmid (pMR3270). We thank Jakob Winther for providing the plasmid pJW373 that encodes the unglycosylated CPY mutant and Peter Walter for providing the antibody to Kar2p. The authors wish to thank Peter Philippsen for providing pFA6a-HIS3MX6.
This work was supported by Public Health Services grants GM43768 (R. Gilmore) and GM47385 (P.A. Silver).
Abbreviations used in this paper
- 5-FOA
5-fluoro-orotic acid
- AP
acid phosphatase
- CPY
carboxypeptidase Y
- Hsp70
heat shock protein 70 family
- OST
oligosaccharyltransferase
- ug-CPY
unglycosylated CPY
- UPR
unfolded protein response
Footnotes
Address all correspondence to Reid Gilmore, Department of Biochemistry and Molecular Biology, University of Massachusetts Medical School, 55 Lake Avenue North, Worcester, MA 01655-0103. Tel.: (508) 856-5894. Fax: (508) 856-6231. E-mail: reid.gilmore@banyan.ummed.edu
Dr. Schlenstedt's present address is Medizinische Biochemie, Gebaeude 44, Universitaet des Saarlandes, D66421 Homberg, Germany.
References
- Baxter BK, James P, Evans T, Craig EA. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiaeendoplasmic reticulum. Mol Cell Biol. 1996;16:6444–6456. doi: 10.1128/mcb.16.11.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Pringle JR. Use of a screen for synthetic lethal and multicopy suppressor mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. . Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg H, Silver PA. A homologue of the bacterial heat-shock gene DnaJ that alters protein sorting in yeast. Nature. 1991;349:627–630. doi: 10.1038/349627a0. [DOI] [PubMed] [Google Scholar]
- Braakman I, Helenius J, Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO (Eur Mol Biol Organ) J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JEA, Wilbanks SM, Scheller RH. The cysteine string secretory vesicle protein activates Hsc70 ATPase. J Biol Chem. 1996;271:25989–25993. doi: 10.1074/jbc.271.42.25989. [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63-BIP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Corsi AK, Schekman R. The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. . J Cell Biol. 1997;137:1483–1493. doi: 10.1083/jcb.137.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO (Eur Mol Biol Organ) J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- De Silva AM, Balch WE, Helenius A. Quality control in the endoplasmic reticulum: folding and misfolding of vesicular stomatitis virus G protein in cells and in vitro. J Cell Biol. 1990;111:857–866. doi: 10.1083/jcb.111.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc Natl Acad Sci USA. 1995;92:1287–1291. doi: 10.1073/pnas.92.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Sanders SL, Feldheim DA, Schekman R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 1991;349:806–808. doi: 10.1038/349806a0. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Chappell TG, Rothman JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. The ERO1gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- Gaut JR, Hendershot LM. Mutations within the nucleotide binding site of immunoglobulin-binding protein inhibit ATPase activity and interfere with release of immunoglobulin heavy chain. J Biol Chem. 1993;268:7248–7255. [PubMed] [Google Scholar]
- Hamilton TG, Flynn GC. Cer1p, a novel Hsp70-realted protein required for posttranslational endoplasmic reticulum translocation in yeast. J Biol Chem. 1996;271:30610–30613. doi: 10.1074/jbc.271.48.30610. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hiller MH, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Jämsä E, Simonen M, Makarow M. Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment with a reducing agent. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- Jones JS, Prakash L. Yeast Saccharomyces cerevisiaeselectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R. Functional characterization of the Ost3p subunit of the Saccharomyces cerevisiae oligosaccharyltransferase: disruption of the OST3gene causes biased underglycosylation of acceptor substrates. J Cell Biol. 1995a;130:567–577. doi: 10.1083/jcb.130.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaoglu D, Silberstein S, Kelleher DJ, Gilmore R. The Saccharomyces cerevisiaeoligosaccharyltransferase. a large hetero-oligomeric complex in the endoplasmic reticulum. Cold Spring Harbor Symp Quant Biol. 1995b;60:83–92. doi: 10.1101/sqb.1995.060.01.011. [DOI] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R. The highly conserved Stt3 protein is a subunit of the yeast oligosaccharyltransferase and forms a subcomplex with Ost3p and Ost4p. J Biol Chem. 1997;272:32513–32520. doi: 10.1074/jbc.272.51.32513. [DOI] [PubMed] [Google Scholar]
- Kohrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- Koshland D, Kent JC, Hartwell LH. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething M-J, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- Kranz JE, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latterich M, Schekman R. The karyogamy gene KAR2and novel proteins are required for ER-membrane fusion. Cell. 1994;78:87–98. doi: 10.1016/0092-8674(94)90575-4. [DOI] [PubMed] [Google Scholar]
- Lawrence CW. Classical mutagenesis techniques. Methods Enzymol. 1991;194:273–281. doi: 10.1016/0076-6879(91)94021-4. [DOI] [PubMed] [Google Scholar]
