Abstract
We have identified CALNUC, an EF-hand, Ca2+-binding protein, as a Golgi resident protein. CALNUC corresponds to a previously identified EF-hand/calcium-binding protein known as nucleobindin. CALNUC interacts with Gαi3 subunits in the yeast two-hybrid system and in GST-CALNUC pull-down assays. Analysis of deletion mutants demonstrated that the EF-hand and intervening acidic regions are the site of CALNUC's interaction with Gαi3. CALNUC is found in both cytosolic and membrane fractions. The membrane pool is tightly associated with the luminal surface of Golgi membranes. CALNUC is widely expressed, as it is detected by immunofluorescence in the Golgi region of all tissues and cell lines examined. By immunoelectron microscopy, CALNUC is localized to cis-Golgi cisternae and the cis-Golgi network (CGN). CALNUC is the major Ca2+-binding protein detected by 45Ca2+-binding assay on Golgi fractions. The properties of CALNUC and its high homology to calreticulin suggest that it may play a key role in calcium homeostasis in the CGN and cis-Golgi cisternae.
The ER is well known to constitute a major storage compartment for intracellular Ca2+ that upon appropriate signaling is released into the cytosol and plays a fundamental role in cell physiology. In addition to its role in signaling, it has become evident that Ca2+ is also essential for many functions that take place within compartments of the exocytic pathway. For example, it is required for chaperone functions and quality control in the ER (Booth and Koch, 1989; Lodish and Kong, 1990; Sambrook, 1990; Lodish et al., 1992) and concentration and proteolytic processing of secretory proteins within regulated secretory granules (Chanat and Huttner, 1991; Carnell and Moore, 1994; Canaff and Brechler, 1996). The Golgi region has also been identified as a Ca2+-enriched compartment based on ion microscopy (Chandra et al., 1991; Zha and Morrison, 1995). Recently, it has been demonstrated directly by electron energy loss spectroscopy (EELS),1 a new technique that allows high resolution calcium mapping of intracellular compartments (Grohovaz et al., 1996), that Golgi cisternae have a high Ca2+ level extending across the entire cis-trans axis of the Golgi (Pezzati et al., 1997). The question is how is this high Ca2+ concentration maintained in the Golgi? In the case of the ER, Ca2+ storage is believed to be maintained by multiple calcium-binding proteins including calnexin, calreticulin, GRP78 (BiP), GRP94, ERp72, protein disulfide isomerase, reticulocalbin, and ERC55 (Pozzan et al., 1994; Meldolesi and Pozzan, 1998) of which the most important appears to be calreticulin (Mery et al., 1996). Much less is known about the internal milieu of the Golgi. To date, only a single Golgi luminal Ca2+-binding protein has been identified, Cab45, which, interestingly, has high homology to the ER Ca2+-binding proteins reticulocalbin and ERC55 (Scherer et al., 1996).
In this paper we have identified a second Golgi Ca2+-binding protein which we call CALNUC with significant sequence homology to another ER Ca2+-binding protein, calreticulin. We identified CALNUC in a yeast two-hybrid screen using the heterotrimeric G protein Gαi3 as bait. CALNUC corresponds to a known protein called nucleobindin (Miura et al., 1992; Wendel et al., 1995). Nucleobindin was thought to be a transcription factor based on its ability to bind DNA fragments in vitro, thus the name nucleobindin (Miura et al., 1992). Nucleobindin was first identified in culture supernatant of a B lymphocyte cell line established from mice prone to the autoimmune disorder, systemic lupus erythematosis (Kanai et al., 1986; Miura et al., 1992), and was later isolated as a minor protein constituent from bone extracellular matrix (Wendel et al., 1995). Recombinant nucleobindin was also shown to bind Ca2+, and the first of its two EF hands was required for binding (Miura et al., 1994). The localization of nucleobindin has been problematic. It has been variously suggested to be a nuclear protein (Wang et al., 1994), a secreted protein (Miura et al., 1992; Wendel et al., 1995), and a resident ER protein, the latter based on its interaction with the cyclooxygenase isoenzymes 1 and 2 (Ballif et al., 1996).
Because of the intriguing diverse properties of this molecule, such as the EF-hand/calcium–binding domains, its homology to calreticulin, and its ability to interact with the Gαi subfamily of heterotrimeric G proteins (Mochizuki et al., 1995), we set out to characterize nucleobindin, hereafter referred to as CALNUC, and especially to define its localization in the hope of shedding light on its function. To our surprise, we found CALNUC both in cytosolic fractions and associated with Golgi membranes. The Golgi-associated form proved to be a Golgi resident protein concentrated in the cis-Golgi network (CGN) and cis-Golgi cisternae facing the Golgi lumen.
Materials and Methods
Antibodies
Rabbit antiserum raised against the rat homologue of nucleobindin (calvaria calcium-binding protein) was obtained from D. Heinegard (University of Lund, Sweden; Wendel et al., 1995). Rabbit antisera to GM130 (Nakamura et al., 1995), α-adaptin, calnexin (Wada et al., 1991), and TGN38 were obtained, respectively, from Drs. Graham Warren (Imperial Cancer Research Foundation, London, UK), S. Schmid (Scripps Research Institute, La Jolla, CA), J.J.M. Bergeron (McGill University, Montréal, Canada), and G. Banting (University of Bristol, UK). Rabbit antiserum to α-Mannosidase II (α-Man II) was prepared as described (Velasco et al., 1993) and mouse mAb 53FC3 against α-Man II was obtained from Dr. Brian Burke (Burke et al., 1982; University of Alberta, Canada). Affinity-purified anti–β-COP IgG, raised against the EAGE peptide (Duden et al., 1991) was characterized earlier (Hendricks et al., 1993). mAb G1/93 against ERGIC-53 was provided by Dr. H.-P. Hauri (Biozentrum, University of Basel, Switzerland).
Construction of the Rat GC Cell Library and Yeast Two-Hybrid Screening
Double stranded cDNA was generated from 7 μg poly(A)+ RNA isolated from rat GC pituitary cells (subcloned from GH3 cells) using a random primed cDNA synthesis kit (Stratagene, Inc., San Diego, CA). EcoRI and XhoI oligo synthesized adapter linkers were ligated onto the cDNA at the 5′ and 3′ end, respectively, and the library was inserted into the Gal4 activation domain pACT2 “prey” vector (CLONTECH Laboratories, Palo Alto, CA). The total library contains ∼2 × 106 independent clones.
The complete rat cDNA of Gαi3 was cloned into the Gal4 DNA-binding domain pGBT9 “bait” vector (CLONTECH Laboratories) as described (De Vries et al., 1995). The pGBT9Gαi3 “bait” vector was transformed into yeast strain HF7c (CLONTECH Laboratories). The transformed yeast colonies were selected on tryptophan (−Trp) selective plates, and after 6 d the plasmids from surviving colonies were analyzed for the presence of pGBT9Gαi3.
For interaction screening in the yeast two-hybrid system (Chien et al., 1991), 50 μg of the rat GC-cell cDNA library in the pACT2 vector was transformed into yeast HF7c(pGBT9Gαi3) strain (Schiestl and Gietz, 1989). Approximately 106 colonies were plated onto selective medium, and colonies that survived were scored for β-galactosidase (β-gal) activity by a colony lift assay (CLONTECH Laboratories). Plasmid DNA from the HIS+/β-gal+ colonies was purified by transforming into Escherichia coli HB101 by electroporation. These plasmids were retransformed into the HF7c strain alone or with various control plasmids, including the original pGBT9Gαi3 “bait” plasmid. Positive clones were grouped based on restriction analysis, and 24 fragments of different sizes were selected and sequenced from the 5′ or the 3′ end of the inserts by automated sequencing.
In Vivo Interactions
To construct a complete coding sequence of CALNUC, the insert of a clone containing the 5′-end was ligated to the insert of a 3′-end via an internal EcoRI site, and the reconstituted full-length insert was ligated into the pACT2 vector. Deletion mutants of CALNUC (CALNUC227–372, CALNUC320–372, and CALNUC227–320) were generated by PCR using CALNUC-specific primers (sequences available upon request), subcloned into the pACT2 vector, and cotransformed with pGBT9Gαi3.
Full-length pACT2-CALNUC was cotransformed with the following Gα subunits in pGBT9 vectors: rat Gαi3 (De Vries et al., 1995), rat Gαi2, and mouse Gαq, (obtained from P. Insel, University of California San Diego), rat Gαi1 (from T. Kosasa, University of Texas, Southwestern Medical Center, Dallas, TX), rat Gαs (from H. Bourne, University of California San Francisco), rat Gαo from E. Neer (Brigham and Women's Hospital, Boston, MA), and rat Gαz, (from E. Ross, University of Texas, Southwestern Medical Center). Interactions were analyzed qualitatively by a colony lift assay for β-gal using 5-bromo-4-chloro-3-indolyl-β-d-galactoside (Guarente, 1983).
Database Searches
Online BLAST searches were performed in the GenBank database (National Institutes of Health, Bethesda, MD) via the National Center for Biotechnology Information's (NCBI) home page on the World Wide Web. PROSITE (Geneworks; IntelliGenetics, San Francisco, CA), BLASTP and Psort were used for protein analysis. To compare the primary structure of CALNUC, calreticulin (CRT), and Cab45 their amino acid (aa) sequences were aligned using GeneWorks 2.5.1 software (IntelliGenetics, San Francisco, CA).
Expression and Purification of Glutathione-S-Transferase–CALNUC Fusion Protein
For the production of recombinant glutathione-S-transferase (GST) fusion protein, CALNUC cDNA was subcloned into the pGEX-KG vector (Pharmacia Biotechnology, Inc., Piscataway, NJ), and transformed into E. coli (Y1090) cells. GST-CALNUC fusion protein was affinity purified from bacterial lysates on glutathione–Sepharose and used for immunization of rabbits, in vitro pull-down assays, and 45Ca2+ overlay.
