Abstract
We have identified a novel pathway for protein import into the nucleus. We have shown that the previously identified but uncharacterized yeast protein Nmd5p functions as a karyopherin. It was therefore designated Kap119p (karyopherin with M r of 119 kD). We localized Kap119p to both the nucleus and the cytoplasm. We identified the transcription elongation factor TFIIS as its major cognate import substrate. The cytoplasmic Kap119p exists as an approximately stoichiometric complex with TFIIS. RanGTP, not RanGDP, dissociated the isolated Kap119p/TFIIS complex and bound to Kap119p. Kap119p also bound directly to a number of peptide repeat containing nucleoporins in overlay assays. In wild-type cells, TFIIS was primarily localized to the nucleus. In a strain where KAP119 has been deleted, TFIIS was mislocalized to the cytoplasm indicating that TFIIS is imported into the nucleus by Kap119p. The transport of various substrates that use other karyopherin-mediated import or export pathways was not affected in a kap119Δ strain. Hence Kap119p is a novel karyopherin that is responsible for the import of the transcription elongation factor TFIIS.
Keywords: yeast, nucleus, import, karyopherin, transcription factor
Several pathways for macromolecular transport into and out of the nucleus have been identified in yeast and in higher eukaryotic cells (for review see Wozniak et al., 1998). Distinguishing features of these pathways are specific signal sequences for import (collectively termed nuclear localization sequences [NLSs]1) or export (collectively called nuclear export sequences [NESs]) and their cognate signal recognition factors, collectively termed karyopherins (or Kaps) (also termed, importins, exportins, transportins, p97, PTACs, RanBPs, NLS receptor). The Kaps belong to a structurally related family of proteins that contain armadillo (ARM) or the related HEAT motifs (Malik et al., 1997).
All of the known import pathways share common intermediates (for review see Ohno et al., 1998; Pemberton et al., 1998). First, substrate/Kap complexes assemble in the cytoplasm followed by Kap-mediated docking of the transport substrate/Kap complex to the nuclear pore complex (NPC). This docking appears to be mediated by a subset of nucleoporins (or Nups), a collective term for NPC proteins. Transport across the gated central channel of the NPC is governed by the small GTPase Ran (Melchior et al., 1993; Moore and Blobel, 1993) and several Ran-interacting proteins, including the cytoplasmic Ran-binding protein, RanBP1; p10 or NTF2, a RanGDP-binding protein; a Ran-GTPase–activating protein, RanGAP; and a RanGDP/ GTP exchange factor, RanGEF (for review see Corbett and Silver, 1997). Although many partial reactions involving Nups, Kaps, Ran, and Ran-interacting proteins have been delineated, the precise sequence of events leading to nuclear import remains to be elucidated.
The first nuclear import pathway (now often referred to as the “classical” import pathway) that was biochemically defined (for review see Wozniak et al., 1998) uses a monopartite NLS (short stretch of basic residues) or bipartite NLS (two short stretches of basic residues connected by a short linker) (Dingwall and Laskey, 1991). These NLSs interact with a dimeric Kap, Kapα/Kapβ1 (Kap60p/Kap95p in yeast). In this case Kapα interacts with the NLS (Conti et al., 1998) and Kapβ1 interacts with the NPC (Görlich et al., 1995; Imamoto et al., 1995; Moroianu et al., 1995; Rexach and Blobel, 1995; Radu et al., 1995a ). Kapβ1 also interacts with RanGTP (Rexach and Blobel, 1995). The NLS/Kapα/ Kapβ1 complex is disrupted by RanGTP (Rexach and Blobel, 1995), possibly lending directionality to import as RanGTP is thought to be concentrated in the nucleus (Bischoff and Ponstingl, 1991).
Several lines of evidence suggested the existence of multiple nuclear import pathways. Early indications of multiple, noncompeting pathways of nuclear import came from microinjection experiments (Michaud and Goldfarb, 1991, 1992). In addition, an NLS was defined for the hnRNA-binding protein A1 that bore no resemblance to the basic NLSs (Siomi and Dreyfuss, 1995; Weighardt et al., 1995). This NLS is rich in glycines and is longer than the Kapα/ Kapβ1 dedicated NLS. Finally, the completed yeast genome showed that there were up to thirteen putative homologues of Kap95p (Fornerod et al., 1997a ; Görlich et al., 1997; Wozniak et al., 1998) suggesting that these may also function as Kaps in either nuclear import and/or export.
In yeast, several of these homologues have been shown to function in nuclear transport. A notable difference to the Kapα/Kapβ1 pathway is that none of these Kapβs appear to require a Kapα adapter to interact with their substrates (for review see Pemberton et al., 1998; Wozniak et al., 1998). They are able to interact directly with substrate(s), the NPC, and RanGTP.