- Lussier M, White AM, Sheraton J, di Paolo T, Treadwell J, Southard SB, Horenstein CI, Chen-Weiner J, Ram AF, Kapteyn JC, et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. . Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Interaction between BiP and Sec63 is required for the completion of protein translocation across the ER of Saccharomyces cerevisiae. . J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Sant A, Kohno K, Normington K, Gething M-J, Sambrook JF. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2(BiP) gene by unfolded proteins. EMBO (Eur Mol Biol Organ) J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S, Endo T. The yeast Jem1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. J Biol Chem. 1997;272:12889–12892. doi: 10.1074/jbc.272.20.12889. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Endo T. Reinvestigation of the functions of the hydrophobic segment of Jem1p, a yeast endoplasmic reticulum membrane protein mediating nuclear fusion. Biochem Biophys Res Com. 1998;244:785–799. doi: 10.1006/bbrc.1998.8342. [DOI] [PubMed] [Google Scholar]
- Normington K, Kohno K, Kozutsumi Y, Gething M-J, Sambrook J. S. cerevisiaeencodes an essential protein homologous in sequence and function to mammalian BiP. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Ram AFJ, Wolters A, Ten R, Hoopen, Klis FM. A new approach for isolating cell wall mutants in Saccharomyces cerevisiaeby screening for hypersensitivity to calcofluor white. Yeast. 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- Riederer MA, Hinnen A. Removal of N-glycosylation sites of the yeast acid phosphatase severely affects protein folding. J Bacteriol. 1991;173:3539–3546. doi: 10.1128/jb.173.11.3539-3546.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Rothblatt J, Schekman R. A hitchhiker's guide to the analysis of the secretory pathway in yeast. Methods Cell Biol. 1989;32:3–36. doi: 10.1016/s0091-679x(08)61165-6. [DOI] [PubMed] [Google Scholar]
- Sadler I, Chiang A, Kurihara T, Rothblatt J, Way J, Silver P. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coliheat shock protein. J Cell Biol. 1989;109:2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW. Sec61p and Bip directly facilitate polypeptide translocation into the ER. Cell. 1992;69:353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- Saris N, Holkeri H, Craven RA, Stirling CJ, Makarow M. The Hsp70 homologue Lhs1p is involved in a novel function of the endoplasmic reticulum, refolding and stabilization of heat-denatured protein aggregates. J Cell Biol. 1997;137:813–824. doi: 10.1083/jcb.137.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Harris S, Risse B, Lill R, Silver PA. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/ Kar2p via a conserved domain that specifies interactions with Hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacker RJ, Hansen WJ, Freeman BC, Alnemri E, Litwack G, Toft DO. Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry. 1996;35:14889–14898. doi: 10.1021/bi961825h. [DOI] [PubMed] [Google Scholar]
- Scidmore MA, Okamura HH, Rose MD. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein S, Collins PG, Kelleher DJ, Rapiejko PJ, Gilmore R. The α subunit of the Saccharomyces cerevisiaeoligosaccharyltransferase complex is essential for vegetative growth of yeast and is homologous to mammalian ribophorin I. J Cell Biol. 1995;128:525–536. doi: 10.1083/jcb.128.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein S, Gilmore R. Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 1996;10:849–858. [PubMed] [Google Scholar]
- Simons JF, Ferro-Novick S, Rose MD, Helenius A. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol. 1995;130:41–50. doi: 10.1083/jcb.130.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar I, te Heesen S, Aebi M. New phenotype of mutations deficient in glucosylation of the lipid-linked oligosaccharide: cloning of the ALG8locus. Proc Natl Acad Sci USA. 1994;91:5977–5981. doi: 10.1073/pnas.91.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratford M. Another brick in the wall? Recent developments concerning the yeast cell envelope. Yeast. 1994;10:1741–1752. doi: 10.1002/yea.320101307. [DOI] [PubMed] [Google Scholar]
- Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO (Eur Mol Biol Organ) J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- Szabo A, Langer T, Schroder H, Bukau B, Flanagan J, Hartl FU. The ATP hydrolysis-dependent reaction cycle of the Escherichia coliHsp70 system-DnaK, DnaJ, and GrpE. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Heesen S, Aebi M. The genetic interaction of kar2 and wbp1 mutations. Distinct functions of binding protein BiP and N-linked glycosylation in the processing pathway of secreted proteins in Saccharomyces cerevisiae. . Eur J Biochem. 1994;222:631–637. doi: 10.1111/j.1432-1033.1994.tb18906.x. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Misra LM, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. . Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Phillipsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Winther JR, Stevens TH, Kielland-Brandt MC. Yeast carboxypeptidase Y requires glycosylation for efficient intracellular transport, but not for vacuolar sorting, in vivo stability, or activity. Eur J Biochem. 1991;197:681–689. doi: 10.1111/j.1432-1033.1991.tb15959.x. [DOI] [PubMed] [Google Scholar]