Preparation, Affinity Purification, and Characterization of Anti-CALNUC IgG
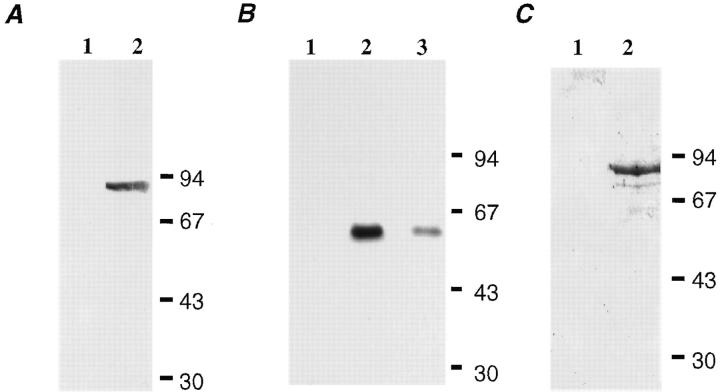
Antibodies were raised in rabbits against purified full-length GST-CALNUC. For affinity purification, purified recombinant CALNUC was coupled to cyanogen bromide-activated Sepharose 4B (Pharmacia Biotechnology, Inc.). Antibodies were then bound to the coupled beads and eluted sequentially with 0.1 M glycine/HCl (pH 2.5) and 0.1 M triethylamine (pH 11.5). The affinity-purified antibody recognized 5 ng recombinant GST-CALNUC at a dilution of 0.67 μg/ml (see Fig. 1 A) and specifically recognized CALNUC in 50 μg NRK cell lysate at a concentration of 0.17 μg/ml (see Fig. 1 B).
Figure 1.
Characterization of GST-CALNUC and affinity-purified anti-CALNUC IgG. (A) 5 ng purified GST-CALNUC was separated by 10% SDS-PAGE, transferred to a PVDF membrane, and immunoblotted with 0.67 μg/ml affinity-purified anti-CALNUC IgG (lane 2) or preimmune serum (lane 1). The antibody was capable of detecting 5 ng GST-CALNUC (90 kD). (B) PNS prepared from NRK cells (50 μg/lane) was immunoblotted with 0.38 (lane 2) and 0.17 (lane 3) μg/ml affinity-purified anti-CALNUC IgG or preimmune serum (lane 1). Endogenous CALNUC (63 kD) could be detected with as little as 0.17 μg/ml of the purified antibody. (C) 45Ca2+-overlay. 50 μg GST (lane 1) or GST-CALNUC (lane 2) was separated by SDS-PAGE, transferred to a PVDF membrane and overlaid with 4 μCi/ml 45CaCl2. After exposure of the washed and dried membrane to Kodak X-Omat AR film, a ∼90-kD signal that corresponds to the fusion protein was detected.
45Ca2+ Overlay
50 μg purified GST-CALNUC or 200 μg Golgi proteins were transferred from SDS gels onto polyvinylidene difluoride (PVDF) membranes (Imobilon P; Millipore Corp., Bedford, MA), followed by incubation of the membranes with 45CaCl2 (Dupont-NEN, Boston, MA) as described (Maruyama et al., 1984). The membranes were processed for autoradiography (Kodak X-Omat Film or a PhosphorImager cassette). Quantitation of the bound Ca2+ was done by densitometry or by direct counting, respectively.
In Vitro Interactions
Wild-type rat Gαi3 cDNA, activated rat Gαi3(Q204L), and inactivated Gαi3(G203A) mutants, obtained from A. Spiegel (National Institutes of Health, Bethesda, MD), were subcloned into pBluescript SK+ (Stratagene, San Diego, CA). In vitro transcription/translation of the various forms of Gαi3 from the T7 promoter in pBluescript SK+ was performed using the TNT-coupled reticulocyte lysate system (Promega Corp., Madison, WI) in the presence of [35S]methionine (in vivo cell labeling grade; Amersham Pharmacia Biotech Inc, Arlington Heights, IL) according to the manufacturer's instructions. Purified GST-CALNUC fusion protein (6 μg) or GST control (6 μg) was immobilized on glutathione–Sepharose beads and incubated with 15,000 cpm 35S-labeled, in vitro-translated Gαi3 (mutant or wild-type) in binding buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 3 mM EDTA, 0.1% NP-40, 1 mM DTT, and protease inhibitors) as previously described (Mochizuki et al., 1995). The mixture was incubated by rotating for 2 h at 4°C. The beads were washed three times in the same binding buffer, resuspended in 25 μl of Laemmli buffer, boiled for 5 min, and then the proteins were loaded on 10% SDS gels and exposed for autoradiography.
Cell Culture
AtT-20/D-16v pituitary cells, obtained from Dr. Richard Mains (Johns Hopkins University, Baltimore, MD), were maintained in DME medium (high glucose) supplemented with 10% (vol/vol) horse serum 2.5% (vol/ vol) FCS (GIBCO BRL, Gaithersburg, MD). Normal rat kidney (NRK) cells were grown in DME supplemented with 5% FCS, 2.5% Nu-IV serum (Collaborative Research, Inc., Waltham, MA), and REF-52, HeLa, and GC cells were maintained in 10% FCS in DME high glucose medium. Vero cells were grown in MEM Earles in 10% FCS. PC12 cells (passage 6 to 20), originally obtained from Dr. L. Greene, were maintained in RPMI 1640 Medium (GIBCO BRL) supplemented with 10% heat-inactivated horse serum, 5% FCS. All culture media contained 100 U/ml penicillin G and 100 μg/ml streptomycin sulfate. Cells were used as confluent monolayers.
Immunocytochemistry
For immunofluorescence, cells were cultured on glass coverslips for 2–3 d, fixed for 1 h with 2% paraformaldehyde (PFA) in 0.075 M phosphate buffer, pH 7.4, and permeabilized with 0.1% Triton X-100 in PBS (10 min). They were then incubated for 1 h at room temperature with affinity-purified rabbit antibodies to CALNUC or mouse mAb to ERGIC-53 or Man II followed by crossed-absorbed FITC- or Texas red–conjugated donkey anti–rabbit or anti–mouse F(ab′)2 (Jackson ImmunoResearch Laboratories, West Grove, PA). For double labeling, cells were incubated simultaneously with rabbit polyclonal anti-CALNUC and mouse mAb ERGIC-53 or Man II followed by incubation with appropriate anti-rabbit and anti-mouse conjugates. Cells were mounted in 25% PBS, 75% glycerol with 1 mg/ml ρ-phenylinediamine, and then examined with a Zeiss Axiophot equipped for epifluorescence (Carl Zeiss Inc., Thornwood, NY).
For immunogold labeling, cultured cells were fixed in 8% PFA, 100 mM phosphate buffer, pH 7.4 (15 min), followed by 4% PFA in phosphate buffer (1 h). Samples were then cryoprotected and frozen in liquid nitrogen as described earlier (McCaffery and Farquhar, 1995). Ultrathin cryosections were prepared and incubated with primary antibodies in 10% FCS/PBS for 2 h at room temperature followed by incubation in 5 or 10 nm gold, goat anti–rabbit or anti–mouse IgG conjugates (Amersham Pharmacia Biotech Inc.) for 1 h at room temperature. Sections were post-fixed (10 min) with 2% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, stained for 30 min in 2% uranyl acetate, adsorption stained for 10 min with 0.2% uranyl acetate, 0.2% methyl-cellulose, and 3.2% polyvinyl alcohol and then examined in a Philips CM10 or a JEOL 1200EX II electron microscope. For immunofluorescence, semithin cryosections (0.5–1.0 μm) were incubated as above except that incubation in primary antibodies was for 2 h at room temperature. For immunofluorescence and immunogold labeling of pituitary, liver, and kidney tissue, organs were perfusion-fixed with PLP fixative (2% paraformaldehyde, 0.75 M lysine, 0.01 M sodium periodate in 0.1 M phosphate buffer, pH 6.2; McLean and Nakane, 1973) and processed as described for cultured cells.
Preparation and Treatments of Membrane Fractions from Cultured Cells
Cells were washed twice with cold PBS and subsequently scraped into homogenization buffer containing protease inhibitors (0.5 mg/ml aprotinin, 0.5 mg/ml pepstatin A, 0.2 mg/ml leupeptin, 1 mM 4-(2-aminoethyl)-benzenesulfonyl-fluoride hydrochloride; ICN Biomedicals, Costa Mesa, CA), and 5 mM EDTA in TBS. They were then gently homogenized by 10 passages through a 28 1/2-G needle and nuclei and unbroken cells were removed by centrifugation (600 g for 3 min). Membrane pellets were prepared by centrifugation of the postnuclear supernatant (PNS) at 100,000 g for 1 h at 4°C in a Beckman TL-100 ultracentrifuge (Beckman Instruments., Inc., Fullerton, CA). The protein concentration of the membrane pellets was obtained by BCA assay (Pierce Chemical Co., Rockford, IL).
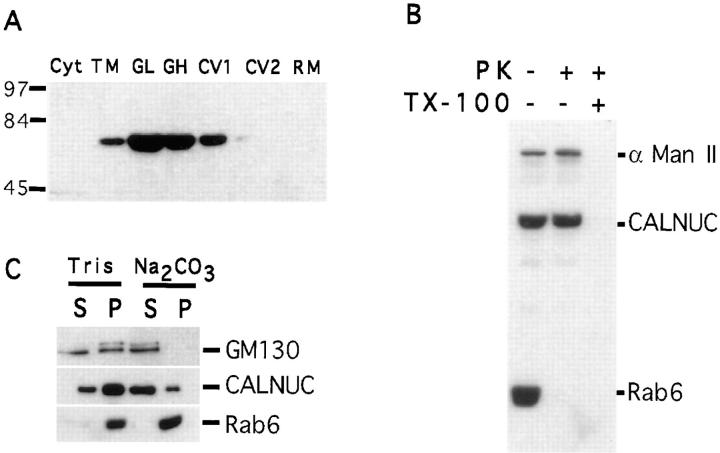
For digestion with proteinase K (PK), membranes (100 μg) were suspended in 200 μl of reaction buffer containing 10 mM Tris-HCl (pH 7.8), 150 mM KCl, 2 mM MgCl2, 2 mM CaCl2, and 0.2 M sucrose followed by incubation with 10 or 20 μg of PK (Boehringer Mannheim GmbH, Mannheim, Germany) at room temperature for 30 min, after which 10 mM PMSF was added to stop the reaction. Proteins were separated by SDS-PAGE and immunoblotted with appropriate antibodies.