With the identification of substrates for eight of these Kap95p homologues (and/or their higher eukaryotic homologues), a framework for the classification of the Kaps has emerged. Three of the Kaps, Crm1p (Xpo1p) (Stade et al., 1997), and the human homologues of Cse1p (CAS) (Kutay et al., 1997) and Los1p (exportin-t) (Arts et al., 1998; Kutay et al., 1998) have been shown to function in a nonoverlapping fashion in nuclear export. Crm1p is responsible for the export of Leu-rich NESs, such as those in Rev and in a polypeptide inhibitor of protein kinase A (Fornerod et al., 1997b ; Ossareh-Nazari et al., 1997; Stade et al., 1997). Kapα is exported from the nucleus by CAS (Kutay et al., 1997) whereas tRNA is exported by exportin-t (Arts et al., 1998; Kutay et al., 1998). Export presumably involves nucleoplasmic formation of a RanGTP-dependent ternary complex of export substrate/Kap/ RanGTP and cytoplasmic dissociation of this ternary complex by RanBP1 and RanGAP (Bischoff and Ponstingl, 1991; Kutay et al., 1997, 1998). Whether the ternary complex interacts with nucleoporins is not known. The other six Kaps appear to function primarily in import (for review see Pemberton et al., 1998; Wozniak et al., 1998), although it cannot be excluded that some Kap(s) function in both import and export. The import of ribosomal proteins is mediated by several Kaps, primarily Kap123p (Rout et al., 1997). However, Kap123p is not essential, and in its absence, Kap121p also can import ribosomal proteins (Rout et al., 1997). In addition, Kap108p has been shown to interact with ribosomal proteins (Rosenblum et al., 1997). In contrast, the mRNA-binding proteins, which are involved in splicing and/or nuclear export of mRNA, are imported by nonoverlapping pathways. In this case Kap104p has been shown to import Nab2p and Hrp1p (Aitchison et al., 1996) and Kap111p has been shown to import Npl3p (Pemberton et al., 1997; Senger at al., 1998). In addition, higher eukaryotic homologues of Kap104p (Kapβ2 or transportin) have been shown to import hnRNA binding protein A1 (Pollard et al., 1996; Bonifaci et al., 1997; Fridell et al., 1997; Siomi et al., 1997). Finally, Kap108p is responsible for the import of Lhp1p, the yeast La protein, which is involved in the biogenesis of RNA polymerase III transcripts (Rosenblum et al., 1997).
In this paper we show that the hitherto uncharacterized yeast protein Nmd5p that exhibits 19% protein sequence identity with Kap95p, functions as a Kap whose major import substrate is the transcription elongation factor TFIIS. Significantly, this is the first identification of a nuclear import pathway whose major substrate is involved in RNA polymerase II transcription.
Materials and Methods
Strains, Growth Conditions, and General Methods
The yeast strains used in this study were Saccharomyces cerevisiae wild-type DF5α (Finley et al., 1987) and its derivative, kap119Δ (MATα, lys2-801, leu2-3, 2-112, ura3-52, his3-Δ200, trp1-1 [am], NMD5::HIS3). Yeast strains were grown at 30°C in YPD or in minimal medium (YNBD). Required supplements were added. Manipulation of yeast cells was performed according to standard methods (Rose et al., 1990). Bacterial transformation was performed essentially as described (Ausubel et al., 1997).
Gene Replacement and Protein A Fusion
To delete the KAP119 gene in wild-type strain DF5α, the HIS3 gene was used as a selective marker for insertion into the genomic locus. Deletion cassettes containing the HIS3 gene and 60 nucleotides of each the 5′ and 3′ untranslated regions of the KAP119/NMD5 open reading frame (ORF) were constructed by PCR. For protein A tagging of Kap119p and TFIIS, a PCR product was generated that contained a protein A, HIS3, and URA3 cassette (Aitchison et al., 1995) flanked by 60 nucleotides directly upstream of the KAP119/NMD5 stop codon and 60 nucleotides downstream of the coding region of the corresponding gene. Yeast cells were transformed by electroporation and the ORF replaced by integrative transformation in a diploid strain. Heterozygous diploids were induced to sporulate, and the meiotic progeny were examined by standard tetrad analysis (Pemberton et al., 1997).
Overlay Assay
Overlay assays were performed essentially as described (Rout et al., 1997). Equal amounts of Escherichia coli lysates were separated by SDS-PAGE and transferred to nitrocellulose. Nitrocellulose strips were incubated with yeast cytosol from strains expressing either Kap119–PrA or Kap95–PrA at 4°C for 14 h. Cytosol from the Kap119–PrA strain was used undiluted, Kap95–PrA cytosol was diluted 1:5 with transport buffer (20 mM Hepes-KOH, pH 7.5, 110 mM KOAc, 2 mM MgCl2, 0.1% Tween 20) containing 5% dried skim milk plus 0.001 vol of a protease inhibitor cocktail (Rout and Kilmartin, 1990). Bound PrA fusion proteins were detected with rabbit IgG (Cappel, Malvern, PA) and anti-rabbit IgG-coupled HRP (Amersham, Arlington, IL) was used as the secondary antibody, and blots were developed using the enhanced chemiluminescence (ECL) system (Amersham). All incubations were performed in transport buffer containing 5% dried skim milk.
Immunofluorescence Microscopy and Immunoblotting
Indirect immunofluorescence microscopy was performed as described by Rout et al. (1997). Yeast cells were fixed with 3.7% formaldehyde for 5 min and cell walls were digested. The PrA tags were detected with rabbit IgG, Nab2p with polyclonal mouse antiserum (Aitchison et al., 1996), and Npl3p using antibody 1E4 (Wilson et al., 1994). CY3-conjugated donkey anti–mouse and anti–rabbit IgG as well as FITC-conjugated donkey anti– mouse IgG were used as 6 μg/ml solutions for visualization. GFP reporter proteins were detected directly in fixed cells. DNA was visualized by 4′,6-diamidino-2-phenylindole (DAPI). Immunoblotting experiments were performed using anti-rabbit IgG-coupled HRP (Amersham) as secondary antibody, and blots were developed using the ECL system (Amersham). Nop1p was detected with monoclonal mouse antiserum D66 (Aris and Blobel, 1998) and 3-phosphoglycerate kinase with monoclonal mouse antiserum 22C5-D8 (Molecular Probes, Eugene, OR).
Cell Fractionation and Immunoisolation
Spheroplasting of yeast cells and preparation of nuclei and postribosomal supernatant were described (Rout and Kilmartin, 1990, 1994; Wilson et al., 1994; Melchior et al., 1995; Aitchison et al., 1996). PrA-tagged Kap119p and TFIIS were immunoisolated according to Aitchison et al. (1996). Proteins were analyzed by SDS-PAGE and stained by Coomassie blue. Bands of interest were excised and prepared for matrix-assisted laser desorption/ ionization time-of-flight (MALDI-TOF) mass spectrometry.
