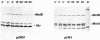Abstract
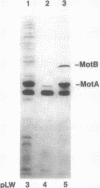
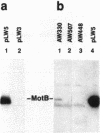
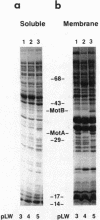
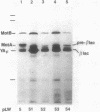
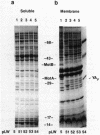
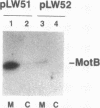
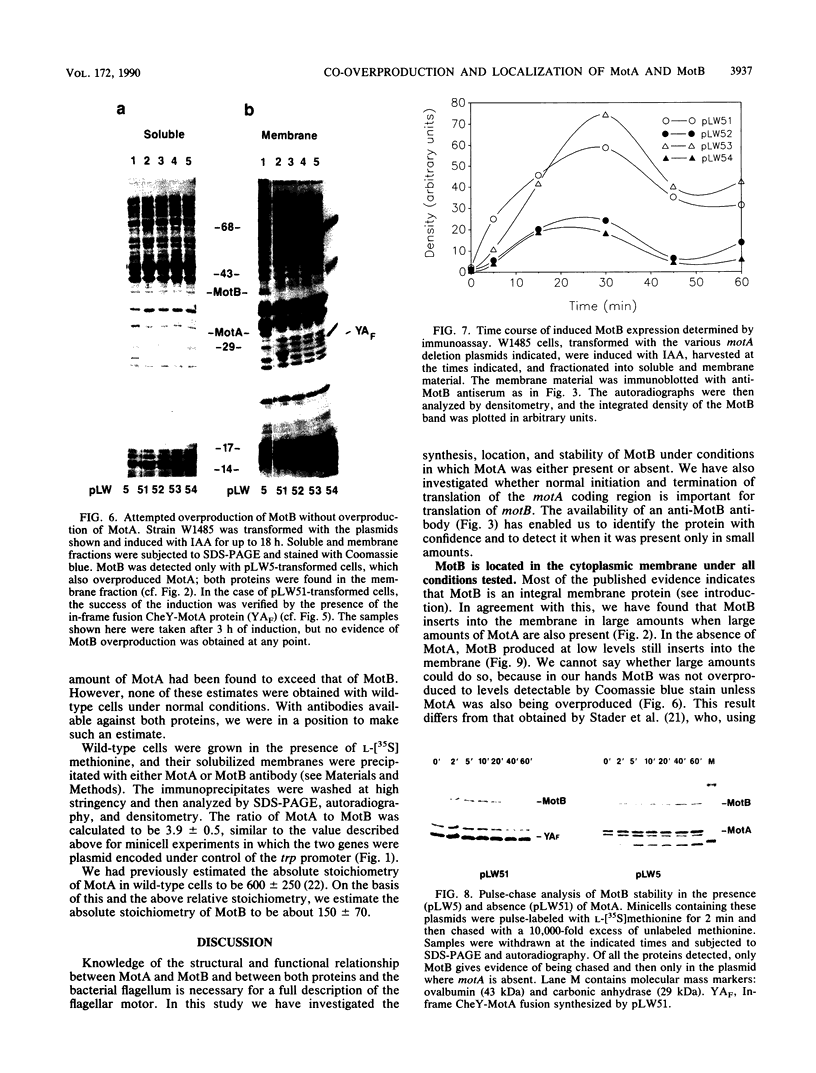
The motility genes motA and motB of Escherichia coli were placed under control of the Serratia marcescens trp promoter. After induction with beta-indoleacrylic acid, the levels of MotA and MotB rose over about a 3-h period, reaching plateau levels approximately 50-fold higher than wild-type levels. Both overproduced proteins inserted into the cytoplasmic membrane. Growth and motility were essentially normal, suggesting that although the motor is a proton-conducting device, MotA and MotB together do not constitute a major proton leak. Derivative plasmids which maintained an intact version of motB but had the motA coding region deleted in various ways were constructed. With these, the levels of MotB were much lower, reaching a peak within 30 min after induction and declining thereafter; pulse-chase measurements indicated that a contributing factor was MotB degradation. The low levels of MotB occurred even with an in-frame internal deletion of motA, whose translational initiation and termination sites were intact, suggesting that it is the MotA protein, rather than the process of MotA synthesis, that is important for MotB stability. Termination at the usual site of overlap with the start of motB (ATGA) was not an absolute requirement for MotB synthesis but did result in higher rates of synthesis than when translation of motA information terminated prematurely. Even in the total absence of MotA, the MotB that was synthesized was found exclusively in the cytoplasmic membrane fraction. In wild-type cells, MotA was estimated by immunoprecipitation to be in about fourfold excess over MotB; a previous estimate of 600 +/- 250 copies of MotA per cell then yielded an estimate of 150 +/- 70 copies of MotB per cell.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong J. B., Adler J. Genetics of motility in Escherichia coli: complementation of paralysed mutants. Genetics. 1967 Jul;56(3):363–373. doi: 10.1093/genetics/56.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D. F., Berg H. C. Restoration of torque in defective flagellar motors. Science. 1988 Dec 23;242(4886):1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- Blair D. F., Berg H. C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990 Feb 9;60(3):439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- Block S. M., Berg H. C. Successive incorporation of force-generating units in the bacterial rotary motor. 1984 May 31-Jun 6Nature. 309(5967):470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- Boyd A., Mandel G., Simon M. I. Integral membrane proteins required for bacterial motility and chemotaxis. Symp Soc Exp Biol. 1982;35:123–137. [PubMed] [Google Scholar]