For alkaline extraction, membranes (100 μg) were suspended in 200 μl–0.2 M Na2CO3 (pH 11.5; Fujiki et al., 1982) containing protease inhibitors, incubated on ice for 30 min, and centrifuged at 100,000 g for 1 h at 4°C. Supernatants were kept on ice for later use, and pellets were resuspended in 200 μl–0.2 M Na2CO3 (pH 11.5), followed by SDS-PAGE and immunoblotting.
Triton X-114 extraction was carried out essentially as described earlier (Bordier, 1991) except that proteins in the aqueous and detergent phases were precipitated with 10% TCA for 30 min on ice followed by SDS-PAGE and immunoblotting.
Preparation of Rat Liver Golgi Fractions
Rat liver fractions were prepared by density gradient centrifugation and characterized as described previously (Saucan and Palade, 1994; Jin et al., 1996). In brief, total microsomes were adjusted to 1.24 M sucrose, loaded onto the bottom of a discontinuous sucrose gradient, and then centrifuged at 82,000 g. All procedures were done at 4°C in the presence of protease inhibitors. The resulting fractions were designated as Golgi light (GL) and Golgi heavy (GH), enriched in Golgi elements, carrier vesicle fractions 1 and 2 (CV1 and CV2), enriched in transport vesicles, and residual microsomes (RM). The protein concentration of each fraction was determined by BCA assay (Bio-Rad Laboratories, Hercules, CA). Fractions were then treated with PK, alkaline pH (11.5), or Triton X-114 essentially as described for membranes from cultured cells except that the Na2CO3-extracted samples were loaded on the top of a 6% sucrose cushion and centrifuged at 400,000 g for 25 min. The supernatants were collected and Golgi membrane pellets were resuspended in Tris-EDTA (10 mM Tris, pH 7.5, 1 mM EDTA) and processed for SDS-PAGE and immunoblotting.
For some experiments liver fractionation was carried out by the method of Leelavathi et al. (1970) as modified by Taylor et al. (1997). In brief, livers of Sprague-Dawley rats were perfused with 20 ml 6% (wt/vol) sucrose in 100 mM KH2PO4/KHPO4, pH 6.8, 5 mM MgCl2 buffer with protease inhibitors, homogenized in the perfusion buffer using a glass/teflon homogenizer and spun at 1,500 g for 10 min. The PNS was loaded onto a discontinuous sucrose gradient (1.3 M, 0.86 M sucrose, PNS, 0.25 M sucrose, 6 ml each) in a SW28 tube (Beckman Instruments., Inc.), centrifuged at 100,000 g for 90 min, and crude fractions designated SI, SII, and SIII were isolated with disposable syringes from the PNS/0.25 M sucrose, 0.86 M sucrose/PNS, and 1.3 M sucrose/0.86 M sucrose interphases, respectively. To prepare Golgi fractions depleted of cargo, rats were injected with 1.5 mg cycloheximide in 150 mM NaCl per kg body weight through the tail vein 90 min before liver excision (Taylor et al., 1997). Fractions were processed for 45Ca2+ overlay or immunoblotting.
SDS-PAGE and Immunoblotting
Proteins were separated on 10% SDS gels under reducing conditions, followed by transfer to PVDF membranes (Millipore Corp.). Membranes were blocked with 5% nonfat dry milk in TBS–0.05% Tween (TBS-T) and blotted with primary antibodies followed by incubation with HRP conjugated goat anti–rabbit IgG (1:3,000 in TBS-T), secondary antibody (Bio-Rad Laboratories), and detection by ECL (Pierce Chemical Co.). Quantitation of protein bands was done by densitometry using ScanAnalysis Software (Biosoft, Cambridge, UK).
Results
Identification of CALNUC as a Ca2+-binding Protein that Interacts with Gαi3
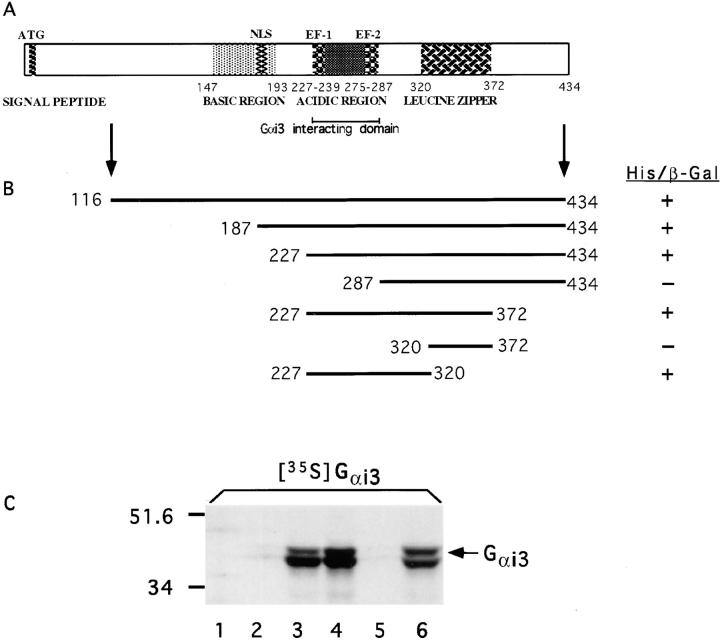
To search for protein(s) that can interact with the heterotrimeric G-protein, Gαi3, we used the yeast two-hybrid system to screen a rat GC cell pituitary library and isolated 24 inserts of different sizes that were sequenced and analyzed. 3 out of the 24 clones sequenced were identified as the rat homologue of human GAIP, a GTPase-activating protein (GAP) identified earlier (De Vries et al., 1995, 1996, 1998), 8 contained novel sequences, and 13 were identified as the rat calvaria calcium-binding protein (Wendel et al., 1995), the rat homologue of human nucleobindin (Miura et al., 1992). Using the BLAST program the sequence we obtained was found to match 100% the rat calvaria calcium-binding protein. Moreover, antibodies raised against the latter protein (Wendel et al., 1995) recognized GST-CALNUC, and GST-CALNUC was found to bind Ca2+ in a 45Ca2+-overlay (Fig. 1 C). Important features of CALNUC are its signal sequence, two EF-hands with an intervening acidic region, leucine zipper, and basic region with a putative nuclear localization signal (NLS; Fig. 2 A). CALNUC lacks a putative transmembrane domain, but examination of its hydrophobicity using the GeneWorks 2.5.1. program revealed a 15-aa hydrophobic region at its COOH terminus.
Figure 2.
(A) Structure of CALNUC. Rat CALNUC is a 434-aa protein with an NH2-terminal signal sequence, a putative DNA-binding domain (BASIC REGION) with a nuclear localization signal (NLS), an acidic region flanked by two EF-hand motifs (EF-1, EF-2), and a leucine zipper motif. (B) aa 227–287 are required for interaction with Gαi3. Those deletion mutants spanning aa 227–287 containing two EF-hands and a central acidic region (CALNUC227–372, CALNUC227–320) bind to Gαi3, whereas, mutants that lack this region (CALNUC287–434, CALNUC320–372) do not. pGBT9Gαi3 “bait” vector was cotransformed with various pACT2-CALNUC mutants into yeast strain HF7c. The transformed colonies were scored for β-gal activity by a colony lift assay. (C) Verification of the site of CALNUC's interaction with Gαi3 by an in vitro binding assay. [35S]Gαi3 (arrows) binds to GST-CALNUC116–434 (lane 6), GST-CALNUC227–372 (lane 4), and GST-CALNUC227–320 (lane 3), but not to GST-CALNUC287-434 (lane 5), GST-CALNUC320–372 (lane 2), or to control beads with GST alone (lane 1). GST-CALNUC fusion proteins bound to glutathione-agarose beads were incubated with 35S-labeled, in vitro translated Gαi3 for 2 h, the bound proteins were separated by 10% SDS-PAGE and detected by autoradiography.
Homology between CALNUC and Calreticulin
Among the known calcium-binding proteins CALNUC was found to have 30% homology with rat CRT. The sequence similarity between rat, human, and bovine CRT and rat and mouse CALNUC was found throughout the two proteins, but it was highest in the P-domain (rat CRT198–297) and C-domain (rat CRT298–416) where it was 36 and 43%, respectively (Fig. 3), and lower (26%) in the N-domain (rat CRT1–197). Interestingly, the P- and C-domains of CRT are thought to contain, respectively, high affinity and low affinity Ca2+-binding sites. Three helix-loop-helix motifs (Type A repeats; Baksh and Michalak, 1996) that are similar to the EF-hand motif in CALNUC (Fig. 3) are found in the P-domain of CRT. The COOH-terminal region of CALNUC, like that of CRT, is also relatively acidic. Since the acidic segments of the C-domain of CRT were reported to represent low affinity, high capacity calcium-binding sites, the same may apply to CALNUC. Thus, the structure of CALNUC suggests that the COOH-terminal region may have low affinity, high-capacity binding sites at CALNUC343–352 and putative higher affinity-binding sites at the sites of the EF-hand motifs. There are also notable differences between the structure of CALNUC and CRT. For example, the three times repeated NPD/E motifs in the P-domain of CRT and the conserved KPEDWD motif found in both CRT and calnexin (Nash et al., 1994) are not found in CALNUC. The COOH-terminal KDEL ER retention signal found in CRT is also absent from CALNUC which is consistent with our finding (see below) that the membrane-associated pool of CALNUC resides in the Golgi, not the ER.
Figure 3.
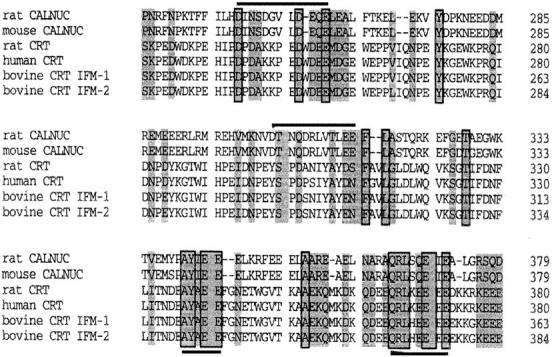
Sequence comparison of rat and mouse CALNUC, and rat, human, and bovine CRT (isoform-1 and -2). CALNUC shares significant sequence homology to the P-domain (rat CRT238–297) and C-domain (rat CRT298–416) of CRT. Regions of aa identity are boxed and regions of aa similarity are shaded. EF-hand loop structures in CALNUC are indicated above (upper line). Two conserved sequences AY(I/A)EE and QRLX(Q/E)E(I/E)E, found in both CALNUC and CRT, are underlined.