In Vitro Dissociation by RanGTP
Recombinant yeast Ran (Gsp1) was prepared and nucleotide exchange performed as described by Floer and Blobel (1996). After nucleotide exchange RanGDP was incubated with Rna1p to convert remaining RanGTP into the GDP-bound state: 50 μM RanGDP and 4 μM RanGAP were incubated in a final volume of 100 μl for 12 h at 21°C. All further RanGDP incubations were performed without previous removal of RanGAP.
PrA-tagged Kap119p and bound TFIIS were coimmunoisolated as described above. After the final wash, IgG-Sepharose–bound Kap119-PrA/ TFIIS was divided into three 50-μl portions and placed into siliconized reaction tubes. The beads were resuspended in 240 μl transport-buffer (20 mM Hepes-KOH, pH 7.5, 150 mM KOAc, 2 mM MgCl2, 1 mM DTT, 0.1% Tween 20, 0.1 mM PMSF, 1 μg/ml leupeptin and 1 μg/ml pepstatin A) and the slurry was incubated in batch with buffer alone, RanGDP, or RanGTP (final concentration 5 μM) in a final volume of 250 μl for 60 min at 21°C. The beads were then collected by centrifugation at 2,000 g for 30 s and unbound proteins were removed in the supernatant and collected for further analysis. Beads were washed twice with 1 ml of transport buffer. Analysis of the bound proteins was performed as described for immunoisolation except that the elution of bound proteins was performed only in two steps with 250 mM and 4,500 mM MgCl2.
Results
Nmd5p (for nonsense-mediated mRNA decay) (GenBank/ EMBL/DDBJ accession number U31375) is a yeast protein that was originally identified in a two-hybrid screen with the cytoplasmic protein Upf1p (up frame shift mutation) as a bait (Atkin et al., 1995; He and Jacobsen, 1995). Upf1p functions in the rapid degradation of mRNAs that contain a premature stop codon (He and Jacobsen, 1995). However, a direct interaction between Upf1p and Nmd5p has not been demonstrated and it is not clear what function, if any, Nmd5p has in Upf1p-mediated mRNA degradation. As we show in this paper that Nmd5p functions as a Kap we suggest the alternative name Kap119p in keeping with previously proposed yeast nomenclature (Enenkel et al., 1995; Aitchison et al., 1996). The amino acid sequence of Kap119p is 19% identical with that of Kap95p and 26% identical with that of Kap108p (Rosenblum et al., 1997, 1998) (data not shown).
Kap119p Is Located in the Nucleus and the Cytoplasm
The KAP119 gene was replaced by KAP119-PrA coding for a chimeric Kap119p that is COOH terminally fused to IgG-binding domains of Staphylococcus aureus protein A. The resulting Kap119–PrA strain showed no difference in its growth rate compared with wild-type cells. By immunofluorescence (Fig. 1 A) and by cell fractionation (Fig. 1 B) the Kap119–PrA was localized to the cytoplasm and the nucleus indicating that the protein might shuttle between these two compartments. Cell fraction analyses (Fig. 1 B) indicated that about one-third of the total Kap119–PrA was localized to the nucleus. Considering that the nucleus occupies less than one-third of the cellular volume, Kap119p appears to be slightly more concentrated in the nucleus than in the cytoplasm.
Figure 1.
Cellular localization of Kap119p. A strain where endogenous Kap119p was replaced by protein A-tagged Kap119p (Kap119-PrA) was used. (A) Yeast cells were visualized by Nomarski (left) and Kap119–PrA was detected by indirect immunofluorescence (middle). Nuclei were visualized by DAPI staining (right). (B) A cell homogenate (T) was fractionated into a cytosolic (C) and a nuclear (N) fraction. Cell equivalent amounts were analyzed by SDS-PAGE and Kap119–PrA, cytoplasmic 3-phosphoglycerate kinase (3-PGK), and nucleolar Nop1p were detected immunologically using either rabbit anti–mouse IgG, monoclonal mouse antiserum D66, or monoclonal mouse antiserum 22C5-D8.
The Transcription Elongation Factor TFIIS Binds Specifically to Kap119p
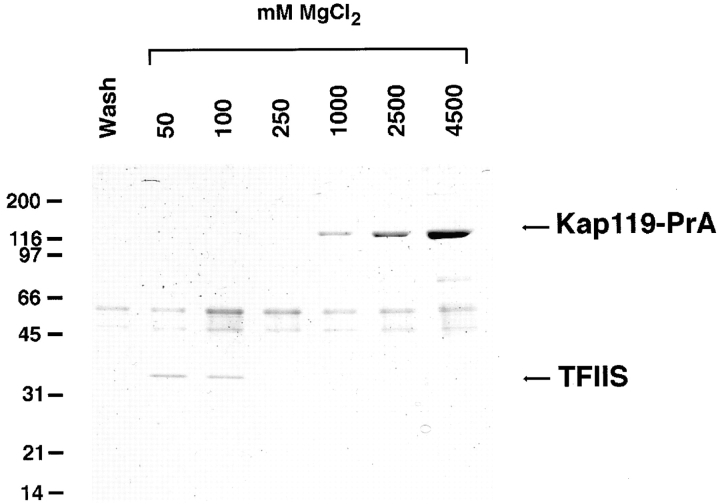
To coimmunoisolate potential transport substrate(s), a cytosol of the Kap119–PrA strain was incubated with IgG Sepharose. After extensive washing, the bound material was eluted with a MgCl2 step gradient ranging from 50 mM to 4.5 M MgCl2. The proteins in the eluted fractions were analyzed by SDS-PAGE and stained with Coomassie blue (Fig. 2). One major protein of ∼35 kD eluted at 50 and 100 mM MgCl2. In agreement with the previously observed elution behavior of transport substrates from PrA-tagged Kaps, the 35-kD band was a strong candidate for an Kap119p-bound transport substrate. As expected (Aitchison et al., 1996; Pemberton et al., 1997; Rosenblum et al., 1997; Rout et al., 1997), the Kap119–PrA eluted between 1.0 and 4.5 M MgCl2. By mass spectrometry, the 35-kD band was identified as TFIIS (also called PPR2, YSII, DST1, STALPHA, or YGL043) (Hubert et al., 1983; Davies et al., 1990; Clark et al., 1991; Nakanishi et al., 1992) by virtue of eight tryptic peptides between 1,000 and 1,915 D. The matching peptides, with 0.5 D tolerance, covered 29% of the TFIIS sequence. The TFIIS gene is not essential (Nakanishi et al., 1992) and codes for a protein of 309-amino acid residues with a calculated M r of 34.8 kD. Homologous proteins have been identified in Drosophila, various mammalian genomes and in the vaccinia virus genome (for review see Kassavatis and Geiduschek, 1993). TFIIS binds to RNA polymerase II (Rappaport et al., 1988), and stimulates cleavage and elongation of nascent RNA in the transcription elongation complex (Izban and Luse, 1992; Nakanishi et al., 1992; Borukhov et al., 1993). As TFIIS contains a zinc-binding domain, it may directly interact with nucleic acid (Agarwal et al., 1991).