- Chun S. Y., Parkinson J. S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988 Jan 15;239(4837):276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- Dean G. E., Macnab R. M., Stader J., Matsumura P., Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984 Sep;159(3):991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto M. Genetic studies of paralyzed mutant in Salmonella. I. Genetic fine structure of the mot loci in Salmonella typhimurium. Genetics. 1966 Sep;54(3):715–726. doi: 10.1093/genetics/54.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Iino T. Structural genes for flagellar hook-associated proteins in Salmonella typhimurium. J Bacteriol. 1985 Aug;163(2):464–471. doi: 10.1128/jb.163.2.464-471.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Macnab R. M. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol. 1990 Mar;172(3):1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. J., Macnab R. M., Okino H., Aizawa S. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J Mol Biol. 1990 Mar 20;212(2):377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- Khan S., Dapice M., Reese T. S. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol. 1988 Aug 5;202(3):575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- Macnab R. M., DeRosier D. J. Bacterial flagellar structure and function. Can J Microbiol. 1988 Apr;34(4):442–451. doi: 10.1139/m88-077. [DOI] [PubMed] [Google Scholar]
- Matsumura P., Silverman M., Simon M. Synthesis of mot and che gene products of Escherichia coli programmed by hybrid ColE1 plasmids in minicells. J Bacteriol. 1977 Dec;132(3):996–1002. doi: 10.1128/jb.132.3.996-1002.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway H. G., Silverman M., Simon M. I. Localization of proteins controlling motility and chemotaxis in Escherichia coli. J Bacteriol. 1977 Nov;132(2):657–665. doi: 10.1128/jb.132.2.657-665.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Matsumura P., Simon M. The identification of the mot gene product with Escherichia coli-lambda hybrids. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3126–3130. doi: 10.1073/pnas.73.9.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Genetic analysis of bacteriophage Mu-induced flagellar mutants in Escherichia coli. J Bacteriol. 1973 Oct;116(1):114–122. doi: 10.1128/jb.116.1.114-122.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stader J., Matsumura P., Vacante D., Dean G. E., Macnab R. M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986 Apr;166(1):244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. L., Macnab R. M. Overproduction of the MotA protein of Escherichia coli and estimation of its wild-type level. J Bacteriol. 1988 Feb;170(2):588–597. doi: 10.1128/jb.170.2.588-597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witholt B., Boekhout M. The effect of osmotic shock on the accessibility of the murein layer of exponentially growing Escherichia coli to lysozyme. Biochim Biophys Acta. 1978 Apr 4;508(2):296–305. doi: 10.1016/0005-2736(78)90332-2. [DOI] [PubMed] [Google Scholar]