Another striking feature of unknown significance shared by CALNUC and CRT are two short conserved aa sequences, AY(I/A)EE and QRLX(Q/E)E(I/E)E, found in the C-domain of CRT (rat CRT337–341 and rat CRT365–372). Further investigation is required to determine the functions of these domains.
When the sequences of CALNUC and Cab45 were compared, they also showed ∼30% similarity throughout, but no striking homology was found except for a short aa sequence AANXE(E/D) identified in both Cab45 (aa 47–52 and CALNUC (aa 73–78).
The Two EF-Hand Motifs of CALNUC and Intervening Acidic Region Are Required for Interaction with Gαi3
Analysis of the CALNUC clones isolated from the yeast two-hybrid screen revealed that the smallest clone that interacted with Gαi3 was CALNUC227–434 (Fig. 2 B). The fact that the clone that coded for CALNUC227–434 interacted but CALNUC287–434 did not points to the importance of aa 227–287, the region containing the two EF-hands, for binding. To further analyze the Gαi3-binding domain several deletion mutants, i.e., CALNUC227–372, CALNUC320–372, and CALNUC227–320, were generated and their ability to interact with Gαi3 assessed in the yeast two-hybrid system. The smallest mutant capable of binding Gαi3 was CALNUC227–320. Once again, this deletion mutant includes both EF-hands and the intervening acidic region. The leucine zipper region, aa 320–372, does not appear to be required because CALNUC227–320 interacted with Gαi3, but CALNUC320–372 did not. From these data we conclude that the two EF-hands and intervening acidic region constitute the site of CALNUC's interaction with Gαi3.
Similar results were obtained using an in vitro binding assay in which 35S-labeled, in vitro–translated Gαi3 was incubated with full-length or truncated GST-CALNUC fusion proteins bound to glutathione-agarose beads: Gαi3 bound to all GST-CALNUC fragments containing aa 227– 287, but not to those lacking this region (Fig. 2 C). Thus, our results demonstrate by two different assays that the region that spans aa 227–287 containing the two EF-hand motifs and intervening acidic aa-rich region, is required for interaction with Gαi3.
CALNUC Interacts with the Gαi and Gαs Family of Gα
We also used the two-hybrid system to test whether CALNUC interacts with other Gα family members. As shown in Table I, CALNUC interacted with all members of the Gαi subfamily (Gαi1, Gαi2, Gαo1, and Gαz) as well as Gαs, but not with Gαq, Gα12, or Gα13. Based on the semi-quantitative β-gal filter assay, the strongest interaction was with Gαi3 and Gαi1 and interactions with Gαs and the other Gαi members were weaker. These results suggest that CALNUC specifically interacts with members of the Gαi and Gs subfamily of heterotrimeric G proteins.
Table I.
Interaction between CALNUC and Gα Subunits in Yeast Two Hybrid Filter Assay
| Bait | β-gal Filter | |||
|---|---|---|---|---|
| Gαi3 | +++ | |||
| Gαi3 (Q204L) | +++ | |||
| Gαi3 (G203A) | +++ | |||
| Gαi1 | +++ | |||
| Gαi2 | ++ | |||
| Gαo1 | + | |||
| Gαz | ++ | |||
| Gαs | ++ | |||
| Gαq | — | |||
| Gα12 | — | |||
| Gα13 | — |
The β-gal filter assay was performed on (Leu−, Trp−) plates, and the color intensity was scored after 8 h. −, no color; +, weak color; ++, intermediate color, +++, strong color. Baits were constructed in pGBT9, and CALNUC prey vector was pACT2-CALNUC.
We further investigated whether the interaction between Gαi3 and CALNUC is nucleotide-dependent using the activated GTP-bound mutant, Gαi3(Q204L), and inactivated, GDP-bound mutant Gαi3(G203A), and found that both mutants bind to CALNUC as efficiently as wild-type Gαi3 (Table I).
CALNUC Is Localized to the Golgi Region by Immunofluorescence
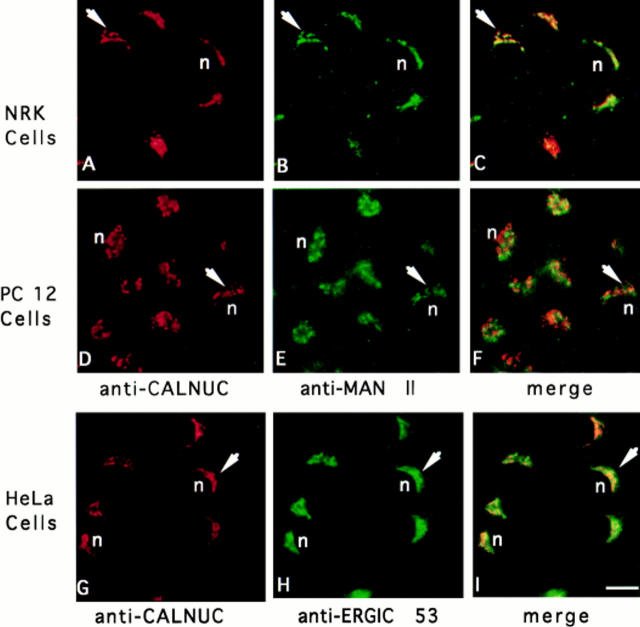
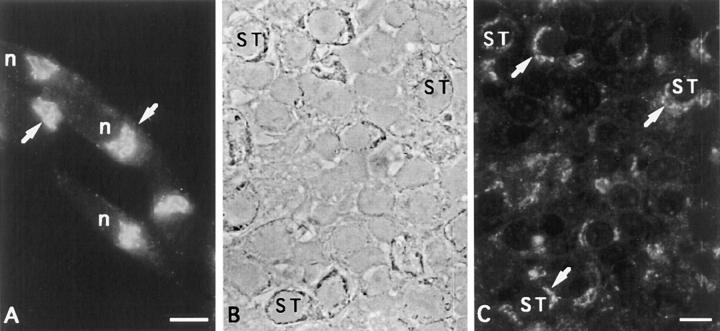
To determine the intracellular distribution of CALNUC we carried out immunofluorescence on a number of rat (GC pituitary cells, PC-12 cells, NRK cells, REF52 fibroblasts), mouse (AtT-20 pituitary cells), human (HeLa), and monkey (Vero) cell lines. To our surprise, we found that in all cells examined, CALNUC was localized to the Golgi region (Figs. 4 and 5 A). After double labeling CALNUC staining partially overlapped with both α-Man II, a Golgi marker, and ERGIC-53, a marker for the ER– Golgi intermediate compartment (ERGIC) and CGN (Fig. 4). We also checked CALNUC's distribution by immunofluorescence in semithin cryosections of rat pituitary (Fig. 5, B and C), liver, and kidney. Again, in all cases CALNUC was localized to the Golgi region. Staining was very similar regardless of whether we used the antiserum made against the native protein, provided by D. Heinegard (Wendel et al., 1995), or the affinity-purified IgG we made against recombinant GST-CALNUC. In some cases (e.g., rat kidney) variable cytoplasmic staining was observed, but the predominant signal was in the Golgi region. CALNUC staining in the Golgi persisted in cells treated for 3 h with cycloheximide (data not shown).
Figure 4.
CALNUC is localized by immunofluorescence in the Golgi region in cultured cells. CALNUC is found in the Golgi region (arrows) in NRK, PC-12, and HeLa cells where it partially overlaps with the Golgi marker, α-Man II, and cis-Golgi/ERGIC marker, ERGIC-53. Cells were fixed in 2% paraformaldehyde in phosphate buffer, permeabilized, and doubly incubated with affinity-purified rabbit polyclonal CALNUC antibody (5 μg/ml) and mouse mAb Man II or ERGIC-53 followed by cross-absorbed Texas red–conjugated donkey anti–rabbit and FITC-conjugated donkey anti–mouse F(ab′)2. Bar, 10 μm.
Figure 5.
Localization of CALNUC to the Golgi region in pituitary cells. (A) AtT-20 cells, prepared as in Fig. 4, showing CALNUC concentrated in the Golgi region near the nucleus (arrows). (B and C) Semithin section of rat pituitary showing phase contrast (B) and immunofluorescence localization (C) of CALNUC in the Golgi region (arrows) of several somatotrophs (ST) as well as other cell types. Preparation was the same as in Fig. 4 except that fixation was by perfusion with PLP. Bar, 10 μm.
CALNUC Is Localized on Golgi Cisternae Concentrated on the Cis Side of the Golgi Stack
To determine whether CALNUC is associated with Golgi membranes or with other organelles located in the Golgi region we carried out immunogold labeling on ultrathin cryosections of AtT-20 and NRK cells (Fig. 6) and rat anterior pituitary (Fig. 7), kidney, and liver tissue. In all cases, labeling was concentrated on membranes of the stacked Golgi cisternae and associated vesicles and was more concentrated on cisternae and vesicles located on one side of the Golgi stack. The latter could be identified as the cis side based on the location of secretion granules (Fig. 7, A and B) and on double labeling with TGN38 (Fig. 7 C). TGN38 was clearly distributed on the opposite or trans side of the Golgi stack (Fig. 7 C). Counts of gold particles found on Golgi membranes after staining for CALNUC indicate that >80% of the gold particles were found on the one to two cis-most Golgi cisternae and associated vesicles that together constitute the cis-Golgi network (CGN; Farquhar and Hauri, 1997). Therefore, the immunocytochemical findings indicate that CALNUC is associated with cis-Golgi cisternae and the CGN.
Figure 6.