Figure 2.
Kap119–PrA pulls out TFIIS from the cytosol. A cytosolic fraction from a Kap119–PrA strain was incubated with IgG– Sepharose. The last wash fraction and fractions subsequently eluted with a step gradient of 50–4,500 mM MgCl2 were analyzed by SDS-PAGE and Coomassie blue staining. The Kap119–PrA elutes between 1,000 and 4,500 mM MgCl2. The major band of ∼35 kD eluting at 50 and 100 mM MgCl2 was identified by mass spectrometry as the transcription factor TFIIS.
Defective Nuclear Transport of TFIIS in a kap119Δ Strain
To investigate whether TFIIS was a nuclear import substrate of Kap119p we prepared a strain lacking KAP119. The kap119Δ strain was viable and grew on YPD plates but was characterized by a slightly extended doubling time compared with the wild-type haploid strain (Fig. 3 A). As TFIIS is primarily localized to the nucleus (Nakanishi et al., 1995) it should be mislocalized to the cytoplasm in the kap119Δ strain if Kap119p functions as its principal cognate Kap. To facilitate localization of TFIIS we protein A–tagged the endogenous TFIIS gene in a wild-type and a kap119Δ strain and investigated its cellular distribution by immunofluorescence and cell fractionation.
Figure 3.

Deletion of KAP119 causes slower growth and leads to mislocalization of TFIIS. (A) Comparison of growth rates of DF5α (wt) and of the KAP119 deletion strain, kap119Δ. Cultures were inoculated at OD 0.1 into rich media, grown at 30°C and samples were taken at the indicated time points. (B) Localization by immunofluorescence in wild-type (wt), kap119Δ, and kap108Δ strains in which the endogenous TFIIS had been replaced by TFIIS–PrA. Whereas TFIIS–PrA was primarily localized to the nucleus in the wt and kap108Δ strains, it was diffusely localized throughout the cell in the kap119Δ strain. (C) A homogenate (T) of kap119Δ and wt cells was fractionated into a cytosolic (C) and a nuclear (N) fraction. Cell equivalent portions were analyzed by SDS-PAGE and TFIIS–PrA, 3-phosphoglycerate kinase (3-PGK) and Nop1p were detected with either rabbit anti–mouse IgG, monoclonal mouse antiserum D66 or monoclonal mouse antiserum 22C5-D8. TFIIS was predominantly localized in the cytoplasm in kap119Δ cells.
By immunofluorescence TFIIS–PrA was indeed largely mislocalized to the cytoplasm in the kap119Δ strain (Fig. 3 B, middle), whereas in the wild-type strain it was primarily localized to the nucleus (Fig. 3 B, top). As Kap108p is the closest relative of Kap119p in yeast (26% identical) (Rosenblum et al., 1997), we also determined the localization of TFIIS–PrA in a kap108Δ strain. We found that TFIIS– PrA was properly localized to the nucleus and was not mislocalized to the cytoplasm (Fig. 3 B, bottom), indicating that Kap108p is not involved in the nuclear import of TFIIS.
The immunofluorescence results were further substantiated by cell fractionation of wild-type and kap119Δ cells and comparison of the distribution of TFIIS relative to marker proteins (Fig. 3 C). Compared with the wild-type strain a clearly reduced amount of TFIIS–PrA was detected in the nuclear fraction of kap119Δ. We conclude that Kap119p functions as cognate Kap for the import of TFIIS into the nucleus.
Kap119p Is the Principal Kap for Import of TFIIS
We tested whether cytosolic TFIIS binds only to Kap119p or whether it can also bind to other Kaps in a strain where Kap119p was deleted. Cytosol from a wild-type or a kap119Δ strain was incubated with IgG–Sepharose, bound protein eluted by MgCl2 step gradients, analyzed by SDS-PAGE, and then stained with Coomassie blue. In the wild-type strain a major band of ∼120 kD eluted a 100 mM MgCl2 (Fig. 4, left). Mass spectrometric analysis showed that this band was Kap119p. The TFIIS–PrA eluted between 1.0 and 4.5 M MgCl2. Judging from the staining intensity of the Kap119p and TFIIS–PrA bands, it appears that most of the cytosolic TFIIS–PrA was associated with Kap119p, suggesting that Kap119p is a major binding partner of TFIIS in the cytosol. Interestingly, in the cytosol prepared from the kap119Δ strain no clear candidates for additional Kaps were observed (Fig. 4, right) suggesting that TFIIS was not associated with another Kap in a strain deleted for Kap119p. This result is in agreement with the observed mislocalization of TFIIS reported above. We therefore conclude that Kap119p is the principal Kap for import of TFIIS.
Figure 4.
TFIIS–PrA pulls out Kap119p in a wt strain but does not appear to pull out an alternative Kap in a kap119Δ strain. Cytosolic fractions were prepared from wt strain or from a kap119Δ strain and analyzed as described in Fig. 2. The major band of ∼120 kD that eluted from the IgG–Sepharose at 100 mM MgCl2 in the wt cytosol experiment (left) was identified by mass spectrometry as Kap119p. Note that no visible bands at the 100 mM MgCl2 step were detected in the kap119Δ cytosol experiment (right) in the region above 90 kD where most Kapβs migrate.
Kap119p Mediates Nuclear Import of TFIIS by a Distinct and Nonoverlapping Pathway
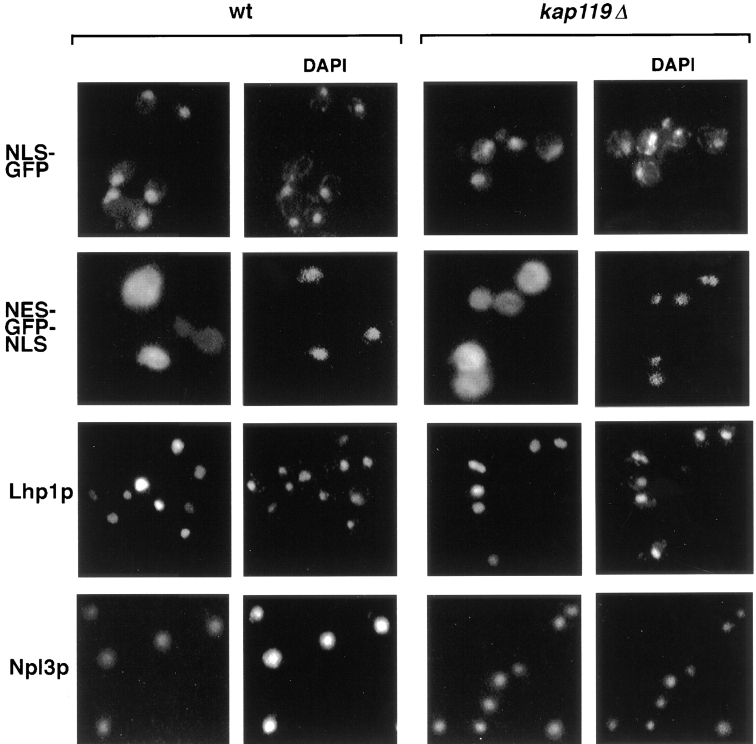
To determine whether deletion of KAP119 generally affected transport, we localized several substrates, whose transport is known to be mediated by a distinct cognate Kap, to see whether they were also mislocalized in a kap119Δ strain. We localized NLS–green fluorescent protein (GFP) (Shulga et al., 1996), NES–GFP–NLS (Stade et al., 1997), Lhp1p–GFP (Rosenblum et al., 1998), Npl3p (Wilson et al., 1994), and Nab2p (Aitchison et al., 1996) in both wild-type and kap119Δ strains. These substrates are transported by Kap60p/Kap95p, Kap124p (Crm1p), Kap108p, Kap111p, and Kap104p, respectively. As shown in Fig. 5, the localization of these substrates (data not shown for Nab2p) appeared similar in both strains. Hence deletion of Kap119p does not generally affect transport nor does Kap119p-mediated import specifically impact any of these other pathways.
Figure 5.
Various substrates that are transported by other Kaps are not mislocalized in a kap119Δ strain. The cellular distribution in wt and kap119Δ strains were directly examined for an NLS–GFP reporter (Shulga et al., 1996), for an NLS–GFP–NES reporter (Stade et al., 1997) and an Lhp1p–GFP reporter (Rosenblum et al., 1998), in fixed cells. Monospecific antibodies and indirect immunofluorescence were used to detect Nlp3p (Wilson et al., 1994). Nuclei were visualized by DAPI staining.
RanGTP Dissociates TFIIS from and Binds to Kap119p In Vitro
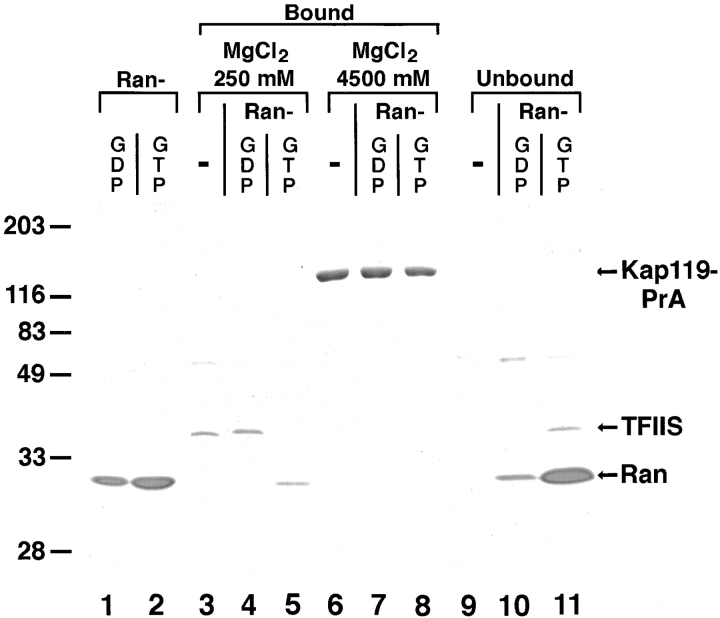
RanGTP has been reported to dissociate several import substrate/Kap complexes (Rexach and Blobel, 1995; Izaurralde et al., 1997; Siomi et al., 1997). To test whether RanGTP would dissociate TFIIS from Kap119p, we incubated the Sepharose-bound Kap119–PrA/TFIIS complex with recombinant RanGTP. After extensive washing, the Sepharose was split into three equal pools. One pool was further incubated with buffer alone while the two others were incubated with either RanGDP or RanGTP. Nearly all of the bound TFIIS (Fig. 6, lanes 3–5) was released from Kap119-PrA (Fig. 6, lanes 9–11) after incubation with RanGTP (Fig. 6, compare lane 5 with 11), but not after incubation with buffer (Fig. 6, compare lane 3 with 9) or with RanGDP (Fig. 6, compare lane 4 with 10). A portion of the added RanGTP remained bound to Kap119– PrA (Fig. 6, lane 5) whereas RanGDP did not bind (Fig. 6, lane 4). Hence, incubation of the complex with RanGTP resulted in dissociation of TFIIS and binding of RanGTP to Kap119–PrA. Importantly, identical amounts of Kap119–PrA were eluted from each pool.