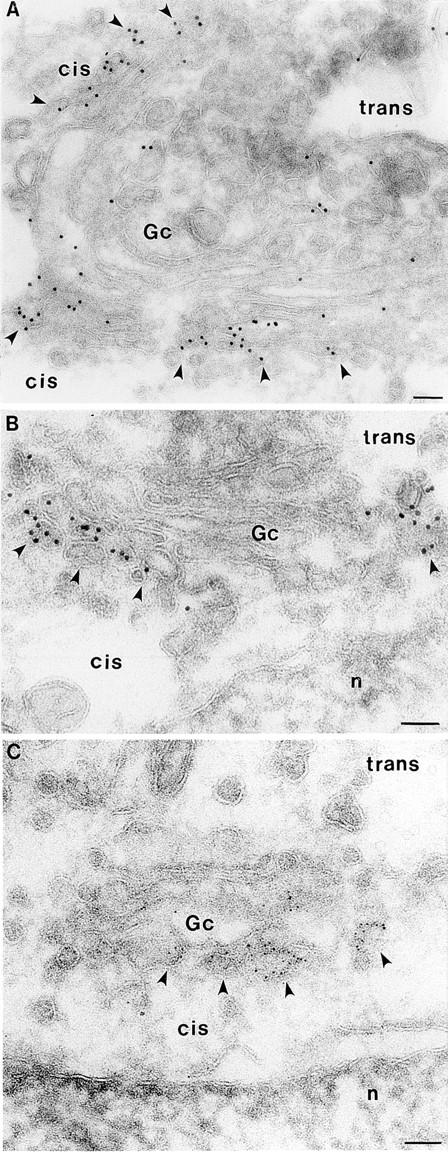
Localization of CALNUC to Golgi cisternae in NRK cells. Gold particles are found largely or exclusively on 1–2 cisternae (Gc) on one side of the Golgi stack (arrowheads). Typically, >80% of the label is on the side facing the nucleus (n) assumed to be the cis side of the Golgi stack. Cells were fixed in 8% paraformaldehyde (PFA; 15 min) and 4% PFA (1 h). Ultrathin cryosections were prepared and incubated sequentially with affinity-purified anti-CALNUC IgG and 5- or 10-nm colloidal gold conjugated to goat anti–rabbit IgG. Sections were postfixed and stained as described in Materials and Methods. Bar, 0.1 μm.
Figure 7.
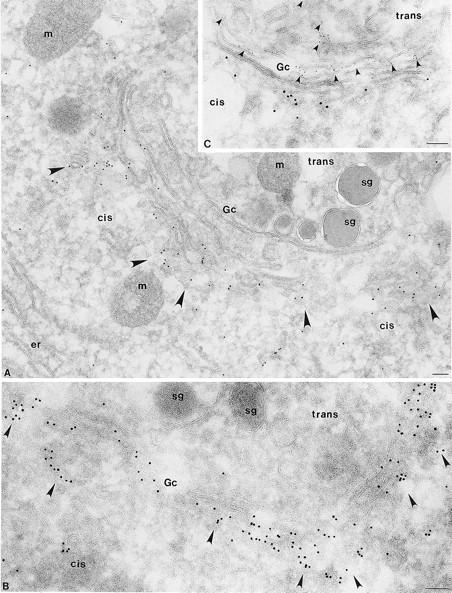
CALNUC is concentrated in cis-Golgi cisternae and the CGN in pituitary cells. (A and B) By immunogold labeling CALNUC (10 nm gold) is concentrated in cis-Golgi cisternae and associated vesicles (arrowheads) in a secretory cell from the rat pituitary. Counts of gold particles reveal that ∼80% of the gold is found on the 1–2 cis-most cisternae and associated vesicles of the CGN. Little gold is found over ER (er), mitochondria (m) or secretion granules (sg). (C) Double immunogold labeling for CALNUC and TGN38 in a cell from the rat pituitary. CALNUC (10 nm gold) is localized exclusively to cis-Golgi cisternae and associated vesicles. TGN38 (5 nm gold) marks the TGN located on the opposite (trans) side of the Golgi stack. There is little or no overlap between the two. Preparation and staining was the same as in Fig. 6 except that fixation was by perfusion with PLP fixative, and “C” was doubly stained for CALNUC and TGN38 followed by gold-conjugated anti–rabbit and anti–mouse gold conjugates. Bar, 0.1 μm.
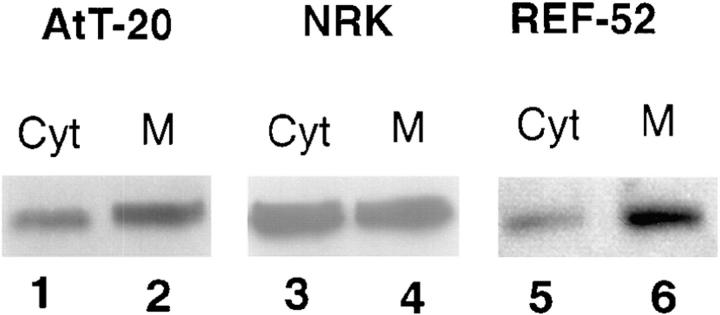
CALNUC Is Found in Both Membrane and Cytosolic Fractions
The results of immunofluorescence and immunogold labeling suggested that CALNUC is associated primarily with Golgi cisternae. To obtain further information on its localization, we assessed CALNUC's distribution in membrane (100,000 g pellet) and cytosolic (100,000 g supernatant) fractions prepared from AtT-20, NRK, and REF-52 cells. In all cases, CALNUC was found in both membrane and cytosolic fractions, with the percent detected in the membrane fraction varying among different cell lines, i.e., 90% in REF-52 cells, 80% in AtT-20 cells, and 50% in NRK cells (Fig. 8). The CALNUC found in cytosolic fractions could represent a true cytosolic pool or a soluble pool released from the Golgi lumen during homogenization. However, it seems unlikely that all the CALNUC found in the cytosolic fraction could be released from the Golgi lumen because in NRK cells only 30% of the Cab45, a soluble luminal Golgi protein, was released into the cytosol under the same conditions (data not shown). Further study is required to clarify the relationship between the CALNUC associated with Golgi membranes and that found in cytosolic fractions.
Figure 8.
CALNUC is found in both membrane and cytosolic fractions. Membrane (M) and cytosolic (Cyt) fractions were prepared by centrifugation of PNS at 100,000 g for 1 h. Membrane pellets were resuspended in homogenization buffer to the same volume as supernatants. 50-μl membrane or cystolic fractions were separated by 10% SDS-PAGE and immunoblotted with affinity-purified rabbit anti-CALNUC. CALNUC (63 kD) is detected in both membrane (M) and cytosolic (Cyt) fractions of all cell types tested. The percent of the total CALNUC associated with membrane fractions is 80% in AtT-20 cells (lane 2), 50% in NRK cells (lane 4), and 90% in REF-52 cells (lane 6).
The Membrane Pool of CALNUC Is Tightly Associated with Golgi Membranes and Faces the Golgi Lumen
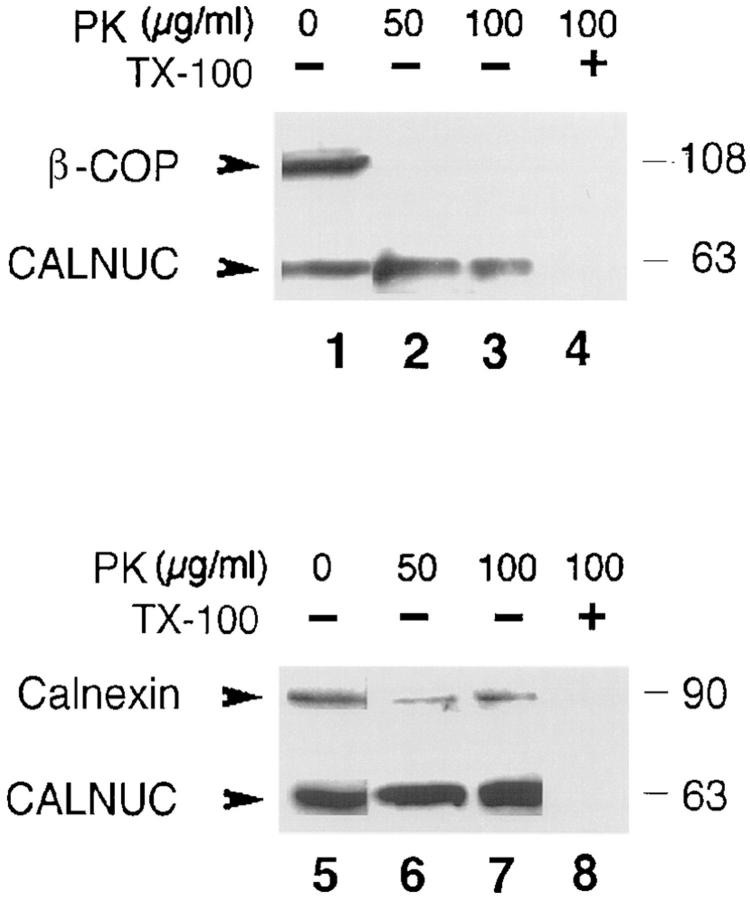
The aa sequence of CALNUC contains a signal peptide and lacks putative membrane-spanning domains suggesting it is likely to be a soluble luminal protein or a peripheral membrane protein. To determine the topography of the membrane-associated pool of CALNUC we digested 100,000 g pellets prepared from AtT-20 cells with proteinase K (PK). β-COP, a peripheral coat protein (Duden et al., 1991) was completely digested by increasing amounts of PK (Fig. 9), whereas CALNUC and calnexin (Fig. 9), an integral membrane protein that faces the ER lumen (Wada et al., 1991), were resistant to PK digestion, suggesting that CALNUC is a luminal protein.
Figure 9.
Membrane-associated CALNUC is a luminal protein. Membranes (100,000 g pellet) prepared from AtT-20 cells (see Fig. 8) were treated with the indicated concentrations of proteinase K (PK) at room temperature for 30 min in the presence and absence of detergent followed by immunoblotting with polyclonal antibodies to β-COP, calnexin, or CALNUC. β-COP, a peripheral coat protein, was completely digested by 50 (lane 2) or 100 (lane 3) μg/ml PK whereas CALNUC (lanes 2, 3, 6, and 7) and calnexin (lanes 6 and 7), a membrane protein that faces the ER lumen, were resistant to PK digestion. In the presence of detergent (1% Triton X-100) both CALNUC and calnexin were digested (lanes 4 and 8).