Figure 6.
RanGTP but not RanGDP dissociates TFIIS from and binds to Kap119p. Incubation of Sepharose-bound Kap119–PrA/ TFIIS complex with recombinant RanGTP results in dissociation of TFIIS. Three times the previously used amount of Kap119– PrA cytosol was used for immunoisolation. Equal amounts of IgG-Sepharose–bound Kap119–PrA/TFIIS were incubated for 60 min at 21°C with either buffer (−), with RanGDP (5 μM) or with RanGTP (5 μM). The bound and unbound fractions from a subsequent two step elution with 250 and 4,500 mM MgCl2 were analyzed by SDS-PAGE and Coomassie blue staining. Lanes 1 and 2 represent equivalent amounts of purified RanGDP and RanGTP that were used for incubation. Note that the recovery of RanGDP in the unbound fraction was less than 100%. Numbers on the left indicate position of M r markers.
Kap119p Binds to Repeat Motif-containing Nucleoporins
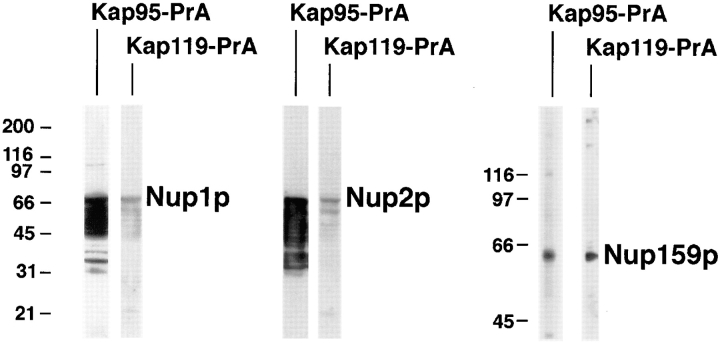
During the course of nuclear import, the import complex needs to dock at the NPC. This docking is thought to be mediated by a subset of nucleoporins that bear peptide-repeat motifs, including Nup1p, Nup2p, and Nup159p (Radu et al., 1995b ). Peptide repeat-containing domains of Nup1p, Nup2p, and Nup159p (Rexach and Blobel, 1995; Kraemer et al., 1995) were expressed in E. coli. Proteins of bacterial lysates were separated by SDS-PAGE, transferred to nitrocellulose, and then incubated with cytosol of the Kap119–PrA or Kap95–PrA strain and detected by luminol-based chemiluminescence (Fig. 7). As previously shown for Kap95–PrA (Iovine et al., 1995; Kraemer et al., 1995; Rexach and Blobel, 1995; Pemberton et al., 1997; Rosenblum et al., 1997), Kap119–PrA also bound to the tested nucleoporins: binding of Kap119–PrA to Nup1p and Nup2p was weaker than that of Kap95–PrA, whereas the extent of binding to Nup159p was similar for both PrA-tagged Kaps. These results indicate that Kap119p shares overlapping binding sites for peptide repeat containing Nups with other Kaps.
Figure 7.
Kap119–PrA binds to various nucleoporins. Proteins of lysates from E. coli strains expressing the peptide repeat regions of Nup1p (amino acid residues 432–816) (Rexach and Blobel, 1995), Nup2p (amino acid residues 186–561) (Rexach and Blobel, 1995), or Nup159p (amino acid residues 441–876) (Kraemer et al., 1995) were separated by SDS-PAGE, transferred to nitrocellulose, incubated with cytosol from either Kap95–PrA or Kap119–PrA strains and then with HRP-labeled rabbit IgG. M r markers for the Nup1p and Nup2p experiments are shown to the left of the Nup1p strips and for the Nup159p experiment to left of the Nup159p strips. Note that Kap95–PrA interacts much more strongly with Nup1p and Nup2p and their degradation products than does Kap119–PrA.
Discussion
As mentioned in the introduction, substrates for nine Kaps had been previously identified. These Kaps function in export and import, and transport several classes of proteins and tRNA. With the experiments presented here, we have identified an additional class of import substrates, represented by TFIIS, which is an RNA polymerase II transcription elongation factor (Davies et al., 1990; Nakanishi et al., 1992). We have shown that Kap119p is the principal nuclear import factor for TFIIS and that TFIIS is largely mislocalized in strains devoid of KAP119, indicating that other Kaps are incapable of efficient import of TFIIS. Although, TFIIS appears to be the major substrate of Kap119p in the growth conditions used here, we cannot exclude the possibility that other proteins are also imported (or exported) by Kap119p. In fact, after submission of this article it was reported that Kap119p was required for the nuclear import of the MAP kinase Hog1p that occurs as an initial response to high osmolarity in the growth medium (Ferrigno et al., 1998). In addition, because of the high cytoplasmic ratio of RanGDP/RanGTP (for review see Corbett and Silver, 1997) export substrates would not be expected to be Kap associated in the cytoplasm, and we would be unable to identify them using the approach we have taken here.