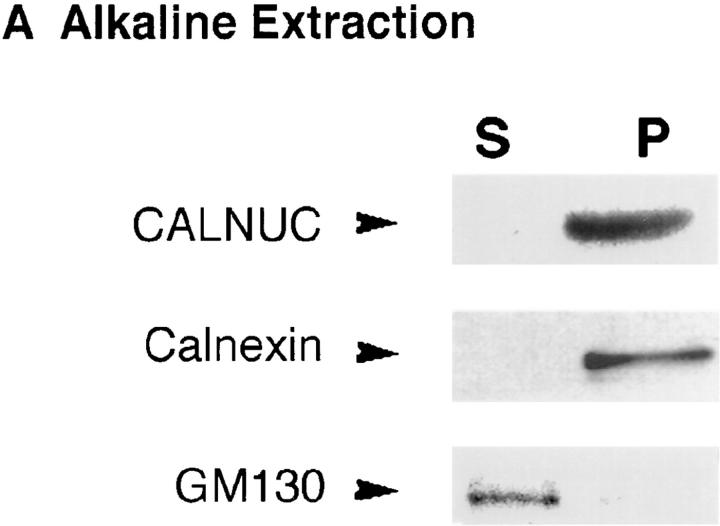
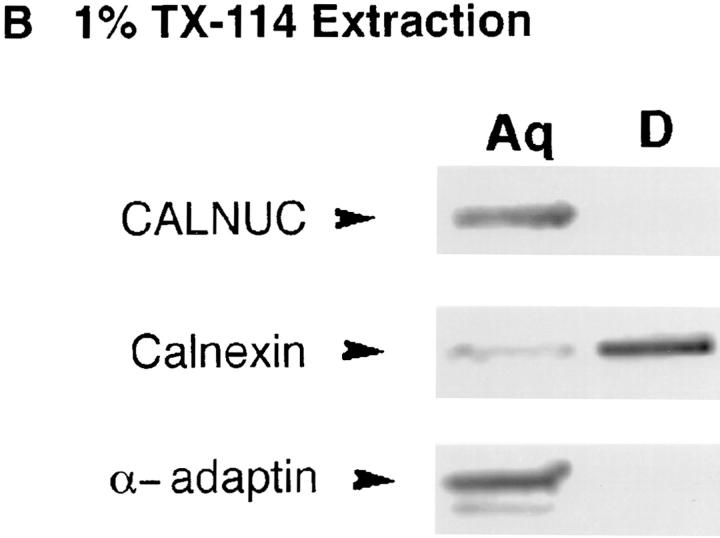
Next, AtT-20 cell membranes were subjected to alkaline extraction (pH 11.5) with Na2CO3. As shown in Fig. 10 A, most (90%) of the GM130, a Golgi-associated peripheral membrane protein (Nakamura et al., 1995), was removed but all of the calnexin, an ER integral membrane protein (Wada et al., 1991), and most of the CALNUC remained associated with the membrane pellet. When the time of alkaline extraction was extended to 1 h, ∼10% of the CALNUC was removed, but the majority (80–90%) remained membrane associated (data not shown). When AtT-20 cell membrane pellets were subjected to Triton X-114 extraction, CALNUC and α-adaptin, a peripheral membrane protein (100 kD) facing the cytosol (Robinson, 1997), were found entirely in the aqueous phase (Fig. 10 B). By contrast most of the calnexin partitioned into the detergent phase. These results together with the localization studies suggest that CALNUC is a peripheral membrane protein tightly associated with the luminal surface of Golgi membranes.
Figure 10.
Characterization of membrane-associated CALNUC. (A) Alkaline extraction: membranes prepared from AtT-20 cells (see Fig. 8) were incubated with 0.2 M Na2CO3 (pH 11.5) for 30 min and immunoblotted with antibodies against CALNUC, calnexin or GM130. Both CALNUC and calnexin, an integral membrane protein, remained associated with the membrane pellet (P), whereas GM130, a peripheral membrane protein, was released into the supernatant. (B) Triton X-114 phase separation: AtT-20 cell membrane fractions were treated with 1% Triton X-114. After phase partitioning, proteins in the aqueous (Aq) and detergent (D) phases were separated by SDS-PAGE and immunoblotted with the indicated antibodies. CALNUC and α-adaptin, a peripheral coat protein, partitioned into the aqueous phase, whereas calnexin was found in the detergent phase.
CALNUC Is Enriched in Golgi Fractions of Rat Liver
To obtain further biochemical evidence for the association of CALNUC with Golgi membranes, we carried out immunoblotting for CALNUC on Golgi fractions prepared from rat liver by a procedure designed to separate Golgi fractions from carrier vesicles and residual microsomes (Saucan and Palade, 1994; Jin et al., 1996). Unlike in the cultured cell lines, CALNUC was detected in membrane fractions but not in cytosolic fractions even when up to 400 μg of cytosolic protein was loaded on the gels (data not shown). As shown in Fig. 11 A, CALNUC was most enriched in the two Golgi fractions (GL and GH). To determine the topology of CALNUC, the GH fraction was treated with PK in the presence or absence of detergent (Fig. 11 B). CALNUC was resistant to PK digestion in the absence of detergent, but was completely digested in the presence of detergent as was Man II, a Golgi membrane protein that faces the Golgi lumen. Rab6, a membrane anchored protein that faces the cytosol, was digested under both conditions. When Golgi membranes were subjected to alkaline extraction (pH 11.5), ∼80% of the CALNUC was released into the supernatant (Fig. 11 C), but 20% remained associated with Golgi membranes even after treatment for up to 1 h. From these results we conclude that CALNUC is a peripheral membrane protein tightly associated with the luminal surface of Golgi membranes.
Figure 11.
CALNUC is enriched in Golgi fractions from rat liver. (A) CALNUC is detected mainly in Golgi light (GL) and Golgi heavy (GH) fractions. CALNUC was not detected in cytosolic (Cyt), carrier vesicle 2 (CV2), or residual microsome (RM) fractions. A small amount was seen in CV1. Total microsomes (TM) were adjusted to 1.24 M sucrose, loaded onto the bottom of a discontinuous sucrose gradient and centrifuged at 82,000 g for 1 h. Fractions were collected, and 50 μg protein from each was separated by SDS-PAGE and immunoblotted with affinity-purified anti-CALNUC IgG. (B) CALNUC is resistant to proteinase K (PK) digestion. Rab6, a membrane anchored protein facing the cytoplasm was digested, whereas CALNUC and Man II, a membrane protein facing the Golgi lumen, were protected from PK digestion. In the presence of detergent (Triton X-100) both Man II and CALNUC were digested. (C) Alkaline extraction: when Golgi membranes (GL and GH) were treated with 200 mM Na2CO3, ∼80% of the CALNUC was released into the supernatant (S) and the remainder remained associated with the membrane pellet (P). Rab6, a membrane-anchored protein, was not released, and all of the GM130, a Golgi peripheral membrane protein, was released into the supernatant after this treatment.
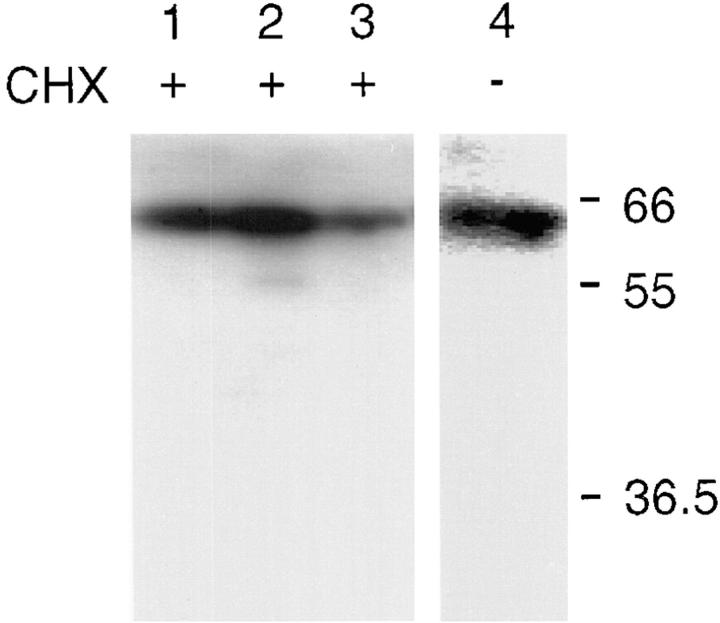
To establish whether CALNUC is a Golgi resident protein or a cargo protein in transit, we cleared rat hepatocytes of proteins in transit by treatment with cycloheximide for 90 min, using the procedure recently introduced to distinguish endogenous Golgi proteins from cargo (Taylor et al., 1997), followed by immunoblotting of Golgi fractions for CALNUC. We found that CALNUC was readily detected in Golgi fractions from cycloheximide (CHX) treated rats at levels indistinguishable from controls (Fig. 12), thus demonstrating that CALNUC is a bona fide, Golgi resident protein.
Figure 12.
Membrane-associated CALNUC is a Golgi resident protein. 100 μg each of Golgi fractions S-I (lightest fraction, lane 1), S-II (lane 2), and S-III (heaviest fraction, lane 3) obtained from livers of rats treated with CHX (lanes 1–3) for 90 min or pooled Golgi fractions from controls (lane 4) were separated by SDS-PAGE and immunoblotted for CALNUC as described in Materials and Methods. Endogenous CALNUC can be identified as a 63-kD band in both controls and Golgi fractions depleted of proteins in transit by CHX treatment.
CALNUC Is a Major Calcium-binding Protein in Golgi Fractions
To directly demonstrate that endogenous CALNUC associated with Golgi membranes is capable of binding Ca2+, we carried out 45Ca2+ overlays on pooled Golgi fractions. We found that 45Ca2+ bound to one major band at 63 kD (Fig. 13, lane 2) that corresponded in mobility to CALNUC (Fig. 13, lane 1). These results indicate that CALNUC is the major Ca2+-binding protein in Golgi fractions as determined by 45Ca2+ overlay.
Figure 13.

45Ca2+-overlay on pooled Golgi fractions (S-I, S-II, and S-III), demonstrating Ca2+-binding to endogenous CALNUC. Golgi proteins (200 μg) were separated by SDS-PAGE and transferred onto a PVDF membrane. The membrane was overlaid with 45CaCl2 (200 μCi/ml), and calcium binding was quantitated on a PhosphorImager. Lane 1, Immunoblot of pooled Golgi fractions indicating the position of CALNUC. Lane 2, A single band, 63 kD, with the same mobility as CALNUC binds 45Ca2+ in this assay.
Discussion
We have identified, characterized, and localized CALNUC, an EF-hand, calcium-binding protein, and shown that it is a resident protein of the CGN and cis-Golgi cisternae. CALNUC corresponds to the rat homologue of a previously identified mouse and human protein called nucleobindin, so named because it bound to DNA in vitro (Miura et al., 1992) and was thought to be located in the nucleus (Wang et al., 1994). We demonstrate here that CALNUC is localized to the Golgi region by immunofluorescence and that it is concentrated in cis cisternae and/or the CGN by immunoelectron microscopy in all tissues and cell lines examined. We further found that CALNUC is a bona fide resident Golgi protein as it is enriched in Golgi fractions prepared from rat livers depleted of cargo. Moreover, we demonstrated by 45Ca2+ overlay that both recombinant CALNUC (see Fig. 1) and endogenous CALNUC (see Fig. 13) are capable of binding Ca2+. In fact, the only band detected in Golgi fractions by 45Ca2+ overlay was CALNUC.
Interestingly, CALNUC shows high homology to CRT and has several properties in common with CRT in that both are Ca2+-binding proteins (Miura et al., 1994; Baksh and Michalak, 1996), both have been found to be secreted in autoimmune diseases (Kanai et al., 1993; Sontheimer et al., 1996), and both have been said to be localized to compartments of the exocytic pathway (Opas, 1996; Krause and Michalak, 1997). It is intriguing that CALNUC shows the highest homology to the P- and C-domains of CRT that are thought to represent, respectively, high affinity, low capacity and high capacity, low affinity binding sites for Ca2+ (Baksh and Michalak, 1996). Besides its role in binding Ca2+, CRT, as well as calnexin, has been shown to be a glucose lectin that functions as an ER chaperone involved in ER quality control by binding incompletely processed proteins with exposed glucose groups (Parlati et al., 1996; Helenius et al., 1997). However, neither the KPEDWD motif found in both CRT and calnexin that binds integrins (Krause and Michalak, 1997), nor the COOH-terminal KDEL ER retention/retrieval signal is present in CALNUC. We discovered that two motifs of unknown function, AY(I/A)EE and QRLX(Q/E)E(I/E)E, found in CALNUC are shared with CRT. These regions may reflect the common role of those two proteins in binding Ca2+ or another, as yet undisclosed function.
A problem that remains unanswered is the significance of CALNUC's interaction with Gα subunits. CALNUC has previously been shown to interact with Gαi2 in the yeast two-hybrid system (Mochizuki et al., 1995), and we found here that it can interact with Gαi3 and other members of the Gαi subfamily as well as Gs but not other Gα subunits in the yeast two-hybrid system. We assume that cytosolic CALNUC interacts with Gα subunits since Gα subunits are mainly anchored through lipid anchors to the cytoplasmic surface of membranes (Mumby, 1997). It seems unlikely that the Golgi luminal pool would have the opportunity to bind Gα subunits as CALNUC and Gα subunits are on opposite sides of membranes. In this paper we focused on characterization of the luminal, Golgi membrane–associated pool of CALNUC, but the relationship between soluble and membrane-associated CALNUC and the interaction of CALNUC with Gα subunits remain intriguing problems for the future.
Another unanswered problem is how the membrane pool of CALNUC is bound to the luminal surface of Golgi membranes. In cultured cells we found that CALNUC has a pI of 4.9 by isoelectric focusing (our unpublished observation), is tightly anchored to Golgi membranes and cannot be extracted by extended alkaline treatment. CALNUC lacks a putative transmembrane domain and is likely to represent a peripheral membrane protein which, like p62 (Jones et al., 1993), is so tightly bound to one or more Golgi proteins that it cannot be readily extracted by high pH treatment. However, we cannot rule out that CALNUC could be anchored by a lipid anchor such as palmitate or by a 15-aa hydrophobic region found at its COOH terminus. In the case of synaptobrevin and other SNARES it has been shown that 12 hydrophobic aa at the COOH terminus is sufficient for membrane anchoring (Kutay et al., 1993; Whitley et al., 1996).
CALNUC is only the second Golgi calcium-binding protein identified to date, with Cab45 being the first (Scherer et al., 1996). Cab45 differs from CALNUC in that it is a soluble Golgi resident protein found in the Golgi lumen and is completely extracted after high pH treatment. It also has a putative glycosylation site that is lacking in CALNUC.
What is the role of calcium-binding proteins in the Golgi? It has recently been shown that high level of Ca2+ in the Golgi is comparable to that of the ER and is seen all across the stacked cisternae, cis to trans (Pezzati et al., 1997). Surprisingly, the concentration in the TGN is somewhat lower. We can speculate, based on studies on the ER, that Ca2+-binding to CALNUC can serve one of two functions: (a) It may serve to maintain the high calcium levels required for Golgi functions such as sorting, lectin binding, and concentration of cargo into regulated secretory granules, or (b) binding of Ca2+ may cause conformational changes in CALNUC that render it competent for specific, as yet unknown interactions. In addition, it is possible that CALNUC may bind other divalent cations. CRT has been shown to bind zinc and iron as well as calcium (Baksh and Michalak, 1996). Specific Golgi functions may have specific divalent cation requirements. For example, it has been shown that UDP-GalNAc:polypeptide N-acetylgalactosamine transferase, the enzyme that adds the initial galactose to mucin-type glycoproteins, requires Mn2+ both in vitro (Sugiura et al., 1982) and in vivo (Kaufman et al., 1994).
This study opens a number of new and intriguing questions concerning the functions of CALNUC in Ca2+ homeostasis in the Golgi, the requirements for Ca2+ for Golgi functions, and whether heterotrimeric G proteins are involved in Ca2+ homeostasis in the Golgi.
Abbreviations used in this paper
- α-Man II
α-Mannosidase II
- β-gal
β-galactosidase
- aa
amino acids
- CGN
cis-Golgi network
- CHX
cycloheximide, CRT, calreticulin
- CV
carrier vesicle fraction
- Cyt
cytosol
- EELS
electron energy loss spectroscopy
- ERGIC
ER–Golgi intermediate compartment
- GAP
GTPase-activating protein
- GH
Golgi heavy
- GL
Golgi light
- GDT
glutathione-S-transferase
- NLS
nuclear localization signal
- PFA
paraformaldehyde
- PK
proteinase K
- PNS
postnuclear supernatant
- PVDF
polyvinylidene difluoride
- RM
residual microsomes
- TBS-T
TBS–0.05% Tween
- TM
total membranes
Footnotes
Helen Le-Niculescu is a graduate student in the Molecular Pathology Graduate Program at UCSD and is a Markey Foundation Fellow. This work was supported by National Institutes of Health grants DK 17780 and CA 58689 to Marilyn G. Farquhar.
P. Lin and H. Le-Niculescu contributed equally to this work.
Dr. Hennemann's present address is the Institute of Cell Biology, Cancer Research, University of Essen, Virchowstr. 173, D-45122 Essen, Germany.
References
- Baksh, S., and M. Michalak. 1996. Basic characteristics and ion binding to calreticulin. In Calreticulin. M. Michalak, editor. R.G. Landes Company, Georgetown. 11–26.
- Ballif BA, Mincek NV, Barratt JT, Wilson ML, Simmons DL. Interaction of cyclooxygenases with an apoptosis- and autoimmunity-associated protein. Proc Natl Acad Sci USA. 1996;93:5544–5549. doi: 10.1073/pnas.93.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth C, Koch GLE. Perturbation of cellular calcium induces secretion of luminal ER proteins. Cell. 1989;59:729–737. doi: 10.1016/0092-8674(89)90019-6. [DOI] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1991;256:1604–1607. [PubMed] [Google Scholar]
- Burke B, Griffiths G, Reggio H, Louvard D, Warren G. A monoclonal antibody against a 135-K Golgi membrane protein. EMBO (Eur Mol Biol Organ) J. 1982;1:1621–1628. doi: 10.1002/j.1460-2075.1982.tb01364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaff L, Brechler V. Secretory granule targeting of atrial natriuretic peptide correlates with its calcium-mediated aggregation. Proc Natl Acad Sci USA. 1996;93:9483–9487. doi: 10.1073/pnas.93.18.9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell L, Moore HP. Transport via the regulated secretory pathway in semi-intact PC12 cells: role of intra-cisternal calcium and pH in the transport and sorting of secretogranin II. J Cell Biol. 1994;127:693–705. doi: 10.1083/jcb.127.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanat E, Huttner WB. Milieu-induced, selective aggregation of regulated secretory proteins in the trans-Golgi network. J Cell Biol. 1991;115:1505–1519. doi: 10.1083/jcb.115.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, Kable EPW, Morrison GH, Webb WW. Calcium sequestration in the Golgi apparatus of cultured mammalian cells revealed by laser scanning confocal microscopy and ion microscopy. J Cell Sci. 1991;100:747–752. doi: 10.1242/jcs.100.4.747. [DOI] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries L, Elenko E, Hubler L, Jones TL, Farquhar MG. GAIP is membrane-anchored by palmitoylation and interacts with the activated (GTP-bound) form of Gαisubunits. Proc Natl Acad Sci USA. 1996;93:15203–15208. doi: 10.1073/pnas.93.26.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries L, Elenko E, McCaffery JM, Fischer T, Hubler L, McQuistan T, Watson N, Farquhar MG. RGS-GAIP, a GAP for Gαiheterotrimeric G proteins, is located on clathrin coated vesicles. Mol Biol Cell. 1998;9:1124–1134. doi: 10.1091/mbc.9.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries L, Mousli M, Wurmser A, Farquhar MG. GAIP, a protein that specifically interacts with the trimeric G protein Gαi3, is a member of a protein family with a highly conserved core domain. Proc Natl Acad Sci USA. 1995;92:11916–11920. doi: 10.1073/pnas.92.25.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R, Griffiths G, Frank R, Argos P, Kreis TE. β-COP, a 110 kd protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to β-adaptin. Cell. 1991;64:649–665. doi: 10.1016/0092-8674(91)90248-w. [DOI] [PubMed] [Google Scholar]
- Farquhar, M.G., and H.-P. Hauri. 1997. Protein sorting and vesicular traffic in the Golgi apparatus. In The Golgi Apparatus. E.G. Berger and J. Roth, editors. Birkhauser, Basel. 63–129.