Although a slight amount of TFIIS still localized to nuclei in kap119Δ cells (the reasons for this Kap119p independent nuclear localization remain to be clarified but could be due to passive diffusion and/or piggyback import together with subunits of RNA polymerase II) TFIIS was not efficiently imported by other Kaps and deletion of Kap119p did not affect the import of substrates by other pathways. Further, TFIIS, being involved in elongation of RNA polymerase II transcripts (for review see Kerppola and Kane, 1991; Kassavatis and Geiduschek, 1993), represents a novel import substrate. TFIIS is the first RNA polymerase II accessory protein that has been shown to require a dedicated Kap for specific import. It has been shown that proteins involved in mRNA processing/export require their dedicated Kaps for import (Aitchison et al., 1995; Pemberton et al., 1997; Senger at al., 1998), so it is not surprising that factors involved in mRNA production would need distinct Kaps as well. Interestingly, Kap119p and Kap108p (Rosenblum et al., 1997) are the most similar pair of yeast Kaps (26% identical). We have not detected any direct overlap in their functions with regard to the import of TFIIS and Lhp1p. However, these two main substrates for the Kap119p and Kap108p pathways may have evolutionarily-related functions, TFIIS being involved in RNA polymerase II transcription (Nakanishi et al., 1992; Kassavatis and Geiduschek, 1993) and Lhp1p interacting with RNA polymerase III transcripts (Yoo and Wolin, 1997).
Two Kaps have been identified in higher eukaryotes that are similar to Kap119p, RanBP7 from Xenopus and RanBP8 from humans (Görlich et al., 1997). In addition, TFIIS is highly conserved between yeast and humans (for review see Kassavatis and Geiduschek, 1993). Although yeast TFIIS appears to contain a consensus bipartite NLS (K66KMISSWKDAINKNKRSR83) our data here suggest that it is not imported by Kap60p/Kap95p. A consensus NLS is not discernible in the human TFIIS sequence. It remains to be seen if RanBP7 or RanBP8 import higher eukaryotic homologues of TFIIS. In addition, it is not yet clear whether evolution has maintained all the identified yeast Kapβs, or has diversified them even more (as seems to be the case with Kapβ2 [Siomi et al., 1997] [Bonifaci, N., G. Blobel, and A. Radu, unpublished results]), or has delegated some of their functions to an expanded Kapα reservoir (Kohler et al., 1997; Seki et al., 1997; Takeda et al., 1997; Nachury et al., 1998; Pemberton et al., 1998; Rosenblum et al., 1998). The advantages of maintaining a dedicated Kap for import of TFIIS are likely to be the regulation of TFIIS transport.
Judging from their relative staining intensities, it appears that the transport substrates and their cognate Kaps are present in import complexes in nearly stoichiometric amounts (Aitchison et al., 1995; Pemberton et al., 1997; Rosenblum et al., 1997) (Fig. 4). Apart from these complexes being required for import, their stability is likely to be physiologically relevant. The physiological benefits of complex formation between a cytoplasmic transport substrate and its cognate Kap may be to prevent cytoplasmic degradation of the transport substrate or to protect reactive sites on transport substrates that are designed to interact with nucleic acids and/or proteins following import into the nucleus. It is even possible that the cytoplasmic complexes of Kaps and substrates prevent unwanted diffusion of small substrates into the nucleus. Alternatively, the formation of complexes between Kaps and transport substrates may facilitate reversible posttranslational modification(s), such as phosporylation/dephosphorylation, for each round of shuttling between the cytoplasm and nucleoplasm. Although there is presently no evidence for such reversible modifications, they might be useful in regulating the frequency of shuttling.
As we have shown, RanGTP was able to dissociate most of the TFIIS from Kap119p. Due to the nuclear localization of the RanGEF (Ohtsubo et al., 1989; Aebi et al., 1990) and cytoplasmic localization of RanGAP (Hopper et al., 1990; Becker et al., 1995; Corbett et al., 1995), the concentration of RanGTP can be expected to be higher in the nucleus than in the cytoplasm (for review see Corbett and Silver, 1997). As such, RanGTP may be a sensor for the nucleoplasm, consistent with the ability of RanGTP to dissociate the Kap119p import complex. In summary, our data here identify Kap119p as a new member of the Kapβ family that is involved in the import of the transcription factor TFIIS.
Acknowledgments
We thank F. Gharahdaghi of the Rockefeller University Protein/DNA Technology Center (New York, NY) for the mass spectral analysis, M. Rout (Rockefeller University) for Kap95–PrA cytosol, J. Aitchison (University of Alberta, Edmonton, Canada) for the Nab2p antiserum, M. Swanson (University of Florida, Gainesville, FL) for the 1E4 antibody, K. Weis (University of California, San Francisco, CA) for the NES–GFP– NLS reporter, D. Kraemer (Universität Würzburg, Würzburg, Germany) and M. Rexach (Stanford University, Standford, CA) for E. coli expression constructs and M. Floer for Ran and RanGAP and much practical help. We are grateful to K. Yoshida and the other members of the Blobel Lab for stimulating discussions and practical help.
Abbreviations used in this paper
- DAPI
4′,6-diamidino-2-phenylindole
- GFP
green fluorescent protein
- Kap
karyopherin
- NES
nuclear export sequence
- NLS
nuclear localization sequence
- NPC
nuclear pore complex
- Nups
nucleoporins
- PrA
protein A from S. aureus
Footnotes
M. Albertini was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (A1425/2-1) and J.S. Rosenblum from the National Institutes of Health (GM-18720).
References
- Aebi M, Clark MW, Vijayraghavan U, Abelson J. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1 which is involved in the control of chromosome condensation. Mol Gen Genet. 1990;224:72–80. doi: 10.1007/BF00259453. [DOI] [PubMed] [Google Scholar]
- Agarwal K, Baek KH, Jeon CJ, Miyamoto K, Ueno A, Yoon H. Stimulation of transcript elongation requires both the zinc finger and RNA polymerase II binding domains of human TFIIS. Biochemistry. 1991;30:7842–7851. doi: 10.1021/bi00245a026. [DOI] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Aris JP, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW, Identification of a nuclear export receptor for tRNA Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- Atkin AL, Altamura N, Leeds P, Culbertson MR. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.J., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1997. Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York. 1.8.1–1.8.7.
- Becker J, Melchior F, Gerke V, Bischoff R, Ponstingl H, Wittinghoffer A. RNA1 encodes a GTPase-activating protein specific for GSP1, the Ran/TC4 homologue of Saccharomyces cerevisiae. . J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc Natl Acad Sci USA. 1991;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. . Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Clark AB, Dykstra CC, Sugino A. Isolation, DNA sequence, and regulation of a Saccharomyces cerevisiaegene that encodes DNA strand transfer protein alpha. Mol Cell Biol. 1991;11:2576–2582. doi: 10.1128/mcb.11.5.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Koepp M, Lee MS, Schlenstedt G, Hopper AK, Silver PA. Rna1p, a Ran/TC4-activating protein is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies CJ, Trgovcich J, Hutchison CA. Homologue of TFIIS in yeast. Nature. 1990;345:298. doi: 10.1038/345298a0. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Ferrigno P, Posas F, Koepp D, Saito H, Silver PA. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO (Eur Mol Biol Organ) J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Floer M, Blobel G. The nuclear transport factor karyopherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur Mol Biol Organ) J. 1997a;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997b;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP-binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobsen A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Traglia HM, Dunst RW. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert JC, Guyonvarch A, Kammerer B, Exinger F, Liljelund P, Lacroute F. Complete sequence of a eukaryotic regulatory gene. EMBO (Eur Mol Biol Organ) J. 1983;2:2071–2073. doi: 10.1002/j.1460-2075.1983.tb01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131:1699–1713. doi: 10.1083/jcb.131.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO (Eur Mol Biol Organ) J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′—5′direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- Kassavatis GA, Geiduschek EP. RNA polymerase marching backward. Science. 1993;259:944–945. doi: 10.1126/science.7679800. [DOI] [PubMed] [Google Scholar]
- Kerppola TK, Kane CM. RNA polymerase: regulation of transcript elongation and termination. FASEB (Fed Am Soc Exp Biol) J. 1991;5:2833–2842. doi: 10.1096/fasebj.5.13.1916107. [DOI] [PubMed] [Google Scholar]
- Kohler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS (Fed Eur Biochem Soc) Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- Kraemer DM, Strambio-de-Castillia C, Blobel G, Rout MP. The essential yeast nucleoporin NUP159 is located on the cytoplasmic side of the nuclear pore complex and serves in karyopherin-mediated binding of transport substrate. J Biol Chem. 1995;270:19017–19021. doi: 10.1074/jbc.270.32.19017. [DOI] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff R, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Malik HS, Eickbush TH, Goldfarb DS. Evolutionary specialization of the nuclear targeting apparatus. Proc Natl Acad Sci USA. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Guan T, Yokoyama N, Nishimoto T, Gerace L. GTP hydrolysis by Ran occurs at the nuclear pore complex in an early step of protein import. J Cell Biol. 1995;131:571–581. doi: 10.1083/jcb.131.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud N, Goldfarb DS. Multiple pathways in nuclear transport: the import of U2 snRNP occurs by a novel kinetic pathway. J Cell Biol. 1991;112:215–223. doi: 10.1083/jcb.112.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud N, Goldfarb D. Microinjected U snRNAs are imported to oocyte nuclei via the nuclear pore complex by three distinguishable targeting pathways. J Cell Biol. 1992;116:851–861. doi: 10.1083/jcb.116.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Ryder UW, Lamond AI, Weis K. Cloning and characterization of hSRP1 gamma, a tissue-specific nuclear transport factor. Proc Natl Acad Sci USA. 1998;95:582–587. doi: 10.1073/pnas.95.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Nakano A, Nomura K, Sekimizu K, Natori S. Purification, gene cloning, and gene disruption of the transcription elongation factor S-II in Saccharomyces cerevisiae. . J Biol Chem. 1992;267:13200–13204. [PubMed] [Google Scholar]
- Nakanishi T, Shimoaraiso M, Kubo T, Natori S. Structure-function relationship of yeast S-II in terms of stimulation of RNA polymerase II, arrest relief, and suppression of 6-azauracil sensitivity. J Biol Chem. 1995;270:8991–8995. doi: 10.1074/jbc.270.15.8991. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Rosenblum JS, Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton LF, Blobel G, Rosenblum JS. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995a;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995b;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Rappaport J, Cho K, Saltzman A, Prenger J, Golomb M, Weinmann R. Transcription elongation factor SII interacts with a domain of the large subunit of human RNA polymerase II. Mol Cell Biol. 1988;8:3136–3142. doi: 10.1128/mcb.8.8.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F. Winston, and P. Hieter. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 198 pp.
- Rosenblum JS, Pemberton LF, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum JS, Pemberton LF, Bonifaci N, Blobel G. Nuclear import and evolution of a multifunctional RNA-binding protein. J Cell Biol. 1998;143:887–900. doi: 10.1083/jcb.143.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., and J.V. Kilmartin. 1994. Preparation of yeast spindle pole bodies. In Cell Biology: A Laboratory Handbook. J.E. Celis, editor. Academic Press, London, UK. 605–612.
- Seki T, Tada S, Katada T, Enomoto T. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem Biophys Res Commun. 1997;234:48–53. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- Senger B, Simons G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO (Eur Mol Biol Organ) J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss GA. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Takeda S, Fujiwara T, Shimizu F, Kawai A, Shinomiya K, Okuno S, Ozaki K, Katagiri T, Shimada Y, Nagata M, et al. Isolation and mapping of karyopherin alpha 3 (KPNA3), a human gene that is highly homologous to genes encoding Xenopusimportin, yeast SRP1 and human RCH1. Cytogenet Cell Genet. 1997;76:87–93. doi: 10.1159/000134521. [DOI] [PubMed] [Google Scholar]
- Weighardt F, Biamonti G, Riva S. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains of hnRNPA1. J Cell Sci. 1995;108:545–555. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- Wilson SM, Datar KV, Paddy MR, Swedlow JR, Swanson M. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. . J Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RW, Rout MP, Aitchison JD. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL. The yeast La protein is required for the 3′ endonucleolytic cleavage that maturates tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]