- Fujiki Y, Hubbard AL, Flower S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohovaz F, Bossi M, Pezzati R, Meldolesi J, Tarelli FT. High resolution ultrastructural mapping of total calcium: electron spectroscopic imaging/electron energy loss spectroscopy analysis of a physically/chemically processed nerve-muscle preparation. Proc Natl Acad Sci USA. 1996;93:4799–4803. doi: 10.1073/pnas.93.10.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–183. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Helenius A, Trombetta ES, Hebert DN, Simons JF. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 1997;7:193–200. doi: 10.1016/S0962-8924(97)01032-5. [DOI] [PubMed] [Google Scholar]
- Hendricks LC, McCaffery M, Palade GE, Farquhar MG. Disruption of endoplasmic reticulum to Golgi transport leads to the accumulation of large aggregates containing β-COP in pancreatic acinar cells. Mol Biol Cell. 1993;4:413–424. doi: 10.1091/mbc.4.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Saucan L, Farquhar MG, Palade GE. Rab1a and multiple other Rab proteins are associated with the transcytotic pathway in rat liver. J Biol Chem. 1996;271:30105–30113. doi: 10.1074/jbc.271.47.30105. [DOI] [PubMed] [Google Scholar]
- Jones SM, Crosby JR, Salamero J, Howell KE. A cystolic complex of p62 and rab6 associates with TGN38/41 and is involved in budding of exocytic vesicles from the trans-Golgi network. J Cell Biol. 1993;122:775–788. doi: 10.1083/jcb.122.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Katagiri T, Mori S, Kubota T. An established MRL/Mp-lpr/lpr cell line with null cell properties produces a B cell differentiation factor(s) that promotes anti-single-stranded DNA antibody production in MRL spleen cell culture. Int Arch Allergy Appl Immunol. 1986;81:92–94. doi: 10.1159/000234114. [DOI] [PubMed] [Google Scholar]
- Kanai Y, Miura K, Uehara T, Amagai M, Takeda O, Tanuma S, Kurosawa Y. Natural occurrence of Nuc in the sera of autoimmune-prone MRL/lpr mice. Biochem Biophys Res Commun. 1993;196:729–736. doi: 10.1006/bbrc.1993.2310. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ, Swaroop M, Murtha-Riel P. Depletion of manganese within the secretory pathway inhibits O-linked glycosylation in mammalian cells. Biochemistry. 1994;33:9813–9819. doi: 10.1021/bi00199a001. [DOI] [PubMed] [Google Scholar]
- Krause K-H, Michalak M. Calreticulin. Cell. 1997;88:439–443. doi: 10.1016/s0092-8674(00)81884-x. [DOI] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Leelavathi DE, Estes LW, Feingold DS, Lombardi B. Isolation of a Golgi-rich fraction from rat liver. Biochim Biophys Acta. 1970;211:124–138. [Google Scholar]
- Lodish HF, Kong N. Perturbation of cellular calcium blocks exit of secretory protein from the rough endoplasmic reticulum. J Biol Chem. 1990;265:10893–10899. [PubMed] [Google Scholar]
- Lodish HF, Kong N, Wikstrom L. Calcium is required for folding of newly made subunits of the asialoglycoprotein receptor within the endoplasmic reticulum. J Biol Chem. 1992;267:12753–12760. [PubMed] [Google Scholar]
- Maruyama K, Mikawa T, Ebashi S. Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J Biochem. 1984;95:511–519. doi: 10.1093/oxfordjournals.jbchem.a134633. [DOI] [PubMed] [Google Scholar]
- McCaffery M, Farquhar MG. Localization of GTPases (GTP-binding proteins) by indirect immunofluorescence and immunoelectron microscopy. Methods Enzymol. 1995;257:259–279. doi: 10.1016/s0076-6879(95)57031-4. [DOI] [PubMed] [Google Scholar]
- McLean W, Nakane PF. A new fixative for immunoelectron microscopy. J Histochem Cytochem. 1973;22:1077–1083. doi: 10.1177/22.12.1077. [DOI] [PubMed] [Google Scholar]
- Meldolesi J, Pozzan T. The endoplasmic reticulum Ca2+store: a view from the lumen. Trends Biochem Sci. 1998;23:10–14. doi: 10.1016/s0968-0004(97)01143-2. [DOI] [PubMed] [Google Scholar]
- Mery L, Masaeli N, Michalak M, Opas M, Lew DP, Krause K-H. Overexpression of calreticulin increases intracellular Ca2+ storage and decreases store-operated Ca2+influx. J Biol Chem. 1996;271:9332–9339. doi: 10.1074/jbc.271.16.9332. [DOI] [PubMed] [Google Scholar]
- Miura K, Kurosawa Y, Kanai Y. Calcium-binding activity of nucleobindin mediated by an EF hand moiety. Biochem Biophys Res Commun. 1994;199:1388–1393. doi: 10.1006/bbrc.1994.1384. [DOI] [PubMed] [Google Scholar]
- Miura K, Titani K, Kurosawa Y, Kanai Y. Molecular cloning of nucleobindin, a novel DNA-binding protein that contains both a signal peptide and a leucine zipper structure. Biochem Biophys Res Commun. 1992;187:375–380. doi: 10.1016/s0006-291x(05)81503-7. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Hibi M, Kanai Y, Insel PA. Interaction of the protein nucleobindin with Gαi2, as revealed by the yeast two-hybrid system. FEBS Lett. 1995;373:155–158. doi: 10.1016/0014-5793(95)01031-9. [DOI] [PubMed] [Google Scholar]
- Mumby SM. Reversible palmitoylation of signaling proteins. Curr Opin Cell Biol. 1997;9:148–154. doi: 10.1016/s0955-0674(97)80056-7. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash PD, Opas M, Michalak M. Calreticulin: Not just another calcium-binding protein. Mol Cell Biochem. 1994;135:71–78. doi: 10.1007/BF00925962. [DOI] [PubMed] [Google Scholar]
- Opas, M. 1996. The intracellular distribution and expression of calreticulin. In Calreticulin. M. Michalak, editor. R.G. Landes Company, Georgetown. 31–36.
- Parlati, F., R. Hemming, W.-J. Ou, J.J.M. Bergeron, and D.Y. Thomas. 1996. The roles of calnexin and calreticulin as the endoplasmic reticulum molecular chaperones. In Calreticulin. M. Michalak, editor. R.G. Landes Company, Georgetown. 43–53.
- Pezzati R, Bossi M, Podini P, Meldolesi J, Grohovaz F. High-resolution calcium mapping of the endoplasmic reticulum-Golgi-exocytic membrane system. Electron energy loss imaging analysis of quick frozen-freeze dried PC12 cells. Mol Biol Cell. 1997;8:1501–1512. doi: 10.1091/mbc.8.8.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzan T, Rizzuto R, Volpe P, Meldolesi J. Molecular and cellular physiology of intracellular calcium stores. Physiol Rev. 1994;74:595–636. doi: 10.1152/physrev.1994.74.3.595. [DOI] [PubMed] [Google Scholar]
- Robinson MS. Coats and vesicle budding. Trends Cell Biol. 1997;7:99–102. doi: 10.1016/S0962-8924(96)10048-9. [DOI] [PubMed] [Google Scholar]
- Sambrook JF. The involvement of calcium in transport of secretory proteins from the endoplasmic reticulum. Cell. 1990;61:197–199. doi: 10.1016/0092-8674(90)90798-j. [DOI] [PubMed] [Google Scholar]
- Saucan L, Palade GE. Membrane and secretory proteins are transported from the Golgi complex to the sinusoidal plasmalemma of hepatocytes by distinct vesicular carriers. J Cell Biol. 1994;125:733–741. doi: 10.1083/jcb.125.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer PE, Lederkremer GZ, Williams S, Fogliano M, Baldini G, Lodish HF. Cab45, a novel Ca2+-binding protein localized to the Golgi lumen. J Cell Biol. 1996;133:257–268. doi: 10.1083/jcb.133.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sontheimer, R.D., T.Q. Nguyen, S.-T. Cheng, T.-S. Lieu, and J.D. Capra. 1996. Calreticulin and autoimmunity. In Calreticulin. M. Michalak, editor. R.G. Landes Company, Georgetown. 117–134.
- Sugiura M, Kawasaki T, Yamashina I. Purification and characterization of UDP-GalNAc:polypeptide N-acetylgalactosamine transferase from an ascites hepatoma, AH 66. J Biol Chem. 1982;257:9501–9507. [PubMed] [Google Scholar]
- Taylor RS, Jones SM, Dahl RH, Nordeen MH, Howell KE. Characterization of the Golgi complex cleared of proteins in transit and examination of calcium uptake activities. Mol Biol Cell. 1997;8:1911–1931. doi: 10.1091/mbc.8.10.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco A, Hendricks L, Moremen KW, Tulsiani DR, Touster O, Farquhar MG. Cell type-dependent variations in the subcellular distribution of α-mannosidase I and II. J Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada I, Rindress D, Cameron PH, Ou W-J, Doherty JJ, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJM. SSRα and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- Wang S, Miyauchi M, Koshikawa N, Maruyama K, Kubota T, Miura K, Kurosawa Y, Awaya A, Kanai Y. Antigen expression associated with lymph node metastasis in gastric adenocarcinomas. Pathol Int. 1994;44:844–849. doi: 10.1111/j.1440-1827.1994.tb01682.x. [DOI] [PubMed] [Google Scholar]
- Wendel M, Sommarin Y, Bergman T, Heinegard D. Isolation, characterization, and primary structure of a calcium-binding 63-kD bone protein. J Biol Chem. 1995;270:6125–6133. doi: 10.1074/jbc.270.11.6125. [DOI] [PubMed] [Google Scholar]
- Whitley P, Grahn E, Kutay U, Rapoport TA, von Heijne G. A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J Biol Chem. 1996;271:7583–7586. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- Zha X, Morrison GH. Ion microscopy evidence that La3+ releases Ca2+ from Golgi complex in LLC-PK1cells. Am J Physiol. 1995;269:C923–C928. doi: 10.1152/ajpcell.1995.269.4.C923. [DOI] [PubMed] [Google Scholar]